Abstract
Pasteurella dagmatis and Neisseria canis were repeatedly isolated from the sputum of a poodle-owning patient with chronic bronchiectasis. Commercially available systems failed to identify these unusual organisms: identification was made by 16S rRNA gene sequencing. Difficulties identifying these and five other canine-associated isolates (P. dagmatis [n = 2], Pasteurella canis [n = 2], and N. canis [n = 1]) are discussed.
CASE REPORT
A 66-year-old male poodle owner with chronic obstructive pulmonary disease (COPD) complicated by bronchiectasis presented to his pulmonologist with a worsening cough and abundant sputum production. The bronchiectasis was managed with cyclic trimethoprim-sulfamethoxazole and amoxicillin. The patient had a history of multiple pneumonias caused by a variety of organisms, including Pasteurella multocida 18 months prior (organism unavailable for study). Sputum cultures grew a short gram-negative rod that formed small tan colonies on chocolate and blood agar but did not grow on MacConkey agar. It was catalase, indole, and oxidase positive but did not require X or V factor for growth. Because the Vitek 1 database (bioMérieux Vitek, Inc., Hazelwood, Mo.) for gram-negative organisms failed to conclusively identify the bacterium (identified as 74% Actinobacillus ureae, 22% Vibrio alginolyticus), additional biochemical testing was performed. The organism fermented maltose, sucrose, glucose, and fructose but not lactose or mannitol and was urease positive, ornithine decarboxylase negative, and able to reduce nitrate to nitrite. These biochemical results suggested an identification of Pasteurella dagmatis, which was confirmed by 16S rRNA gene sequencing using MicroSeq (Applied Biosystems Inc., Foster, CA) methodology. We compared the first 500 bp of our organisms with both the MicroSeq databases of over 1,400 organisms and the GenBank database. The patient was treated with ciprofloxacin for 3 weeks but had recurrent symptoms 1 week after discontinuing antibiotics, at which time sputum cultures were positive for P. dagmatis. A third sputum culture 1 month later was again positive for the same organism. The patient was switched to doxycycline therapy for 2 weeks, with subsequent negative sputum cultures 1 month later. Antibiotic susceptibility testing by the Kirby-Bauer disk diffusion technique showed the organism was sensitive to both ciprofloxacin and doxycycline, as well as penicillin, amoxicillin-clavulanic acid, gentamicin, clindamycin, levofloxacin, erythromycin, and trimethoprim-sulfamethoxazole (interpretation with Haemophilus influenzae standards).
Two months after the negative sputum cultures, the patient presented with a mild increase in coughing and sputum production. Sputum cultures grew similar small tan colonies on chocolate and blood agar, but the Gram stain demonstrated a pleomorphic organism with both coccoid and rod forms. This organism was catalase and oxidase positive and did not require X and V factors for growth. It was indole and urease negative and was biochemically inactive except for glucose fermentation, proline, and nitrate. The Vitek gram-negative identification card database could not identify the organism. It was initially reported as a Pasteurella-like bacterium. Because the patient's symptoms were not severe, he was not treated with additional antibiotics. Two months later, a bacterium with the same characteristics grew from a sputum culture. 16S rRNA gene sequencing identified both strains as Neisseria canis. Antibiotic susceptibility testing by the Kirby-Bauer disk diffusion technique showed the organism was sensitive to both ciprofloxacin and doxycycline, as well as penicillin, amoxicillin-clavulanic acid, gentamicin, clindamycin, levofloxacin, erythromycin, and trimethoprim-sulfamethoxazole (interpretation with Neisseria gonorrhoeae standards).
Canine and feline oropharyngeal commensal bacteria have long been recognized as potential pathogens in human bite wounds. Rarely, these organisms are reported to cause systemic disease in humans after casual contact with animals. We report here a case of chronic long-term respiratory tract infection in a COPD patient with two unusual gram-negative bacteria found in canine oral flora, P. dagmatis and N. canis.
Pasteurella species are nonmotile, gram-negative coccobacilli that commonly inhabit the upper respiratory and gastrointestinal tracts of healthy and diseased wild and domestic animals. P. multocida heavily colonizes the oropharynxes of cats (50 to 90%) and dogs (50 to 66%) and is the most commonly isolated gram-negative organism from dog and cat bite wounds (11, 17). Reports of P. multocida causing systemic infections after only casual exposure to domestic animals are increasing, especially in association with chronic lung and liver disease patients (12). Little is known about human infection with Pasteurella species other than P. multocida, possibly because non-P. multocida strains are not often identified to the species level. P. dagmatis is estimated to account for as few as 3% of bite wounds infected with Pasteurella species (11). P. dagmatis has more recently been reported in systemic infections such as pneumonias (7), peritonitis (2, 16), septicemia (5), and endocarditis (9, 14, 15) after casual animal contact. In the case reported here, P. dagmatis caused an exacerbation of bronchiectasis symptoms in a patient with COPD and casual contact with a canine pet.
Neisseria species are present in up to 68% of dog oral and nasal fluids (3) but are rarely isolated from human dog bite wounds. N. canis was first described after it was isolated in 1962 from the pharynx of a healthy dog (4). However, it has only been reported in humans with infected cat bites (8, 10). Pulmonary infection with a closely related canine bacterium, Neisseria weaveri, has been reported in a number of bite wound patients (1), as well as a single patient with acute bronchiectasis exacerbation similar to our case (13). N. canis appears to have caused mild disease in the COPD patient described here, which may be a reason why it is rarely reported as causing human disease. It is also possible that human infection with N. canis is underestimated because of difficulty in identifying this bacterium with the standard clinical microbiology tests.
Routine clinical microbiology laboratories are not usually well equipped to identify veterinary bacteria to the species level. Forsblom et al. (6) characterized aerobic gram-negative bacteria from subgingival sites of dogs with periodontitis. They found that the Pasteurella species isolated could be differentiated using standard biochemicals but the Neisseria spp. had few positive biochemical reactions, making definitive identification possible only by 16S rRNA gene sequencing. Table 1 shows the major difficulties encountered at our institution in identifying these two isolates and five other canine-associated isolates. All were identified by 16S rRNA gene sequencing (MicroSeq 500) with >99% identity to the type strain, except for N. canis from specimen 3, which was 98.3% similar to the type strain.
TABLE 1.
Major difficulties in identifying canine-associated isolates
| Specimen no. | Clinical association | 16S rRNA gene sequence identification | Preliminary identification | Tests that were confusing |
|---|---|---|---|---|
| 1 | This case | Pasteurella dagmatis | Actinobacillus ureae; Pasteurella dagmatis with additional tests | Pasteurella dagmatis is not in Vitek 1 or 2 database |
| Neisseria canis | Unknown Haemophilus or Pasteurella species | Gram stain was interpreted as a coccobacillus; thus, we used identification tables for Haemophilus-Pasteurella-Actinobacillus; species was not in databases | ||
| 2 | Dog bite | Pasteurella dagmatis | Pasteurella species; Pasteurella dagmatis with additional tests | Pasteurella dagmatis is not in Vitek 1 or 2 database |
| Pasteurella canis | Unknown Pasteurella species | Biochemical data were not in our databases | ||
| 3 | Foot ulcer, dog owner | Neisseria canis | Actinobacillus sp. | Gram stain was interpreted as coccobacillus; identification tables were those for Haemophilus-Actinobacillus; species was not in databases |
| 4 | Face abscess | Pasteurella canis | Pasteurella pneumotropica | Identification was performed on the API 20E strip; Pasteurella canis was not in database |
| 5 | Cellulitis and osteomyelitis, diabetic foot, dog owner | Pasteurella dagmatis | Actinobacillus ureae; Pasteurella dagmatis with additional tests | As in first case, a positive urease test makes identification as Actinobacillus ureae probable |
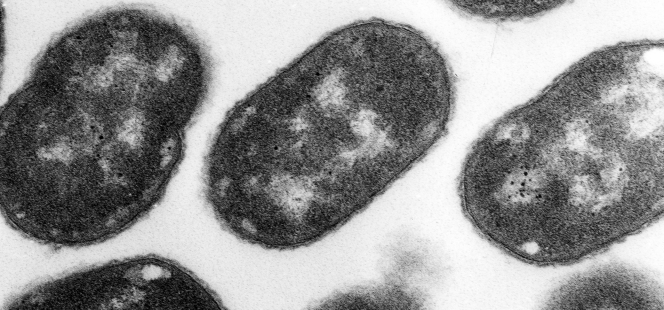
Pasteurella species are not morphologically distinctive, and most commercially available bacterial identification systems do not contain species other than P. multocida in their databases. Additional identification to the species level may require a panel of biochemical testing or 16S rRNA gene sequencing. Our three isolates of P. dagmatis were biochemically typical (urease positivity distinguishes them from other Pasteurella spp.) of the species and were correctly identified by classical biochemical testing but were initially misidentified as the commercial system did not include this species in the database. The identification of our two strains of N. canis was more difficult. Because of the rod-like forms seen on Gram staining and confirmed by electron microscopy (Fig. 1), which were unlike the diplococci classic for most Neisseria species, we mistakenly used identification tables for the Haemophilus-Actinobacillus-Pasteurella group. In addition, N. canis is not in the databases of most commercial systems. The absence of indole production may be useful in differentiating Neisseria from Pasteurella. Only with 16S rRNA gene sequencing was this isolate definitively identified as N. canis.
FIG. 1.
Electron micrograph of an N. canis colony. Rod forms are not uncommon.
Why were these organisms so difficult to eradicate in this patient? Our patient had five documented infections with the unusual canine flora over a period of 7 months. However, since these organisms appeared quite susceptible by disk diffusion testing to the antibiotics used to treat this patient, one might postulate that continued exposure to the animal may have resulted in repeated colonization and/or infection.
Gram-negative bacteria such as P. dagmatis and N. canis can chronically colonize and sometimes infect the respiratory tracts of patients with chronic lung disease and casual contact with oral secretions from canine pets. These organisms can prove difficult to identify without the aid of a panel of biochemical tests and 16S rRNA gene sequencing. Confusion with other gram-negative rods and coccobacilli can occur. Eradication of these bacteria in patients with chronic lung disease may be difficult in patients continuously exposed to their pets' oral secretions.
Acknowledgments
We acknowledge the excellent work of Sally Mizuki and the clinical microbiology laboratory staff at the VA Medical Center, Seattle, Wash.
REFERENCES
- 1.Anderson, B. M., A. G. Steigerwalt, S. P. O'Connor, D. G. Hollis, R. S. Weyant, R. E. Weaver, and D. J. Brenner. 1993. Neisseria weaveri sp. nov., formerly CDC group M-5, a gram-negative bacterium associated with dog bite wounds. J. Clin. Microbiol. 31:2456-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley, B. D., M. Noone, A. D. Dwarakanath, and H. Malnick. 2004. Fatal Pasteurella dagmatis peritonitis and septicaemia in a patient with cirrhosis: a case report and review of the literature. J. Clin. Pathol. 57:210-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailie, W. E., E. C. Stowe, and A. M. Schmitt. 1978. Aerobic bacterial flora of oral and nasal fluids of canines with reference to bacteria associated with bites. J. Clin. Microbiol. 7:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, U. 1962. Ueber das Vorkommen von Neisserien bei einigen Tieren. Z. Hyg. 148:445-457. [Google Scholar]
- 5.Fajfar-Whetstone, C. J. T., L. Coleman, D. R. Biggs, and B. C. Fox. 1995. Pasteurella multocida septicemia and subsequent Pasteurella dagmatis septicemia in a diabetic patient. J. Clin. Microbiol. 33:202-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsblom, B., E. Sarkiala-Kessel, A. Kanervo, M. L. Vaisanen, I. M. Helander, and H. Jousimies-Somer. 2002. Characterization of aerobic gram-negative bacteria from subgingival sites of dogs—potential bite wound pathogens. J. Med. Microbiol. 51:207-220. [DOI] [PubMed] [Google Scholar]
- 7.Furie, R. A., R. P. Cohen, B. J. Hartman, and R. B. Roberts. 1980. Pasteurella multocida infection: report in urban setting and review of the spectrum of human disease. N.Y. State J. Med. 80:1597-1602. [PubMed] [Google Scholar]
- 8.Guibourdenche, M., T. Lambert, and J. Y. Riou. 1989. Isolation of Neisseria canis in mixed culture from a patient after a cat bite. J. Clin. Microbiol. 27:1673-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gump, D. W., and R. A. Holden. 1972. Endocarditis caused by a new species of Pasteurella. Ann. Intern. Med. 76:275-278. [DOI] [PubMed] [Google Scholar]
- 10.Hoke, C., and N. A. Vedros. 1982. Characterization of atypical aerobic gram-negative cocci isolated from humans. J. Clin. Microbiol. 15:906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holst, E., J. Rollof, L. Larsson, and J. P. Nielson. 1992. Characterization and distribution of Pasteurella species recovered from infected humans. J. Clin. Microbiol. 30:2984-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbert, W. T., and M. N. Rosen, I. I. 1970. Pasteurella multocida infections in man unrelated to animal bite. Am. J. Public Health 60:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panagea, S., R. Bijoux, J. E. Corkill, F. Al Rashidi, and C. A. Hart. 2002. A case of lower respiratory tract infection caused by Neisseria weaveri and review of the literature. J. Infect. 44:96-98. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbach, K. A., J. Poblete, and J. Larkin. 2001. Prosthetic valve endocarditis caused by Pasteurella dagmatis. South. Med. J. 94:1033-1035. [PubMed] [Google Scholar]
- 15.Sorbello, A. F., J. O'Donnell, J. Kaiser-Smith, et al. 1994. Infective endocarditis due to Pasteurella dagmatis: case report and review. Clin. Infect. Dis. 18:336-338. [DOI] [PubMed] [Google Scholar]
- 16.Wallet, F., F. Toure, A. Devalckenaere, D. Pagniez, and R. J. Courcol. 2000. Molecular identification of Pasteurella dagmatis peritonitis in a patient undergoing peritoneal dialysis. J. Clin. Microbiol. 38:4681-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber, D. J., J. S. Wolfson, M. N. Swartz, and D. C. Hooper. 1984. Pasteurella multocida infection. Report of 34 cases and review of the literature. Medicine 63:133-153. [PubMed] [Google Scholar]