Abstract
Measurement of human immunodeficiency virus type 1 (HIV-1) plasma RNA levels using Roche AMPLICOR version 1.5 (HIV RNA) is an integral part of monitoring HIV-infected patients in industrialized countries. These assays are currently unaffordable in resource-limited settings. We investigated a reverse transcriptase (RT) assay as a less expensive alternative for measuring viral burden that quantifies RT enzyme activity in clinical plasma samples. A comparison of RT and HIV RNA assays was performed on 29 paired plasma samples from patients living in the United States and 21 paired plasma samples from patients living in Cameroon. RT levels correlated significantly with plasma HIV RNA viral loads in plasma from U.S. patients (r = 0.898; P < 0.001) and Cameroonian patients, a majority of whom were infected with HIV-1 clade type CRF02_AG (r = 0.669; P < 0.01). Among 32 samples with HIV viral load of >2,000 copies/ml, 97% had detectable RT activity. One Cameroon sample had undetectable RNA viral load but detectable RT activity of 3 fg/ml. The RT assay is a simple and less expensive alternative to the HIV RNA assay. Field studies comparing these assays in resource-limited settings are warranted to assess the practicality and usefulness of this assay for monitoring HIV-infected patients on antiretroviral therapy.
Human immunodeficiency virus (HIV) plasma viral load measurement is an integral part of monitoring infected patients in industrialized countries to determine the need to initiate or change antiretroviral (ARV) therapy. A commonly used Food and Drug Administration-approved assay, the Roche AMPLICOR version 1.5 (HIV RNA), is unaffordable for most patients in resource-limited settings. This HIV RNA assay uses nucleic acid amplification by PCR of conserved gene sequences, including non-clade B sequences (4, 9). One alternative to measuring RNA copies is to measure the activity of the virus-encoded reverse transcriptase enzyme (RT). An RT assay kit, ExaVir Load version 2, is currently available and distributed in Africa, but no published studies to date have correlated version 2 of the RT assay with HIV RNA viral load. We describe this correlation in plasma samples from HIV-infected patients from New York and Cameroon.
MATERIALS AND METHODS
Plasma samples from HIV-infected patients in New York City and Cameroon. New York City.
Twenty-nine anonymous plasma samples from HIV-infected persons attending New York University Medical Center in 2004 were studied. Specimens were presumed to contain predominantly clade B HIV-1 virus, although clade analysis was not performed on these samples. While information on clinical symptoms or antiretroviral therapy history was not available for this anonymous sample, a large proportion of patients attending this clinic receive antiretroviral therapy. All samples were processed within 6 h and stored at −70°C.
Cameroon.
Twenty-one anonymous plasma samples from Bamenda, Cameroon, were obtained as part of a larger study (3). All patients were antiretroviral treatment naïve. Blood was collected in blood bags containing the anticoagulant citrate acid dextrose, which was shipped at room temperature by express courier approximately 1 to 3 days after collection (3). Upon arrival, plasma was aliquoted and stored at −70°C. Sequence and phylogenetic analysis was performed on a subset of the cohort demonstrating that the predominant clade type was the circulating recombinant form, CRF02_AG (3, 5).
RT assay.
RT activity was quantified using the RT activity kit (ExaVir Load [version 2]; Cavidi Tech-AB, Uppsala, Sweden), as recommended by the manufacturer (7). One-milliliter plasma samples were incubated for 1 h at room temperature with a protein-reactive reagent [100 μl of 220 mM 5,5′-dithio-bis(2-nitrobenzoic acid) in 0.8 M Tris, pH 8.1] (7) to destroy cellular polymerases present in the plasma while preserving intact RT contained within virions. Next, 1.5 ml of an ion-exchange gel (Fractogel EMD TMAE Hicap; in 209 mM morpholineethanesulfonic acid [pH 5.0], 275 mM KI, and 0.5 g/liter heparin) (7) was added to each tube and incubated for 90 min at room temperature on an end-over-end shaker. The gel immobilizes the HIV particle by binding to the lipid membrane of the virion. After thorough mixing, samples were transferred to 10-ml plastic filtration columns mounted in a vacuum manifold. Vacuum is applied to isolate the ion-exchange gel particles bound to virions trapped by the filter. The fixed gel was washed four times with 9 ml of wash buffer to remove RT activity-blocking antibodies and antiviral drugs. A further two washes with 9 ml of gel were performed to create a suitable environment for RT release after virion lysis. Using 500 μl of lysis buffer, lysates containing RT were collected into individual test tubes and used directly in the RT activity assay described below.
RT activity was assayed by adding lysates to a 96-well microtiter plate which had a poly(A) nucleotide chain covalently bound to the base of each well. A mix of 5′ bromodeoxyuridine triphosphate (BrdUTP) was added, and the plate was incubated at 33°C for 48 h.
The plate was washed to terminate the reaction and a BrdU-binding antibody conjugated to alkaline phosphatase (AP) was added and incubated at 33°C for 90 min.
Following a second wash, the AP substrate p-nitrophenyl phosphate was added and the resulting yellow color change was measured to quantify the amount of incorporated BrdUTP. Following addition of the AP substrate each plate was read at three time points: 10 min, 2 to 3 h, and 5 to 6 h or overnight at 15 to 24 h. The results were expressed in femtograms HIV-1 RT activity/milliliter plasma. The lower limit of detection was <1 fg/ml. Results were calculated using ExaVir Load Analyzer software that automatically determines the amount of RT activity in samples using a standard curve generated from an 11-point dilution of a known amount of recombinant HIV-1 RT. The results are also given in units of HIV-1 RNA equivalent copies/milliliter using a conversion factor derived from a Swedish cohort comprised mainly of subtype B virus. We report only reverse transcriptase activity in this paper. Assay precision was determined on duplicates of five samples, and assay reproducibility was determined on seven samples run on different days.
Plasma HIV RNA assay.
HIV RNA load was determined using the ultrasensitive format of the AMPLICOR HIV-1 Monitor version 1.5 assay (Roche-Diagnostics, Branchburg, N.J.) in identical plasma samples to those used for the RT assay. The lower limit of detection was 50 copies/ml and the upper limit of detection was 100,000 copies/ml (4).
Statistical analysis.
For purposes of analysis, samples with <1 fg/ml of activity were arbitrarily assigned a value of 0.9 fg/ml, and samples with viral load of ≥100,000 copies/ml were arbitrarily assigned a viral load of 100,000 copies/ml. The Spearman rank correlation coefficient was used to compare results of the two assays. Continuous variables were analyzed using Student's t test. Correlation of variation was calculated on duplicate samples to determine reproducibility of the RT assay. All statistical analyses were conducted using SPSS for Windows version 11.0 (SPSS, Inc., Chicago, Ill.).
RESULTS
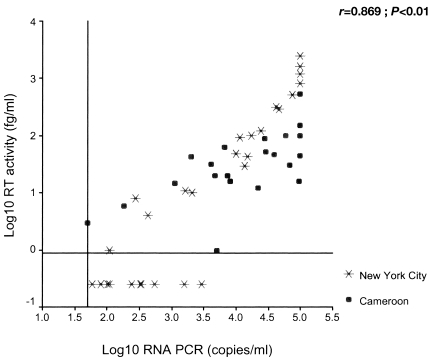
Fifty plasma samples from the New York and Cameroon cohorts were tested using both RT and HIV RNA assays (Table 1). Overall concordance was 76% (38 of 50 samples tested positive in both assays; no sample tested negative in both assays). A positive result in the RT assay alone was found in one sample. Eleven (22%) samples tested positive with the HIV RNA assay alone and had an HIV viral load below 3,000 copies/ml (median, 326; range, 60 to 2,955), and all were from the New York cohort. Among 32 samples from either cohort with an HIV viral load of >2,000 copies/ml, 3% had undetectable RT activity whereas among 18 samples with a viral load of ≤2,000 copies/ml, 56% had undetectable RT activity. A strong correlation was found between RT activity and HIV RNA viral load among all clade B and non-clade B samples (r = 0.869; P < 0.01) (Figure 1).
TABLE 1.
HIV-1 RNA viral load by PCR and reverse transcriptase (RT) activity for Cameroon and New York City
| Cohort and sample no. | HIV RNA PCR (copies/ml) | RT activity (fg/ml) |
|---|---|---|
| Cameroon | ||
| 1 | 50 | 3 |
| 2 | 181 | 6 |
| 3 | 1,110 | 15 |
| 4 | 1,976 | 43 |
| 5 | 4,142 | 32 |
| 6 | 4,741 | 20 |
| 7 | 5,053 | 1 |
| 8 | 6,554 | 62 |
| 9 | 7,467 | 20 |
| 10 | 8,044 | 16 |
| 11 | 21,831 | 12 |
| 12 | 27,575 | 88 |
| 13 | 29,097 | 52 |
| 14 | 39,180 | 46 |
| 15 | 59,609 | 100 |
| 16 | 68,117 | 31 |
| 17 | 94,530 | 16 |
| 18 | 100,000 | 536 |
| 19 | 100,000 | 44 |
| 20 | 100,000 | 152 |
| 21 | 100,000 | 100 |
| Total | 21 | 21 |
| New York | ||
| 1 | 60 | <1 |
| 2 | 80 | <1 |
| 3 | 102 | <1 |
| 4 | 109 | 1 |
| 5 | 109 | <1 |
| 6 | 243 | <1 |
| 7 | 273 | 8 |
| 8 | 326 | <1 |
| 9 | 327 | <1 |
| 10 | 336 | <1 |
| 11 | 435 | 4 |
| 12 | 547 | <1 |
| 13 | 1,556 | <1 |
| 14 | 1,645 | 11 |
| 15 | 2,067 | 10 |
| 16 | 2,955 | <1 |
| 17 | 10,148 | 49 |
| 18 | 11,365 | 92 |
| 19 | 13,544 | 29 |
| 20 | 14,820 | 43 |
| 21 | 17,141 | 100 |
| 22 | 24,232 | 122 |
| 23 | 41,638 | 311 |
| 24 | 46,567 | 292 |
| 25 | 75,758 | 514 |
| 26 | 100,000 | 818 |
| 27 | 100,000 | 2,400 |
| 28 | 100,000 | 1,165 |
| 29 | 100,000 | 1,577 |
| Total | 29 | 29 |
FIG. 1.
Correlation of HIV load based on RT activity and HIV RNA assay (log10 values). Cameroon and New York City cohorts (n = 50). The lower limit of detection were 1.7 log10 RNA copies/ml (50 copies/ml) and −0.05 log10 RT activity/ml (0.9 fg/ml). The upper limit of detection for the HIV RNA assay was 5 log10 RNA copies/ml (100,000 copies/ml), and samples above this detection limit were attributed arbitrary values of 5 log10 RNA copies/ml. All data are shown and included in the correlation; r, Spearman correlation coefficient.
New York City cohort.
Eighteen (62%) of the 29 plasma samples from the New York City cohort tested positive for both RT activity and HIV RNA. The median HIV RNA viral load was 2,067 copies/ml (range, 60 to >100,000 copies/ml); the median RT activity was 10 fg/ml (range, <1 to 2,400 fg/ml). Among samples with HIV RNA PCR levels of ≥50 and ≤3,000 copies/ml, 11 (69%) had undetectable RT activity. Among the 13 samples with HIV RNA PCR levels of >3,000 copies/ml, all had detectable RT activity. A strong correlation was found between RT activity and HIV RNA viral load in the New York cohort (r = 0.898; P < 0.01).
Cameroon cohort.
Twenty of the 21 samples from the Cameroon cohort tested positive using both RT and HIV RNA assays. The one discordant sample had undetectable viremia (<50 copies/ml) and detectable RT activity (3 fg/ml). Overall, the median viral load in this cohort was 21,831 copies/ml (range, <50 to >100,000 copies/ml), and the median RT activity was 32 fg/ml (1 to 536 fg/ml). A significant correlation was found between RT activity and HIV RNA viral load among the HIV-1 non-clade B samples (r = 0.669; P < 0.001).
Duplicates of five Cameroon samples were assayed in the same run and had a mean of 1.87 fg/ml with a coefficient of variation of 7.9%. Duplicates of six Cameroon samples were assayed in two different runs and had a mean of 1.60 fg/ml with a coefficient of variation of 8.6%.
DISCUSSION
This is the first study to evaluate version 2 of the RT assay in clade B or non-clade B plasma samples. We found a significant correlation between the RT and HIV RNA assays in estimating viral load in patient plasma samples from New York City and Cameroon. The correlation between the two assays was strong in the New York cohort, consistent with findings of previous studies conducted in persons infected with clade B virus (8) and moderate but significant in the non-clade B cohort. The number of samples in the New York cohort with RNA viral loads of <2,000 copies is striking. The most likely explanation for this observation is that the majority of patients in the New York cohort were receiving antiretroviral treatment, although the specific proportion of treated patients was unknown.
The weaker correlation between RT activity and HIV RNA viral load in the Cameroon cohort may be a result of several factors. Most significantly, the long transport time of the Cameroon samples may have affected the RT and the RNA PCR assays to different degrees, therefore adversely affecting their correlation. It is noteworthy that although the manufacturer of the RT assay recommends that all blood samples be processed within 6 h of acquisition and frozen at −20°C before analysis, in fact, the RT assay performed well and correlated closely with HIV RNA viral loads despite the delay between blood draw and processing. This suggests that the RT assay is robust and may be useful in the field setting where collection and processing time may vary. Another possibility for the weaker correlation in the Cameroon cohort is that the sensitivity of the second-generation HIV RNA assay may be reduced for recombinant HIV genomes such as those frequently found in Cameroon. Of note, however, one previous study evaluating three viral load methods for detecting circulating recombinant strains such as AG found Amplicor version 1.5 to be accurate and reliable (1).
Published studies evaluating the ExaVir Load RT assay are few, and none have evaluated version 2 (2, 7). Braun et al. compared the RT assay, version 1, with RNA PCR in 322 plasma samples including 66 non-clade B subtypes (2). A strong correlation was found between the two assay (r = 0.850; P < 0.0001) independent of subtype. The sensitivity of the RT assay, version 1, was poor for samples with viral load of 10,000 copies/ml or below (2). Malmsten et al. compared the RT assay, version 1, to RNA PCR (Roche Amplicor version 1.5) in 390 plasma samples from patients living in Sweden (7). The correlation was strong (r = 0.90, P < 0.0001); however, RT activity was detectable in only 50% of samples with a viral load between 2,500 and 6,900 copies/ml (2). The RT assay was also evaluated by the manufacturer and researchers in Uganda in a cohort of patients from Uganda (8). This study compared the RT assay, version 1, and RNA viral load in a cohort of 46 patients from Uganda, and they report that while correlation was good (R2 = 0.85) overall, RT activity was detected in only 47% of samples with a viral load of less than 55,000 copies/ml (8).
Improvements have been made to the first version of the ExaVir Load RT assay to increase sensitivity. These changes included improved wash of the gels containing immobilized virions, improved virion lysis to increase the RT enzyme yield, use of larger lysate volume, and a longer RT reaction time (approximately 40 h) (6). The manufacturer reports that in 262 clinical samples, the detection limit of version 2 of the ExaVir RT assay was 400 RNA copies/ml, below which only 19% of 52 samples had detectable RT activity. However, among 93 samples containing 51 to 1,500 RNA copies/ml, only 39 (42%) had detectable RT activity (6). Similarly, our study found 6 (40%) of 15 samples containing 50 to 1,500 RNA copies/ml had detectable RT activity but that 31 (97%) of 32 samples containing >2,000 copies/ml had detectable RT activity. Therefore, the detection limit of the RT assay in our study is approximately 2,000 copies/ml, higher than that reported by the manufacturer (6).
Low cost and technical simplicity are two advantages of the RT assay. The setup for the RT assay includes the ExaVir Load start-up kit ($3,000 U.S.) and the assay kit itself. The start-up kit includes a sample box and lid, column holder, waste collector, sample collector, tube rack, vacuum pump, waste container, buffer dispenser, 4 3-liter wash buckets, various plastic bottles and racks, and ExaVir Load Analyzer software. Other equipment that is required but not provided include an enzyme-linked immunosorbent assay reader, an end-to-end mixing table, an incubator, pipettes (including multichannel pipettes), and a −20°C refrigerator. Each ExaVir Load kit contains reagents and consumables for analysis of 28 samples, one negative control, and one positive control. Disadvantages of this assay include the requirement for 1 ml of plasma (of particular concern for pediatric patients), lack of standard positive and negative controls in the kit, and the need for 3 days to complete the assay. The cost per ExaVir Load kit currently is approximately $843.75 U.S. when purchased from local Kenyan distributors, which is approximately $28.13 per test, not including start-up costs and technician time. While considerably less expensive than the HIV RNA assay, which is approximately $100 U.S. per test when performed in Kenya, currently the RT assay may be too costly to recommend for routine use in resource-limited settings.
Measurements taken by HIV RNA PCR and RT assays represent different aspects of viral replication. Plasma HIV RNA assays measure virion-associated RNA while the RT assay measures the activity of a virion-associated enzyme required for replication. Measuring enzyme activity as marker for viral replication has several advantages in the international setting aside from cost. The reverse transcriptase enzyme is well conserved across different genetic clade types, therefore the RT assay would be expected to function well in areas of the world where non-clade B strains, including complex recombinant strains, are common. Our data suggest that the RT assay is able to detect RT activity in plasma containing complex recombinant non-B subtype HIV virus. In addition, RNA-based methods do not differentiate between functional and defective virions and therefore may not reflect true viral replication activity, whereas the RT assay quantifies actual reverse transcriptase activity.
In the search for more affordable alternatives for viral load measurement for monitoring of HIV-1-infected patients living in resource-limited settings, the RT assay appears promising. RT assay results for both U.S. and Cameroon cohorts correlate well with the HIV RNA assay and demonstrate adequate sensitivity above 2,000 copies/ml. However, the usefulness of the RT assay in monitoring patients on antiretroviral therapy in a resource-limited setting is unknown. Field trials of the RT assay in such settings will be important to assess the cost, ease, and applicability of this assay in routine HIV care.
Acknowledgments
We thank Paul Nickolopalous for assistance with the Roche Amplicor assay; Maura Laverty for administrative support; Gary Corrigan from Cavidi-Tech for technical support; Sherri Burda and Constance Williams for processing of the Cameroon specimens; Charles Gonzalez for assistance in obtaining plasma samples from the New York City cohort; Steven Oliver for laboratory assistance; and Joel Ernst and Judith Aberg for their critical reading of the manuscript.
New York University Centers for AIDS Research Discretionary Funds was the source of funding.
REFERENCES
- 1.Amendola, A., L. Bordi, C. Angeletti, U. Visco-Comandini, I. Abbate, G. Cappiello, M. A. Budabbus, O. A. Eljhawi, M. I. Mehabresh, E. Girardi, A. Antinori, G. Ippolito, and M. R. Capobianchi. 2002. Under-evaluation of HIV-1 plasma viral load by a commercially available assay in a cluster of patients infected with HIV-1 A/G circulating recombinant form (CRF02). J. Acquir. Immun. Defic. Syndr. 31:488-494. [DOI] [PubMed] [Google Scholar]
- 2.Braun, J., J. C. Plantier, M. F. Hellot, E. Tuaillon, M. Gueudin, F. Damond, A. Malmsten, G. E. Corrigan, and F. Simon. 2003. A new quantitative HIV load assay based on plasma virion reverse transcriptase activity for the different types, groups and subtypes. AIDS 17:331-336. [DOI] [PubMed] [Google Scholar]
- 3.Burda, S., F. A. J. Konings, C. A. U. Williams, C. Anyangwe, and N. P. Nyambi. 2004. HIV-1 CRF09_cpx circulates in the north east province of Cameroon where CRF02_AG infections predominate and recombinant strains are common. AIDS Res. Hum. Retrovir. 20:1358-1363. [DOI] [PubMed] [Google Scholar]
- 4.Erali, M., and D. R. Hillyard. 1999. Evaluation of the ultrasensitive Roche Amplicor HIV-1 monitor assay for quantitation of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 37:792-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konings, F. A., P. Zhong, M. Agwara, L. Agyingi, L. Zekeng, J. M. Achkar, L. Ewane Saa, E. Afane Ze, T. Kinge, and P. N. Nyambi. 2004. Protease mutations in HIV-1 non-B strains infecting drug-naive villagers in Cameroon. AIDS Res. Hum. Retrovir. 20:105-109. [DOI] [PubMed] [Google Scholar]
- 6.Malmsten, A., X. W. Shao, S. Sjodahl, E. L. Fredriksson, I. Petterson, T. Leitner, C. F. Kallander, E. Sandstorm, and J. S. Gronowitz. 2005. Improved HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J. Med. Virol. 76:291-296. [DOI] [PubMed] [Google Scholar]
- 7.Malmsten, A., X. W. Shao, K. Aperia, G. E. Corrigan, E. Sandstrom, C. F. Kallander, T. Leitner, and J. S. Gronowitz. 2003. HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J. Med. Virol. 71:347-359. [DOI] [PubMed] [Google Scholar]
- 8.Mermin, J., R. Downing, G. Anyaegani, A. Malmsten, J. Gronowitz, and G. Corrigan. 2002. Evaluating the Cavidi reverse transcriptase assay to monitor response to antiretroviral therapy, abstr. MoPe3106. Presented at the XIV International AIDS Conference, Barcelona, Spain.
- 9.Schmitt, Y. 2001. Performance characteristics of quantification assays for human immunodeficiency virus type 1 RNA. J. Clin. Virol. 20:31-33. [DOI] [PubMed] [Google Scholar]