Abstract
Although enterococci expressing acquired vancomycin resistance phenotype have been reported increasingly worldwide, they have been rarely reported in France. From August to December 2004 we faced an outbreak of vancomycin-resistant Enterococcus faecium (VRE) isolates in the nephrology department at Bicêtre Hospital (K.-Bicêtre, France). The expression of the glycopeptide resistance varied among the 26 VRE isolates, with vancomycin MICs ranging from 12 to >256 μg/ml, whereas teicoplanin MICs ranged from 4 to 48 μg/ml. However, several strains appeared to be susceptible to glycopeptides according to disk diffusion testing and expressed resistance only after subculture with glycopeptides. In addition, a heterogeneous expression of glycopeptide resistance was also observed. This so-called VanD-like phenotype of resistance (low-level resistance to vancomycin and mostly susceptibility to teicoplanin) was surprisingly associated with a vanA gene. Plasmid extraction and mating-out experiments indicated that the vanA gene was located on a 200-kb self-transferable plasmid. Pulsed-field gel electrophoresis identified mostly dissemination of a single clone, whereas diffusion of the VanA-positive plasmid in different genomic backgrounds had also occurred. The vanA gene was part of a vanA-type operon for expression of resistance located on a Tn1546-like transposon. Sequencing of this transposon identified insertion of insertion sequence IS16 in the vanY gene that encodes a d,d-carboxypeptidase that might explain in part the peculiar VanD-type phenotype of resistance. This report is the first description of a VRE outbreak in France and underlines the difficulty in detecting this organism due to variability on the expression of the glycopeptide resistance trait, if any.
Over the last two decades Enterococcus spp. have emerged as important nosocomial pathogens (4, 37). Since their initial discovery from patients in France and the United Kingdom in 1988 (23, 35), vancomycin-resistant enterococci (VRE) have been reported worldwide (4, 37). Whereas hospital outbreaks with clonally related VRE have been reported extensively in the United States (4), the prevalence of VRE remains low in French hospitals (11, 18, 33). Six glycopeptide resistance phenotypes (VanA to VanE and VanG) have been described (37). Except for VanC, which is a naturally encoded phenotype of resistance, the phenotypes correspond to acquired resistance to glycopeptides. The VanA and VanB phenotypes are the most frequently encountered phenotypes; VanD has been found in a few strains of Enterococcus faecium, VanE has been found in two Enterococcus faecalis strains, and VanG has been found in a few E. faecalis strains from Australia and Canada (4, 37).
The VanA phenotype is characterized by acquired inducible and high-level resistance to vancomycin and teicoplanin, whereas the VanB phenotype is characterized by variable levels of resistance to vancomycin with in vitro susceptibility to teicoplanin (teicoplanin MIC of 0.5 μg/ml) (4, 37). The VanD phenotype is characterized by low-level resistance to vancomycin (vancomycin MIC of 64 to 128 μg/ml) and with susceptibility or intermediate resistance to teicoplanin (teicoplanin MIC of 8 to 16 μg/ml). Recently, VRE with a vanA genotype that were susceptible to teicoplanin, and hence have a VanB phenotype, have been detected in Japan, Korea, and Taiwan (12, 14, 22).
The VanA phenotype is determined by seven van genes present on Tn1546-type transposons, located immediately downstream of genes designated orf1 and orf2, which are associated with transposition functions (23, 37). These transposons are often located on plasmids that facilitate horizontal transfer among enterococcal strains (1, 2, 4, 19, 28, 37).
Three van genes are essential for expression of the vancomycin resistance phenotype: vanA, which encodes the d-Ala-d-Lac ligase; vanH, which encodes a dehydrogenase that produces d-lactate from pyruvate; and vanX, which encodes a d,d-dipeptidase that destroys d-Ala-d-Ala, which is the product of the chromosomally encoded ligase, Ddl, and which forms the terminus of glycopeptide-susceptible pentapeptides. Upstream of these genes are two regulatory genes, vanR and vanS, which encode a two-component signal transduction system that senses glycopeptides in the environment and induces expression of resistance genes (37). Two nonessential genes are located downstream of the vanRSHAX cluster, vanY, which encodes a d,d-carboxypeptidase that contributes to vancomycin resistance by cleaving the C-terminal d-Ala residue of the precursor, and vanZ, which encodes a peptide that confers low-level teicoplanin resistance (2, 4). In addition to these genes, insertion sequence (IS)-like elements have been found in the vanA gene cluster of several isolates (17, 24, 25, 36). These ISs were inserted mostly in the intergenic regions, but insertion in coding regions has been rarely observed (17, 24, 25).
We present here a detailed molecular analysis of VRE isolates from the Bicêtre Hospital (K-Bicêtre [southern suburbs of Paris]), giving rise to an outbreak from August 2004 to December 2004. This is the first report of a VanD phenotype associated with a vanA genotype and the first description of a heterogeneous expression of vancomycin resistance in E. faecium. Furthermore, we report an IS element, IS16, that has never been reported to be associated with the vanA gene, inserted in the coding region of the vanY gene that might be involved in the peculiar phenotype of resistance to glycopeptides.
MATERIALS AND METHODS
Hospital settings, outbreak, and surveillance.
Bicêtre Hospital is a 1,000-bed tertiary-care teaching facility with 800 adult beds and 200 child beds. The infection/colonization state was evaluated by the clinicians according to the French National recommendations, which derived from those of the Centers for Disease Control and Prevention (7, 13).
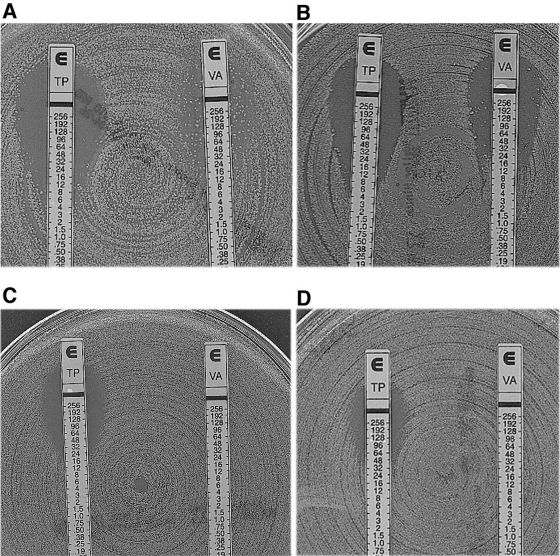
The VRE outbreak was identified in September 2004 when three VRE infections were reported in 10 days in the nephrology ward. These strains were characterized by a heterogeneous vancomycin resistance phenotype associated with susceptibility or intermediate resistance to teicoplanin. This heterogeneous expression of resistance was characterized by a low vancomycin MIC (Table 1), as seen on the E-test with colonies growing inside the inhibition ellipse (Fig. 1), similar to what is known for methicillin-resistant Staphylococcus aureus (5). Four other VRE infections were retrospectively identified in August by searching in database of the microbiology department: three in the nephrology ward and one in the internal medicine ward that were thought to be unrelated at that time. Going further back in the databases of the Microbiology Department (until 1996), no other vancomycin-resistant E. faecium isolate had been identified in the hospital.
TABLE 1.
Patients and strain characteristics
| Patientm | Isolate | Onseta (days) | Sexb | Age (yr) | Infection statusc | Underlying diseased | Treatmente | MICf (μg/ml) of VA/TEC | PFGEn | PCRg |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vanA | IS16 | IS16-vanY | ||||||||||
| I | 8 | 20 | M | 85 | UTI | Prostatic cancer | None | 256/16 | A | + | + | + |
| II | 13 | 15 | F | Peritonitis | Lymphoma | LZD, SXT | 12/4 | A | + | + | + | |
| III | 11 | 20 | F | 64 | Bacteremia | Transplantation + ESRD | None | 24/6 | A | + | + | + |
| IV | 9 | 7 | F | 77 | UTI | Diabetes + ARF | None | 24/6 | A | + | + | + |
| 10h | 21 | F | 77 | C | Diabetes + ARF | - | 16/8 | A | + | + | + | |
| V | 5 | 21 | M | 80 | Peritonitis | ESRD | LZD, SXT | 256/8 | A | + | + | + |
| VI | 4 | 22 | F | 67 | UTI | Diabetes + ARF | None | 24/8 | A | + | + | + |
| VII | 1 | 4 | F | 63 | UTI | Transplantation + ESRD | None | 12/6 | A | + | + | + |
| VIII | 3 | 15 | F | 68 | C | ESRD | - | 24/6 | A | + | + | + |
| IX | 14 | 10 | F | 75 | C | Myeloma | - | 24/8 | A | + | + | + |
| X | 2 | 11 | F | 48 | C | Transplantation | - | 12/6 | A | + | + | + |
| XI | 12 | 8 | F | 45 | C | Transplantation | - | 256/48 | A | + | + | − |
| XII | 20i | 23 | F | 54 | C | Transplantation | - | 256/32 | B | + | + | − |
| 21 | 23 | F | 48 | C | Transplantation | - | 32/8 | A | + | + | + | |
| XIII | 17 | 10 | 59 | C | Transplantation + polycystic kidney disease | - | 24/8 | A | + | + | + | |
| XIV | 15 | 5 | M | 55 | C | Nephroangiosclerosis | - | 256/16 | A | + | + | + |
| XV | 16 | 24 | F | 39 | C | Transplantation | - | 96/16 | A | + | + | + |
| XVI | 18 | 15 | F | 44 | C | Immunoglobulin A nephropathy | - | 32/6 | A | + | + | + |
| XVII | 19 | 21 | F | 69 | C | Amyloid kidney | - | 24/12 | A | + | + | + |
| XVIII | 23 | 4 | M | 75 | C | Myeloma | - | 16/6 | A | + | + | + |
| XIX | 22 | 4 | M | 65 | C | Liver cirrhosis | - | 32/16 | A | + | + | + |
| XX | 24 | 23 | F | 45 | C | Transplantation | - | 32/12 | A | + | + | + |
| XXI | 28 | 2 | M | 69 | C | Transplantation + diabetes | - | 256/6 | A1 | + | + | + |
| XXII | 25 | 14 | F | 30 | C | ESRD | - | 16/6 | A | + | + | + |
| XXIII | 27 | 5 | M | 77 | C | ESRD | - | 256/48 | A | + | + | − |
| XXIV | 29 | 24 | F | 62 | C | ARF | - | 32/12 | A | + | + | + |
| C1j | 6 | 256/48 | C | + | − | − | ||||||
| C2k | 7 | 48/0.5 | D | − | − | − | ||||||
| C3l | 26 | 0.25/0.25 | E | - | - | - | ||||||
Onset corresponds to the number of days of hospitalization prior to VRE isolation.
M, male; F, female.
UTI, urinary tract infection; C, colonization.
Reason for hospital admission. ARF, acute renal failure; ESRD, end-stage renal disease.
Treatment is only indicated for VRE infections. LZD, linezolid; SXT, trimethoprim-sulfamethoxazole; None, no antibiotic. -, Not relevant (since colonized patients were not treated for VRE).
MICs of vancomycin (VA) and teicoplanin (TEC).
PCR results for vanA gene, IS16, and IS16 inserted within the vanY gene.
This isolate was resistant to trimethoprim-sulfamethoxazole.
This isolate was resistant at low levels to gentamicin and to trimethoprim-sulfamethoxazole.
CI, BM4147 VanA prototype.
C2, E. faecium V883 VanB prototype.
C3, E. faecium Gil (vancomycin susceptible).
The numbering in Roman numerals corresponds to the patients listed in chronological order. For two patients, two strains were identified with a different antibiotic susceptibility profile. The strain numbering in Arabic numerals corresponds to the order in which the strains were identified as vanA genotype.
Pulsotypes are according to Tenover et al. (34).
FIG. 1.
Teicoplanin (TP) and vancomycin (VA) MICs determined by E-test for E. faecium isolate 11 (A), E. faecium isolate 13 (B), a colony of E. faecium isolate 13 grown in the inhibition zone next to the E-test strip (C), and E. faecium isolate 12 that had a homogeneous and high level of resistance to vancomycin and teicoplanin (D).
A prospective surveillance study was initiated in order to determine the epidemiology of VRE, and infection control measures were strengthened according to the recommendations of the Hospital Infection Control Practice Advisory Committee. From September to December 2004 rectal swab cultures were taken once a week from all patients hospitalized in the nephrology ward and for all newly admitted patients in the nephrology ward. Of 180 patients that have been screened during that period, 20 were positive for the epidemic VRE clone. Patients admitted to the nephrology ward were informed about the ongoing VRE outbreak and about the implemented rectal screening procedure, and the screening results were individually communicated to each screened patient. None of the patients admitted to the ward refused the screening.
Implementation of stringent hand disinfection and environmental disinfection policies and of patient isolation contained this outbreak but still did not eradicate VRE colonization from the unit.
Bacterial strains and VRE isolation.
VRE isolation from clinical specimens was carried out by standard microbiological techniques, as previously described (6). Rectal swabs for patient colonization screening were first grown in a bile esculin broth for 24 h at 37°C prior to plating on bile esculin-containing agar plates supplemented with 6 μg of vancomycin/ml (9). For each patient, duplicate isolates were excluded unless differences were observed in their antibiotic susceptibility pattern.
Species identification was performed by using species-specific PCR for E. faecalis and E. faecium (10) in addition to conventional tests and the API ID 32 STREP test (bioMérieux, Marcy l'Étoile, France). E. faecium BM4147 and E. faecalis V583 were used as a vanA- and vanB-containing reference strains, respectively (29). E. faecium Gil corresponds to a susceptible strain isolated at the Bicêtre Hospital and that was not related to the outbreak. Rifampin- and fusidic acid-resistant E. faecium BM76 was used as recipient in conjugation experiments (30).
Antimicrobial agents and MIC determinations.
Routine antibiograms were determined by a disk diffusion method on Mueller-Hinton agar plates (Bio-Rad, Marnes-La-Coquette, France) and interpreted according to the method of NCCLS (27). MICs of vancomycin and teicoplanin, were determined by the E-test method (AB BIODISK, Solna, Sweden) on Mueller-Hinton agar according to the manufacturer's instructions. All plates were incubated at 37°C for 18 h. MIC results were interpreted according to Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) guidelines (27). The presence of β-lactamase was detected by using cefinase-containing disks (BD Diagnostic Systems, Le Pont-de-Claix, France).
Plasmid content and conjugation assays.
Direct transfer of the vancomycin resistance determinant from E. faecium clinical isolates to E. faecium BM76 was attempted by solid mating-out assays at 37°C as previously described (30). Transconjugant selection was performed onto TS agar plates containing rifampin (60 μg/ml; Aventis, Paris, France), fusidic acid (20 μg/ml; Léo Pharma, St Quentin-en-Yvelines, France), and vancomycin (10 μg/ml; DakoTA Pharm, Le Plessis-Robinson, France).
Plasmid DNA of the parental E. faecium strains and of transconjugants were prepared by using an alkaline lysis protocol (32). Plasmid DNAs were analyzed by electrophoresis on a 0.8% agarose gel containing 0.15 μg of ethidium bromide/ml. Escherichia coli NCTC 50192 harboring 154-, 66-, 38-, and 7-kb plasmids was used as a plasmid-containing reference strain (8).
PCR and sequencing.
DNAs were extracted by boiling as described previously (29). Standard PCR amplification of vanA, vanB, and vanD genes was performed as previously described (3, 10, 29). Whole-cell DNA of isolates was purified as described previously (6). A total of 500 ng of whole-cell DNA was used in standard PCR mixtures in a GeneAmp 2700 thermal cycler (Applied Biosystems, Les Ulis, France). Tn1546-like elements were amplified by using PCR primers as previously described (26, 36).
IS16 PCR amplification was carried out with the following laboratory designed primers: IS16U (5′-AGAAGAACGGCAATCACAAAGA-3′) and IS16L (5′-TCAACCTCATCAAAAGCACAAT-3′), generating a 1.3-kb internal product. PCR screening for IS16 insertion in the vanY gene was done with primers IS16U and 9580R (36) (Fig. 2), generating a 1.4-kb fragment, and primers 8913F (5′-GTAAATCCAGTAGGGCGAAAT-3′) and 9580R located on each side of the insertion site of IS16, yielding a 2.1-kb fragment when IS16 was present and a 0.7-kb fragment when IS16 was absent.
FIG. 2.
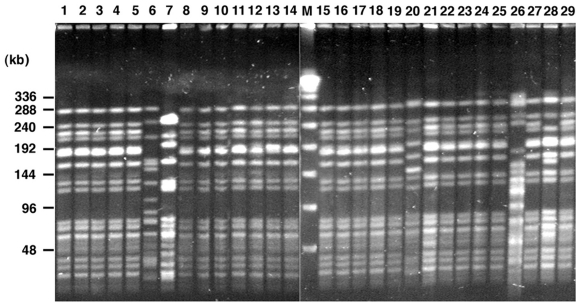
Analysis of SmaI-digested genomic DNA by PFGE. Lanes 1 to 29 represent the 26 VRE strains studied and three control strains (lanes 6, 7, and 26) as indicated in Table 1. Lane M, bacteriophage lambda concatemers were used as molecular size markers (Bio-Rad).
Amplicons were purified by using a QiaQuick PCR purification kit (QIAGEN) prior to their sequence determination with an ABI Prism 3100 sequencer (Applied Biosystems, Les Ulis, France). Nucleotide sequence alignments were carried out at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Genetic modifications in the van genes were studied by comparing the obtained sequences to those of prototype Tn1546 (23).
PFGE typing.
Whole-cell DNAs embedded in 1% agarose plugs (Bio-Rad) were digested with SmaI restriction enzyme (Amersham Pharmacia Biotech) and separated in a 1% pulsed-field-certified agarose gel (Bio-Rad) by using a CHEF DRII system (Bio-Rad), as described previously (6). Pulsed field gel electrophoresis (PFGE) was run at 14°C, with a 6-V/cm current, a switch angle of 120°, and switch times of 0.1 to 20 s for 20 h. After migration, gels were stained in a 0.5-μg/ml ethidium bromide solution, and PFGE results were analyzed according to the criteria of Tenover et al. (34).
RESULTS
Bacterial strains.
Twenty-six vancomycin-resistant E. faecium isolates were recovered from August to December 2004 from 24 patients (7 male, 17 female) aged between 30 and 85 years (mean age, 69 years). Twenty-three patients were hospitalized in the department of nephrology, and one was in the department of Internal Medicine. Eighteen patients were carriers, and seven had true VRE infections: one bacteremia, two peritonitis, and four urinary tract infections (Table 1). The duration of the stay in the nephrology department before VRE isolation ranged from 2 to 24 days (geometric mean, 13.5 days). Most of the patients were kidney-transplanted patients (33%), and the most frequent underlying disease was end-stage renal disease.
Antimicrobial susceptibility testing.
Most of the E. faecium isolates had a variable phenotypic expression of glycopeptide resistance (vancomycin MIC, 12 to 256 μg/ml; teicoplanin MIC, 4 to 32 μg/ml) (Fig. 1 and Table 1). These MICs were compatible with a VanD phenotype (4). In contrast, E. faecium isolates 12, 20, and 27 from patients XI, XII, and XXIII, respectively, had a homogeneous phenotype made of high-level resistance to vancomycin and to teicoplanin compatible with a VanA phenotype (vancomycin MIC, 256 μg/ml; teicoplanin MIC, 48 μg/ml) (Fig. 1D). All of the isolates were also resistant to ampicillin, clindamycin, erythromycin, and levofloxacin and displayed high-level resistance to gentamicin, kanamycin, and streptomycin but remained susceptible to chloramphenicol, linezolid, pristinamycin, rifampin, tetracycline, and trimethoprim-sulfamethoxazole. Differences were observed for isolate 10 from patient IV, which was trimethoprim-sulfamethoxazole resistant, and for isolate 20 from patient XII, which was resistant to trimethoprim-sulfamethoxazole and to gentamicin at a low level (Table 1). All E. faecium isolates were resistant to ampicillin and were β-lactamase negative as determined by a negative cefinase test result.
Heterogeneous expression of vancomycin resistance.
E. faecium isolates that displayed variable levels of vancomycin resistance were susceptible to or of intermediate resistance to teicoplanin (Table 1 and Fig. 1). Interpretation of MICs for vancomycin was difficult since most of these isolates presented heterogeneous expression of resistance, illustrated by growth of colonies in the elliptic inhibition zone when performing E-test susceptibility testing (Fig. 1A and B). This heterogeneous phenotype of resistance was not easily detectable after 24 h of culture and became clearly visible only after 48 h of growth (Fig. 1A). Colonies that grew inside the inhibition ellipse once retested displayed a homogeneous phenotype of resistance to vancomycin (Fig. 1C). Several isolates, although of intermediate susceptibility to teicoplanin, had homogeneous patterns of resistance to vancomycin (Fig. 1C). However, this homogeneous pattern of resistance was unstable and was reversible to an heterogeneous pattern of resistance after repeated subculture in drug-free medium. Conversely, the growth of heterogeneous isolates in the presence of 10 μg of vancomycin/ml eliminated the glycopeptide-susceptible subpopulation and selected for a homogeneous vancomycin-resistant subpopulation displaying teicoplanin resistance level that was only slightly increased. The homogeneous pattern of resistance persisted in these antibiotic-selected cells but reverted to its former heterogeneous pattern of resistance after subculture in an antibiotic-free medium. These observations corresponded to those observed in vivo, with a level of vancomycin resistance among isolates varying according to the use of vancomycin for treating patients prior to sampling of rectal swabs. Patient XIV (isolate 15) received a vancomycin-containing regimen 1 week prior to systematic screening was started. The VRE isolate had a high-level homogeneous vancomycin resistance phenotype. Rectal swabs performed the following weeks revealed only the heterogeneous phenotype for this same isolate (data not shown).
Detection and sequencing of vanA genes.
The first vancomycin-resistant E. faecium isolates found in clinical samples were considered to be phenotypically VanD-type isolates. However, by using specific PCR primers for vanA, vanB, and vanD genes, they were identified as vanA genotype. Subsequently, the vanA gene was detected by PCR in all isolates. The sequence of the 730-bp amplicon determined for all E. faecium isolates was identical to that published for the prototype vanA gene (20) (GenBank accession number M97297). The strains were PCR negative for the vanB and vanD genes.
Strain typing.
The 26 VRE isolates were analyzed by PFGE, and the results are shown in Table 1 and Fig. 2. The predominant PFGE strain type “A” was present in all patients. For patient XII, a second VRE isolate with a different PFGE strain type (“B”) was found. Our data suggest that the outbreak is monoclonal, but in vivo transfer of the antibiotic resistant determinant might have occurred since one patient had two distinct clones.
Plasmid extraction and mating experiments.
Plasmid DNA extraction from isolates 11, 12, and 20 revealed the presence of a large plasmid 200 kb (data not shown). For these isolates vancomycin-resistant transconjugants were obtained at a frequency of 10−5 to 10−6. No other antibiotic resistance determinant was cotransferred with the glycopeptide resistance determinant (data not shown). The vancomycin and teicoplanin MICs were similar to those for parental strains, i.e., respectively, 48 and 12 μg/ml for transconjugant (Tc) of isolate 11 and 256 and 48 μg/ml for transconjugants Tc12 and Tc20. Transconjugant Tc11 also displayed the heterogeneous expression of the vancomycin resistance of its parent isolate 11, suggesting that this heterogeneous expression of resistance was plasmid related (data not shown).
Amplification and sequencing of Tn1546-like transposons.
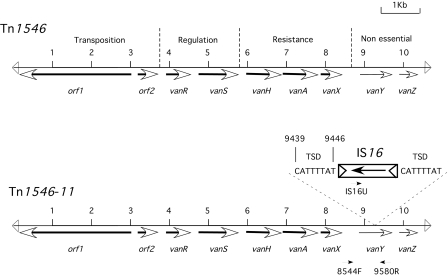
The genetic backgrounds of three E. faecium isolates were investigated in detail: isolate 11 (patient III), which expressed heterogeneous low-level vancomycin resistance (Fig. 1B), and isolate 12 (patient XI) and isolate 20 (patient XII), which both expressed high-level vancomycin resistance but belonged to different PFGE strain types (Fig. 1D). These isolates had a vanA genotype with a genetic organization identical to that described for Tn1546 (Fig. 3), except for the presence of IS16 (31) inserted into the vanY gene of isolate 11 (patient III). IS16 insertion led to an 8-bp target site duplication that disrupted the vanY coding sequence (Fig. 3). This IS16 element was not inserted in the vanY gene of isolates 12 and 20. Point mutations were not found in either the genes involved in resistance to glycopeptides or in the genes involved in its regulation.
FIG. 3.
Schematic representation of Tn1546-like transposons. The orientation of the genes is indicated by an arrow. (A) Structure of the prototype Tn1546 transposon (37); (B) structure of the Tn1546-like transposon characterized from E. faecium isolate 11. Primers used for IS16 mapping are indicated by an arrow. TSD, target site duplication generated by IS16 upon insertion into vanY gene.
Screening for IS16.
PCR screening for IS16 in the vanY gene of the other VRE isolates revealed that this mobile element was present in all of the isolates except isolates 12, 20, and 27, for which a high-level vancomycin and teicoplanin resistance was observed. These isolates contained, however, IS16 inserted somewhere else, as revealed by positive PCR results (Table 1). IS16 was absent from E. faecium BM4147 and from E. faecium BM76. Similarly, the transconjugants Tc11, Tc12, and Tc20 were positive for IS16, but only Tc11 had an IS16 inserted into the vanY gene.
DISCUSSION
The prevalence of VRE remains low among nosocomial acquired pathogens in France (11, 18, 33). A recent study performed in 2000 to 2002 showed that the vancomycin resistance rate among E. faecium isolates in French intensive care units was 0.8% (18). We report here the outbreak in a French hospital of a VRE strain with very uncommon expression of the glycopeptide resistance determinant. Although a few isolates displayed a “true” VanA phenotype (vancomycin MIC, >256 μg/ml; teicoplanin MIC, 48 μg/ml), most of the isolates had a typical VanD phenotype with vancomycin MICs ranging from 12 to 24 μg/ml and teicoplanin MICs ranging from 4 to 8 μg/ml (Table 1). The vancomycin MICs would also fit with a VanB phenotype but the teicoplanin MICs fit only with a VanD phenotype (4, 37). The fact that these isolates also displayed a low-level heterogeneous resistance to vancomycin made their detection very difficult. Indeed, one of the very first VRE isolates was not retained as VRE since its glycopeptide MICs were in the susceptibility range. Only PCR analysis allowed detection of the vanA genotype in that case, demonstrating the usefulness of molecular tools for detecting a van genotype and for the detailed follow-up of such outbreaks. However, retesting these VRE isolates with low-level resistance to vancomycin and teicoplanin with an automated antimicrobial susceptibility test system (VITEK2; bioMérieux) permitted their unambiguous identification as VRE (P. Nordmann, unpublished data).
Colonization and infection with VRE primarily affect moderately to severely ill patients in acute-care hospitals (15, 16). The patients involved in our outbreak had most of the known risk factors, with advanced age and the severity of the underlying illness being the most important (4, 16). Screening for carriers, implementation of reinforced hygiene measures, and cohorting of carriers led to the control of the outbreak but did not allow for eradication of VRE since most of the carriers are chronic patients that required frequent rehospitalizations (21). Several recent investigations suggest that a monoclonal outbreak, if not brought under control, can evolve into polyclonal endemicity (16, 20). Early detection and implementation of control measures has kept the epidemic as a clonal dissemination, as demonstrated by a unique E. faecium PFGE type. However, horizontal gene transfer also occurred, as illustrated by the two isolates with distinct PFGE strain types isolated from a single patient.
VRE strains with the vanA genotype that were susceptible to teicoplanin and thus have the VanB phenotype have been detected in Japan, Taiwan, and Korea (14, 17, 24). Hashimoto et al. (14) suggested that three point mutations in the putative sensor domain of vanS could be responsible for the impaired resistance phenotype to teicoplanin for VRE isolates possessing a vanA gene cluster. Direct sequencing of the vanS gene failed to demonstrate point mutations in the vanS gene in our isolates. Impairment of accessory proteins VanY d,d-carboxypeptidase, which contributes to vancomycin resistance by cleaving the C-terminal D-Ala residue, has also been proposed as a possible explanation for impaired resistance to teicoplanin among VRE isolates possessing the vanA gene cluster (24). Similarly, the vanZ gene that confers low-level resistance to teicoplanin has also been implicated (2, 24).
In our study, a heterogeneous vancomycin resistance, along with susceptibility or intermediate resistance to teicoplanin, was related to an IS16 inserted into the vanY gene. Several IS-like elements have been found in the vanA gene cluster of several E. faecium isolates. These are IS1251, which has been found in the intergenic region of vanS/H; IS1216V-like, which has been found in several noncoding regions of the resistance operon (36); IS1542, which is inserted in the intergenic region of orf2/vanR (17); and IS19 in the intergenic region of vanS/H (17). In addition, IS1476 has been identified in vanY of a clinical isolate of E. faecium resistant to vancomycin from Canada (25). IS16 has never been associated with vanA gene clusters. It was initially characterized from a E. faecalis strain to be part of Tn1547, a 64-kb composite transposon harboring a vanB gene cluster flanked by two distantly related ISs, designated IS256-like and IS16, in a direct orientation (31). Recently, genetic rearrangement of vanY or vanZ or partial or complete deletion of both genes following insertion of IS1216V have been pointed out as being the origin of VanB phenotype and vanA genotype VRE in Korea (17, 24). Similarly, IS1476 that was inserted in vanY gene led to the inactivation of this gene and to reduction of the expression of the downstream-located gene vanZ (25). Our data are consistent with these findings in the sense that the IS16 inserted in the vanY gene may impair the VanY synthesis and probably affects vanZ transcription, resulting in a lower teicoplanin MIC (12, 24, 25).
The heterogeneous vancomycin expression is linked to the plasmid itself, as suggested by the results of the conjugation experiment. It seems that the strains expressing an heterogeneous vancomycin resistance phenotype are made of two subpopulations: susceptible and highly resistant cells. Only rare cells express resistance trait and grow in the presence of high concentrations of drug. Upon vancomycin selection, these cells are selected for a homogeneous expression of resistance. However, this trait is not stable and, after subculture in vancomycin-free medium, the heterogeneous phenotype is restored.
Finally, it should be pointed out that three isolates (isolates 12, 20, and 27) expressed a high-level homogeneous vancomycin resistance phenotype with no IS16 insertion in the vanY gene, as observed for Tn1546 of E. faecium BM4147 (23). Unlike E. faecium BM4147, which lacks the IS16 element, isolates 12, 20, and 27 contained IS16, as revealed by PCR analysis. The transconjugants of isolate 12 and 20 were also positive for IS16. These data suggest that the conjugative plasmid that carried the vanA gene also carried another copy of IS16. Precise excision of IS16 would restore wild-type VanY and VanZ activity and thus result in a high-level homogeneous expression. Further investigations will be necessary to understand the molecular basis of this heterogeneous vancomycin expression, including the role of IS16.
To the best of our knowledge, this is the first reported outbreak of VanD phenotype, vanA genotype enterococci and also the first vanA-related VRE outbreak in France. These strains may become a serious problem in the future since their detection is very difficult.
Acknowledgments
We are very grateful to C. Poyart for the gift of strain BM76 and to F. Claverie-Martin for providing the reference strains used in this study. We thank also J. Sirot for complementary susceptibility testing.
This study was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France; by the Assistance Publique-Hôpitaux de Paris, Paris, France; and by the European Community (6th PCRD, LSHMCT-2003-503-335).
REFERENCES
- 1.Arthur, M., and P. Courvalin. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, C. Molinas, P. Reynolds, and P. Courvalin. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87-92. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D. A., P. Kibsey, D. Roscoe, and M. R. Mulvey. 2004. Enterococcus faecium N03-0072 carries a new VanD-type vancomycin resistance determinant: characterization of the VanD5 operon J. Antimicrob. Chemother. 54:680-683. [DOI] [PubMed] [Google Scholar]
- 4.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers, H. F. 1988. Methicillin-resistant staphylococci. Clin. Microbiol. Rev. 1:173-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colak, D., T. Naas, F. Gunseren, N. Fortineau, D. Ogunc, M. Gultekin, and P. Nordmann. 2002. First outbreak of vancomycin-resistant enterococci in a tertiary hospital in Turkey. J. Antimicrob. Chemother. 50:397-401. [DOI] [PubMed] [Google Scholar]
- 7.Comité Technique National des Infections Nosocomiales. 1999. 100 Recommendations pour la surveillance et la Prévention des Infections Nosocomiales, 2nd ed. Ministére de l'Emploi et de la Solidarité, Secrétariat d'Etat à la Santé et l'Action Sociale, Paris, France.
- 8.Danel, F., M. C. Hall, D. Gur, and D. Livermore. 1995. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1881-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danmap. 2003. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods, and humans in Denmark. [Online.] Ministry of Food, Agriculture, and Fisheries and the Ministry of Health, Copenhagen, Denmark. http://www.dfvf.dk/Files/Filer/Zoonosecentret/Publikationer/Danmap/Danmap_2003.pdf.
- 10.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Antimicrobial Resistance Surveillance System. 2002. EARSS Annual Report 2002: on-going surveillance of S. pneumoniae, S. aureus, E. coli, E. faecium, and E. faecalis. [Online.] EARSS, Bilthoven, The Netherlands. http://www.earss.rivm.nl/PAGINA/DOC/rep2002/annual-report-2002.pdf.
- 12.Eom, J. S., I. S. Hwang, B. Y. Hwang, J. G. Lee, Y. J. Lee, H. J. Cheong, Y. H. Park, S. C. Park, and W. J. Kim. 2004. Emergence of vanA genotype vancomycin-resistant enterococci with low or moderate levels of teicoplanin resistance in Korea. J. Clin. Microbiol. 42:1785-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control. 16:128-140. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, Y., K. Tanimoto, Y. Ozawa, T. Murata, and Y. Ike. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant Enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185:247-254. [DOI] [PubMed] [Google Scholar]
- 15.Hayden, M. K., G. M. Trenholme, J. E. Schultz, and D. F. Sahm. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J. Infect. Dis. 167:1224-1227. [DOI] [PubMed] [Google Scholar]
- 16.Hayden, M. K. 2000. Insights into the epidemiology and control of infection with vancomycin-resistant enterococci. Clin. Infect. Dis. 31:1058-1065. [DOI] [PubMed] [Google Scholar]
- 17.Huh, J. Y., W. G. Lee, K. Lee, W. S. Shin, and J. H. Yoo. 2004. Distribution of insertion sequences associated with Tn1546-like elements among Enterococcus faecium isolates from patients in Korea. J. Clin. Microbiol. 42:1897-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, M. E., D. C. Draghi, C. Thornsberry, J. A. Karlowsky, D. F. Sahm, and R. P. Wenzel. 2004. Emerging resistance among bacterial pathogens in the intensive care unit: a European and North American Surveillance study (2000-2002). Ann. Clin. Microbiol. Antimicrob. 3:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawalec, M., M., Gniadkowski, and M. Hryniewicz. 2000. Outbreak of vancomycin-resistant enterococci in a hospital in Gdansk, Poland, due to horizontal transfer of different Tn1546-like transposon variants and clonal spread of several strains. J. Clin. Microbiol. 38:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, W. J., R. A. Weinstein, and M. K. Hayden. 1999. The changing molecular epidemiology and establishment of endemicity of vancomycin resistance in enterococci at one hospital over a 7-year period. J. Infect. Dis. 179:163-171. [DOI] [PubMed] [Google Scholar]
- 21.Lai, K. K., A. L. Kelley, Z. S. Melvin, P. P. Belliveau, and S. A. Fontecchio. 1998. Failure to eradicate vancomycin-resistant enterococci in a university hospital and the cost of barrier precautions. Infect. Control Hosp. Epidemiol. 19:647-652. [DOI] [PubMed] [Google Scholar]
- 22.Lauderdale, T. L., L. C. McDonald, Y. R. Shiau, P. C. Chen, H. Y. Wang, J. F. Lai, and M. Ho. 2002. Vancomycin-resistant enterococci from humans and retail chickens in Taiwan with unique VanB phenotype-vanA genotype incongruence. Antimicrob. Agents Chemother. 46:525-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium strain. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 24.Lee, W. G., J. Y. Huh, S. R. Cho, and Y. A. Lim. 2004. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob. Agents Chemother. 48:1379-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKinnon, M. G., M. A. Drebot, and G. J. Tyrrell. 1997. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob. Agents Chemother. 41:1805-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miele, A., M. Bandera, and B. P. Goldstein. 1995. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob. Agents Chemother. 39:1772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2004. Methods of dilution antimicrobial susceptibility tests for bacteria that grow aerobically: M7 approved standards, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Palepou, M. F.-I., A. A. Adebiyi, C. H. Tremlett, L. B. Jensen, and N. Woodford. 1998. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J. Antimicrob. Chemother. 42:605-612. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Hernandez, X., S. Mendez-Alvarez, and F. Claverie-Martin. 2002. A PCR assay for rapid detection of vancomycin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 42:273-277. [DOI] [PubMed] [Google Scholar]
- 30.Poyart, C., and P. Trieu-Cuot. 1994. Heterogeneric conjugal transfer of the pheromone-responsive plasmid pIP964 (IncHlyI) of Enterococcus faecalis in the apparent absence of pheromone induction. FEMS Microbiol. Lett. 122:173-180. [DOI] [PubMed] [Google Scholar]
- 31.Quintiliani, R., Jr., and P. Courvalin. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1-8. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schouten, M. A., J. A. Hoogkamp-Korstanje, J. F. Meis, A. Voss, et al. 2000. Prevalence of vancomycin-resistant enterococci in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 19:816-822. [DOI] [PubMed] [Google Scholar]
- 34.Tenover, F. C., R. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA re-striction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 36.Willems, R. J., J. Top, N. Van den Braak, A. Van Belkum, A. Mevius, D. J. Hendriks, G. M. Van Santen-Verheuvel, and J. D. Van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodford, N. 2001. Epidemiology of the genetic elements responsible for acquired glycopeptide resistance in enterococci. Microb. Drug Resist. 7:229-234. [DOI] [PubMed] [Google Scholar]