Abstract
Streptococcus agalactiae, also designated group B streptococcus (GBS), is an important pathogen in neonates, pregnant women, and nonpregnant adults with predisposing conditions. We used multilocus sequence typing (MLST) to characterize 158 GBS isolates that were associated with neonatal and adult invasive disease and that were collected in northern and western Sweden from 1988 to 1997. Five major genetic lineages (sequence type [ST] 19, ST-17, ST-1, ST-23, and ST-9 complexes) were identified among the isolates, including serotype Ia, Ib, and II to V isolates, indicating a highly clonal population structure among invasive GBS isolates. A number of STs were found to contain isolates of different serotypes, which indicates that capsule switching occurred rather frequently. Two distantly related genetic lineages were identified among isolates of serotype III, namely, clonal complex 19 (CC19), and CC17. CC19 was equally common among isolates from adult and neonatal disease (accounting for 10.3% of GBS isolates from adult disease and 18.7% from neonatal disease), whereas CC17 significantly appeared to be associated with neonatal invasive disease (isolated from 21.9% of neonatal isolates but only 2.6% of adult isolates). The distribution of the mobile elements GBSi1 and IS1548 reveals that they can act as genetic markers for lineages CC17 and CC19, respectively.
Streptococcus agalactiae, also designated group B streptococcus (GBS), has remained a leading cause of neonatal mortality and morbidity in the United States and Europe since its recognition as an important pathogen in the mid-1960s (2). The incidence of infection among neonates has ranged from 1.0 to 4.7 per 1,000 live births during the last decade (3). GBS also causes diseases in pregnant women and nonpregnant adults with underlying conditions.
Capsular serotyping has been the classic method used in the descriptive epidemiology of GBS. According to capsular expression, GBS isolates are subclassified into nine serotypes, Ia, Ib, and II through VIII. There are demographic, geographic, and temporal variations with respect to the predominant serotypes present in the human population (19, 23). Nevertheless, investigations from the United States and Western Europe have shown that four serotypes (Ia, II, III, and V) account for 86 to 90% of all clinical isolates (19, 22). Serotype III is of particular interest since it causes a substantial proportion of early-onset neonatal infections and almost all late-onset infections (17).
The population structure of GBS has been studied by a variety of techniques, including multilocus enzyme electrophoresis typing, pulsed-field gel electrophoresis, restriction digestion pattern typing (RDP), and restriction fragment length pattern analysis (25-28, 30). Diverse lineages among serotype III isolates have been distinguished, and lineages have been suggested to differ in their virulence potentials and tissue tropisms. However, in few of these studies were the isolates collected from a well-defined population during a defined time span; thus, the conclusions could be influenced by sampling bias. In this study we utilized a multilocus sequence typing (MLST) scheme, an unambiguous sequence-based typing method, to characterize the invasive isolates collected from northern and western Sweden from 1988 to 1997. The objectives of this study were to analyze the genotype distribution, i.e., the population structure, of GBS isolates in the two geographic areas, approximately 1,000 km apart, and to monitor the population dynamics over a 10-year span. Another intent was to investigate whether a correlation between particular genotypes and infection types existed or not.
MATERIALS AND METHODS
Collection of clinical isolates. The retrospective study was conducted at the Departments of Clinical Bacteriology, Umeå University Hospital, in Umeå (northern Sweden) and at the Sahlgrenska University Hospital in Gothenburg (western Sweden) from 1988 to 1997. A collection of 158 invasive isolates was studied, including isolates recovered from infants and adults. The collection of isolates from the Sahlgrenska University Hospital has been previously described by Berg et al. (4). The Umeå unit serves the county of Västerbotten, which has 250,000 inhabitants; and 87 consecutive isolates were recovered from this region. Isolates were cultured from blood or cerebrospinal fluid and were identified on the basis of colony morphology, beta-hemolysis, Gram staining, and the CAMP test; and the identities were confirmed by Lancefield group antigen determination (Streptex; Murex Biotech, Dartford, United Kingdom). GBS isolates were serotyped by use of a coagglutination typing kit (Essum; Bacterum AB, Umeå, Sweden).
MLST.
MLST was performed as described by Jones et al. (20). Briefly, fragments of the MLST loci were amplified with Taq polymerase (Applied Biosystems, Stockholm, Sweden) and PCR systems (Whatman Biometra, Täby, Sweden). The amplified fragments were purified and sequenced with the BigDye terminator v1.1 (Applied Biosystems). The nucleotide sequences were determined with an ABI prism sequencer (Applied Biosystems) and analyzed by use of the DNASIS program (Hitachi Genetic Systems, Berlin, Germany). Each compiled sequence was determined at least once for each strand. Alleles for seven loci were assigned on the MLST website (http://sagalactiae.mlst.net), and each isolate was defined by the sequence type (ST), which was assigned according to the unique allelic profile. All the new alleles were reamplified and resequenced. Cluster analysis was carried out with START, using the unweighted pair group method with arithmetic mean (UPGMA) to construct the dendrogram from the matrix of pairwise allelic differences between the STs. The program eBURST was used to group isolates into lineages or clonal complexes (CCs), which share six or seven identical alleles (12). To assess whether any of the lineages is associated with adult or neonatal patients, the distribution of each clonal complex and clone (singleton) was examined against the random distribution by the chi-square test (Bonferroni corrected).
Detection of mobile genetic elements by PCR.
The PCR amplifications of mobile genetic elements were carried out with the primers and conditions described previously (16). For amplification of IS1548, primer pair 5′-GGTAAAACGATTCGAGGCAATCGAGG-3′ and 5′-TCTCTTCATCCTTTTGTGCCCGGAC-3′ was used; and for the detection of GBSi1, PCR was carried out with primer pair 5′-ATCTTCTAAGCGTGCTTTAG-3′ and 5′-CACATCAACTGAAGGGATAAG-3′. For the negative control, the DNA template was replaced by distilled water. The amplified products were separated in a 1% agarose gel and were detected by UV transillumination after ethidium bromide staining.
RESULTS
Relatedness of GBS isolates.
The DNA sequences of the seven loci were determined, and allelic profiles were assigned. The 158 isolates were resolved into 29 unique STs. Ten STs were represented by at least four isolates, while 6 STs were represented by two isolates and 13 STs were represented by only a single isolate (Tables 1 and 2). In the Umeå collection, 19 STs were identified among the 87 isolates, and 50 isolates (57.4% of the data set) were represented by one of the following three STs: ST-19 (23%), ST-17 (17.2%), and ST-23 (17.2%). In comparison to these major STs, other STs were uncommon, accounting for 1 to 8% of the data set. Among the 71 isolates from Gothenburg, 21 STs were found; and the major STs were ST-17 (23.9%), ST-19 (15.5%), and ST-1 (12.7%). Eleven STs were present in both geographic regions, whereas 8 were found only in Umeå and 10 were found only in Gothenburg. Six STs identified in Umeå and four STs identified in Gothenburg have not previously been reported to the database. No major difference could be seen in the distribution of the predominant STs over time, with the exceptions of ST-1, which did not appear until the early 1990s, and ST-23, which became rare after 1994 in both areas. ST-4 was the largest transient ST, represented by a total of five isolates in Umeå between 1993 and 1994 and by two isolates from Gothenburg between 1994 and 1996.
TABLE 1.
Characteristics of invasive GBS isolates from Västerbotten (northern Sweden) according to sequence type and disease category
| CC and ST | Allelic profilea | Serotype (no. of isolates) | Frequency (% of data set) | Source (no. of isolates) |
Time span | ||
|---|---|---|---|---|---|---|---|
| Adult | Childb | Neonatec | |||||
| CC9 | |||||||
| 9 | 8, 1, 4, 1, 3, 3, 2 | Ib (6) | 6.9 | 1 | 5 | 1989-1997 | |
| 10 | 9, 1, 4, 1, 3, 3, 2 | II (2), Ib (1) | 3.4 | 3 | 0 | 1988-1996 | |
| 8 | 4, 1, 4, 1, 3, 3, 2 | Ib (1) | 1.1 | 1 | 0 | 1994 | |
| 12 | 10, 1, 4, 1, 3, 3, 2 | III (1) | 1.1 | 1 | 0 | 1994 | |
| 230 | 8, 1, 28, 1, 3, 23, 2 | Ib (1) | 1.1 | 1 | 1991 | ||
| CC19 | |||||||
| 19 | 1, 1, 3, 2, 2, 2, 2 | III (18), II (2) | 23 | 8 | 12 | 1988-1997 | |
| 121 | 32, 1, 3, 2, 2, 2, 2 | III (1) | 1.1 | 0 | 1 | 1989 | |
| 122 | 1, 1, 3, 2, 2, 19, 2 | III (1) | 1.1 | 0 | 1 | 1997 | |
| 124 | 1, 1, 22, 2, 2, 2, 2 | III (1) | 1.1 | 0 | 1 | 1990 | |
| 107 | 15, 1, 3, 2, 2, 2, 2 | III (2) | 2.3 | 0 | 2 | 1992-1997 | |
| 28 | 1, 1, 3, 5, 2, 2, 2 | II (2) | 2.3 | 1 | 1 | 1990-1991 | |
| CC17 | |||||||
| 17 | 2, 1, 1, 2, 1, 1, 1 | III (15) | 17.2 | 4 | 11 | 1988-1997 | |
| 119 | 2, 1, 21, 2, 1, 1, 1 | III (2) | 2.3 | 0 | 2 | 1993 | |
| 120 | 2, 15, 1, 2, 1, 1, 1 | III (2) | 2.3 | 0 | 2 | 1994 | |
| CC1 | |||||||
| 2 | 1, 1, 3, 1, 1, 2, 2 | II (3) | 3.4 | 2 | 1 | 1992-1993 | |
| 1 | 1, 1, 2, 1, 1, 2, 2 | V (5) | 5.7 | 2 | 3 | 1992-1997 | |
| 196 | 1, 1, 3, 1, 1, 12, 2 | IV (1) | 1.1 | 1 | 1995 | ||
| CC23 | |||||||
| 23 | 5, 4, 6, 3, 2, 1, 3 | III (2), Ia (13) | 17.2 | 5 | 1 | 9 | 1988-1997 |
| Singleton | |||||||
| 4 | 1, 1, 4, 1, 1, 3, 4 | Ia (5) | 5.7 | 3 | 2 | 1993-1994 | |
| Total | 87 | 100 | 32 | 2 | 53 | ||
The allelic profiles are present in the following order: adhP, pheS, atr, glnA, sdhA, glcK, and tkt.
Two to 4 years old.
Up to 3 months old.
TABLE 2.
Characteristics of invasive GBS isolates from Gothenburg (western Sweden) according to sequence type and disease category
| CC and ST | Allelic profilea | Serotype (no. of isolates) | Frequency (% of data set) | Source |
Time span | ||
|---|---|---|---|---|---|---|---|
| Adult | Childb | Neonatec | |||||
| CC9 | |||||||
| 12 | 10, 1, 4, 1, 3, 3, 2 | Ib (1), II (3) | 5.6 | 3 | 1 | 1988-1993 | |
| 8 | 4, 1, 4, 1, 3, 3, 2 | Ib (1) | 1.4 | 1 | 0 | 1992 | |
| 9 | 8, 1, 4, 1, 3, 3, 2 | Ib (1) | 1.4 | 1 | 0 | 1995 | |
| 10 | 9, 1, 4, 1, 3, 3, 2 | Ib (1) | 1.4 | 1 | 0 | 1994 | |
| 7 | 10, 1, 2, 1, 3, 2, 2 | V (2) | 2.8 | 0 | 2 | 1994 | |
| CC19 | |||||||
| 19 | 1, 1, 3, 2, 2, 2, 2 | III (11) | 15.5 | 4 | 1 | 6 | 1989-1996 |
| 27 | 1, 1, 3, 4, 2, 2, 2 | III (2) | 2.8 | 2 | 0 | 1991-1994 | |
| 86 | 1, 1, 3, 2, 2, 1, 2 | III (1) | 1.4 | 1 | 0 | 1988 | |
| 107 | 15, 1, 3, 2, 2, 2, 2 | III (4) | 5.6 | 0 | 4 | 1989-1996 | |
| 175 | 1, 1, 3, 3, 2, 2, 2 | III (1) | 1.4 | 0 | 1 | 1996 | |
| CC17 | |||||||
| 17 | 2, 1, 1, 2, 1, 1, 1 | III (17) | 23.9 | 0 | 17 | 1990-1997 | |
| 125 | 2, 1, 1, 2, 19, 1, 1 | III (1) | 1.4 | 0 | 1 | 1995 | |
| 150 | 2, 1, 1, 2, 1, 1, 12 | III (1) | 1.4 | 0 | 1 | 1994 | |
| CC1 | |||||||
| 1 | 1, 1, 2, 1, 1, 2, 2 | V (8), Ib (1) | 12.7 | 5 | 4 | 1992-1996 | |
| 2 | 1, 1, 3, 1, 1, 2, 2 | II (2), IV (2) | 5.6 | 1 | 3 | 1991-1994 | |
| 139 | 1, 1, 4, 1, 1, 2, 2 | II (1) | 1.4 | 0 | 1 | 1994 | |
| 151 | 34, 1, 2, 1, 1, 2, 2 | V (1) | 1.4 | 1 | 0 | 1996 | |
| CC23 | |||||||
| 23 | 5, 4, 6, 3, 2, 1, 3 | Ia (5) | 7 | 1 | 4 | 1990-1994 | |
| 234 | 5, 4, 6, 3, 2, 1, 16 | III (1) | 1.4 | 0 | 1 | 1994 | |
| 88 | 5, 10, 6, 3, 2, 1, 3 | Ia (1) | 1.4 | 0 | 1 | 1995 | |
| Singleton | |||||||
| 4 | 1, 1, 4, 1, 1, 3, 4 | Ia (2) | 2.8 | 0 | 2 | 1994-1995 | |
| Total | 71 | 100 | 21 | 1 | 49 | ||
The allelic profiles are present in the following order: adhP, pheS, atr, glnA, sdhA, glcK, and tkt.
Seven months old.
Up to 3 months old.
Identification of clonal lineages.
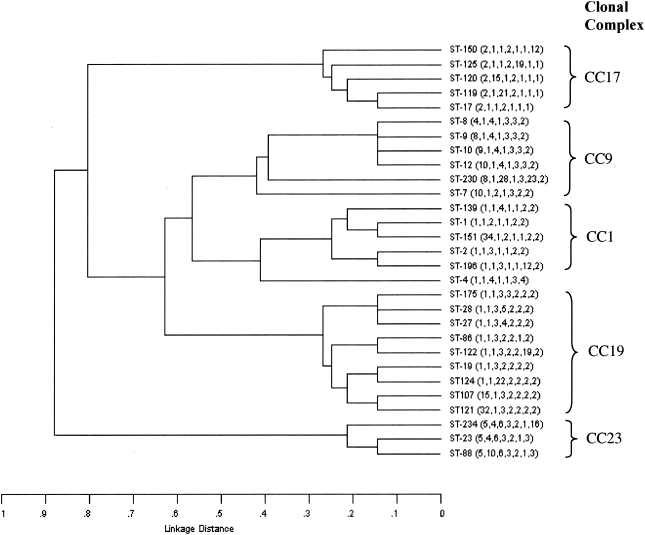
UPGMA was used to construct a dendrogram (Fig. 1) from the matrix of pairwise allelic differences between the 29 STs of all 158 isolates. These 29 STs were also grouped by eBURST into seven lineages, where the STs share six or seven identical alleles. The eBURST analysis revealed five major clonal complexes and two singletons. The ancestors of three CCs were identified as ST-1, ST-19, and ST-17; for the other two clonal complexes the program could not unambiguously predict the founder ST. The clonal complex was designated after its ancestor ST or the representative ST that appeared with the highest frequency. CC19 (n = 46 isolates) was the most predominant, followed by CC17 (n = 38), CC1 (n = 24), CC23 (n = 22), and CC9 (n = 21). Eight single-locus variants (SLVs; which differed from the ancestor ST at one allele) were identified for ST-19, five SLVs were identified for ST1, and four SLVs were identified for ST-17. Most SLVs were unique to the respective geographical area and contained only one or two isolates; ST-107 and ST-2, however, were represented by a few isolates in both areas (Tables 1 and 2). In the UPGMA dendrogram, isolates of CC19, CC9, and CC1 appeared to be relatively closely related, whereas isolates of CC23 and CC17 were more divergent. In support of this, when the group definition was set to five of seven alleles with eBURST, isolates of CC19, CC9, and CC1 fell into a single group, whereas CC23 and CC17 remained as distinct groups. The close relationship between CC19 and CC1 was further supported by the identification of ST-2, which is an SLV of ST-1 and a double-locus variant (DLV) of ST-19. The grouping of ST-7 was a problem, since it differed at two alleles from both ST-1 and ST-12. ST-7 was designated a singleton by eBURST, since it lacked an SLV link to ST-1 or ST-12. Nevertheless, ST-7 appeared to be more closely related to CC9 in the UPGMA dendrogram; therefore, we manually placed it as a member of CC9.
FIG. 1.
UPGMA dendrogram showing the genetic relationship between the 29 STs of 158 invasive isolates. The allelic profile of each ST is shown in parentheses. The CC was grouped by BURST.
CC19 and CC17 were the predominant clonal complexes, together accounting for half of the isolates, whereas CC9 and CC23 were much less common, represented by 10 and 17% of the isolates in the two regions, respectively. A geographical difference in the distribution of the clonal complexes was noted for CC1, which was more common in Gothenburg (21.1%) than in Umeå (10.2%). The distributions of CC19, CC17, and CC9 showed stability over time. In contrast, CC23 almost disappeared after 1994, whereas CC1 did not appear until the early 1990s but has caused a considerable proportion of invasive cases of disease ever since. The fluctuation of lineages over time was more apparent in the western part of Sweden than in the northern region.
Relationship between ST and capsular serotype.
The capsular serotype was determined for all the isolates (Table 1 and 2). Serotype III was predominant (84 isolates), followed by Ia (26 isolates), V (16 isolates), II (15 isolates), Ib (14 isolates), and IV (3 isolates). No isolates of serotype VI, VII, or VIII were identified. In general, the serotypes appeared to be associated with the overall genotype defined by the ST. However, six STs were found to contain isolates of more than one serotype. As stated above, closely related STs constituted a clonal complex, and all clonal complexes except CC17 were shown to contain isolates of more than one serotype. The 38 CC17 GBS isolates all expressed a serotype III capsule. The majority of the 45 isolates in CC19 expressed serotype III, but 4 isolates were found to be serotype II. Two serotypes, Ia (19 isolates) and III (3 isolates), were present among GBS isolates of ST-23. CC9 contained 13 serotype Ib isolates, 5 serotype II isolates, 2 isolates of serotype V, and 1 isolate of serotype III. CC1 also demonstrated a high degree of serotypic variation and contained isolates of serotype V (n = 14), II (n = 6), IV (n = 3), and Ib (n = 1).
When the clonal complex and STs were analyzed with respect to serotype, isolates of serotypes Ib and V were mostly present in a single clonal complex, CC9 and CC1, respectively. In contrast, the genotypes of isolates of serotype Ia, II, and III were more heterogeneous (Table 3). These serotypes were represented by isolates in distantly related clonal complexes or STs, which differed by at least five alleles from each other. However, even though the isolates of serotype III displayed the highest number of STs, the majority of serotype III isolates fell into CC19 (42 isolates) or CC17 (39), and the rest of the serotype III isolates (4) were represented by ST-23 and ST-9. Likewise, the majority of serotype Ia isolates belonged to CC23 (19) or ST-4 (7), which were genetically far apart, according to the dendrogram.
TABLE 3.
Distribution of sequence types within serotypes of 158 invasive GBS isolates
| Sero- type | Total no. of isolates | No. of STs | STs (no. of isolates) |
|---|---|---|---|
| Ia | 26 | 3 | 23a(18), 88 (1), 4 (7) |
| Ib | 14 | 6 | 9 (7), 10a(2), 8 (2), 230 (1), 12a(1), 1a(1) |
| II | 15 | 7 | 12 (3), 2 (3), 10 (2), 19a(2), 28 (2), 2a(2), 139 (1) |
| III | 84 | 17 | 17 (32), 19 (29), 107 (6), 28 (2), 27 (2), 122 (2), 23 (2), 119 (2), 120 (2), 12 (1), 121 (1), 125 (1), 124 (1), 86 (1), 175 (1), 234 (1), 150 (1) |
| IV | 3 | 2 | 2 (2), 196 (1) |
| V | 16 | 3 | 1 (13), 7 (2), 151 (1) |
STs which contain multiple serotypes.
The association between lineage and disease.
The distributions of the lineages among adults and neonates with invasive GBS disease are summarized in Table 4. The five major lineages were present among both the adult and the neonatal patients. However, there was a highly significant association between CC17 and neonatal infections (chi-square test, Bonferroni corrected, P = 0.006), whereas no correlation between other lineages and a specific patient group could be established (chi-square test, Bonferroni corrected, P > 1). Isolates from CC9 and CC1 were nearly evenly represented among the adult and the neonatal isolates from Umeå. However, in the Gothenburg region, six of nine isolates belonging to CC9 were isolated from adult patients, whereas among neonatal patients, six of seven isolates belonged to CC23 (Tables 1 and 2).
TABLE 4.
Distribution of clonal complexes in relation to patient group
| CC or ST | No. (%) of isolates |
Total | |
|---|---|---|---|
| Adult | Neonatal | ||
| CC9a | 12 (7.7) | 8 (5.2) | 20 (12.9) |
| CC19a | 16 (10.3) | 29 (18.7) | 45 (29.0) |
| CC17b | 4 (2.6) | 34 (21.9) | 38 (24.5) |
| CC1a | 12 (7.7) | 12 (7.7) | 24 (15.4) |
| CC23a | 6 (3.9) | 15 (9.7) | 21 (13.6) |
| ST4a | 3 (1.9) | 4 (2.6) | 7 (4.5) |
| Total | 53 (34.2) | 102 (65.8) | 155 |
Chi-square test, Bonferroni corrected, P > 1.
Chi-square test, Bonferroni corrected, P = 0.006.
Distribution of mobile genetic elements among serotype III isolates.
In our previous studies, GBSi1 and IS1548 were found to never coexist among the serotype III isolates, which suggests that these elements may be markers for separate lineages (15, 24). The distributions of IS1548 and GBSi1 among the serotype III isolates were also examined in this study. The presence of IS1548 was indicated by an amplification product of 860 bp. For GBSi1, an insertion of 2,000 bp was detected at the scpB locus, the most common insertion site for GBSi1 (24), whereas a “space” of 150 bp was amplified in other isolates. Strikingly, all CC19 isolates carried IS1548 but none harbored GBSi1, whereas GBSi1 was present in all CC17 isolates. IS1548 was not found among any of the isolates belonging to CC17. In contrast, none of the elements were present in the two serotype III isolates of ST-23 and a single isolate of ST-12.
DISCUSSION
The group B streptococcus is a common cause of neonatal diseases, such as pneumonia, septicemia, and meningitis; but GBS is also known to cause invasive as well as noninvasive infections in adults. However, little is known about the genetic relationship among the isolates, the diversity of virulence, and possible differences in tropism among GBS isolates. We used MLST analysis and screened for the presence of mobile genetic elements among clinical isolates collected in two geographic regions in Sweden during a 10-year time span to investigate the population structure and dynamics among invasive GBS isolates.
By MLST analysis, 158 GBS invasive isolates collected from northern and western parts of Sweden were resolved into 29 STs and grouped into six genetic lineages, including five major clonal complexes and one singleton. The highly clonal structure has also been shown by a number of other methods, such as pulsed-field gel electrophoresis, multilocus enzyme electrophoresis typing, and randomly amplified polymorphic DNA analysis (18, 25-28, 30). However, it is difficult to compare the results among studies by using these traditional approaches. In 2003 (20), a MLST scheme was developed and validated with a global collection of GBS isolates; and 29 STs were identified among 152 isolates, although the representation of STs was not identical to that found in the present study. However, the predominant STs and the genetic lineages found in Sweden were similar to those found in the global collection, suggesting that disease is caused by a limited set of clonal lineages. In comparison to MLST analysis of Streptococcus pyogenes, the diversity of GBS appears to be much lower, even though these two species of streptococci share a highly similar genetic backbone (11, 14). In S. pyogenes, the rate of recombination has been reported to be relatively high in the overall population (21). Numerous epidemiological investigations have shown that the serotype distribution of GBS varies both geographically and over time (17, 21). Such a difference in respect to lineage distribution could also be seen in this Swedish collection. The difference was mainly due to the emergence of CC1 and the disappearance of CC23 over time and the fact that CC1 was more established in Gothenburg than in Umeå. A change of genotype in the population might reflect differences in the fitness of the genotypes, e.g., the spread of an antibiotic-resistant clone, or changes in herd immunity. The latter has been noticed in organisms such as Bordetella pertussis, where a rise in the frequency of unrelated genotypes was pronounced in the postvaccination era (32).
With the consecutive sample of GBS isolates, including six serotypes, we were also able to determine the levels of genetic heterogeneity within serotypes and investigate how often clones or members of a clonal complex express various capsular types. We have shown that GBS serotypes are composed of distinct clones, similar to the finding of Takahashi et al. (31). In this study, the same serotype was seen in isolates that differed at six loci, such as ST-17 and ST-19 isolates, which shared expression of the type III capsule, and ST-23 and ST-4 isolates, which encode a capsule of type Ia. This observation indicates that the horizontal transfer of capsular genes has occurred in the population. Importantly, the level of capsular switching appears to be high, and the exchange of capsule gene clusters between GBS isolates with different genetic backgrounds occurs without restriction to certain genetic lineages. Among the clonal complexes, CC1 showed a high level of serotypic diversity and contained all of the serotypes included in this study except Ia and III. The ancestor ST, ST-1, was also found among isolates of serotypes III, VI, and VIII, according to Jones et al. (20). Changes at the capsular locus are probably driven by the host immune response, and the new genotype-serotype combination may be lost or may persist in the population and diversify if increased fitness is acquired. This might explain the observation of an altered serotype only once or twice and why the stable clones, such as ST-19 and ST-17, of serotype III persist or dominate. Even though serotype V has emerged relatively recently (5), it appears to be subjected to frequent capsule switching. It has been demonstrated that a single gene confers serotype specificity in GBS isolates of capsular types III and Ia (7), and allelic exchange by horizontal genetic transfer and subsequent recombination would lead to a change in serotype. In GBS isolates, the primary shuttle vehicle for horizontal transfer might be a composite transposon, a plasmid, or a phage. The genes that encode the surface proteins ScpB and Lmb have been shown to be transferable by a composite transposon (13), whereas GBSi1 has been proposed to be imported into certain lineages of GBS isolates by a plasmid (24). Although no complete cryptic prophage was identified in the NEM316 genome, a large number of plasmid- and phage-related genes were identified (14). However, serotype switching during natural infection has not been shown to occur as it has in Streptococcus pneumoniae and Neisseria meningitidis, which are naturally transformable (8, 9, 29). Nevertheless, comparison of the genome sequences of GBS and S. pneumoniae revealed that homologues of the genes required for S. pneumoniae transformation competence exist in GBS (14). Further study of the capsular gene clusters might shed light on the nature of the recombination event(s) that gives rise to capsule switching.
Different lineages among GBS isolates of serotype III have previously been identified in a number of epidemiological investigations. In this study, four lineages were identified among GBS isolates of serotype III, in accordance with the lineages found by RDP and MLST analysis (6, 20). ST-19 and ST-17 correspond to the previously designated lineages RDP III-2 and RDP III-3, respectively. The minor lineage RDP III-1 corresponds to ST-23, and RDP III-4 corresponds to ST-7 (a DLV of ST1). However, in contrast to previous findings, isolates from the two major lineages, CC17 and CC19, were equally represented among neonatal patients, whereas the invasive isolates from adults mainly belonged to CC9, CC19, and CC1. CC17 appeared to predominantly cause neonatal infections but not adult infections, whereas CC19 caused invasive diseases among both neonates and adults. Musser et al. (25) and Takahashi et al. (30) suggested the existence of a highly virulent clone as a cause of neonatal invasive diseases. The basis for that conclusion was that while electrophoretic type ET1 (RDP III-3) was underrepresented among carriage isolates, this clone represented the vast majority of isolates from neonatal invasive diseases. Similar studies were also conducted in France and Denmark (18, 26). In the French collection of GBS isolates, multiple virulent clones with the capacity to invade the central nervous system were identified. However, no significant difference in the lineage distribution among disease-associated serotype III isolates and no difference in pathogenic potential were noted. Recently, in agreement with the conclusions from the Danish study, no highly virulent clone was identified among serotype III isolates collected from North America (10). It is important to remember that adult invasive isolates were not examined in any of the aforementioned studies, a fact that might explain the striking difference between the two major lineages reported here for the first time. It is possible that ST-19 represents a highly successful clone and is composed of isolates with an increased capacity to cause diseases among both neonates and adults, whereas CC17 seems to be strongly adapted to neonates. The differences in the distributions of CC17 and of CC19 between the two patient groups might reflect a difference in a number of factors, such as the expression of adhesins and other virulence factors. Clarification of whether ST-19 and ST-17 complexes differ in their virulence potentials requires further studies. So far it is known that ET1/RDP III-3/ST-17 has hyaluronidase activity; shows higher production of sialic acid; exhibits growth inhibition in a chemically defined medium with a high concentration of phosphate; and possesses spb1, which encodes a protein that mediates epithelial cell invasion (1, 16, 18, 25, 30). In addition to these differences, the present study shows that GBSi1 and IS1548 are genetic markers for CC17 and CC19 of serotype III isolates, respectively, in accordance with previous findings (15, 24, 31). Recently, ST-17 and ST-19 were suggested to represent intermediate genotypes between GBS lineages that have a more clear-cut tropism for humans and cattle, even though they are rarely identified among bovine GBS isolates. ST-23, one of two major genotypes from cattle, is a minor ST among human isolates and lacks GBSi1 and IS1548 (6). In the future, comparative genomics and identification of genes important for host tropism may provide insight into both the pathogenesis and the evolution of GBS isolates.
Acknowledgments
This work was supported by grants from the Swedish Research Council (08675), the Västerbottens Landsting, the Medical Faculty at Umeå University, and Insamlings Stiftelsen at Umeå University to M.N. S.-L.L. was supported by a grant from the Kempe Foundation.
Nicola Jones is gratefully acknowledged for technical assistance and discussions. We are also grateful to Elvira Vunic for technical assistance, Birger Trollfors and Stefan Berg for collection of GBS isolates from Gothenburg, and Patrik Ryden and Hans Stenlund for discussion of the statistical calculations.
REFERENCES
- 1.Adderson, E. E., S. Takahashi, Y. Wang, J. Armstrong, D. V. Miller, and J. F. Bohnsack. 2003. Subtractive hybridization identifies a novel predicted protein mediating epithelial cell invasion by virulent serotype III group B Streptococcus agalactiae. Infect. Immun. 71:6857-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, B. F., and D. M. Okada. 1977. The emergence of group B streptococci in infections of the newborn infant. Annu. Rev. Med. 28:355-369. [DOI] [PubMed] [Google Scholar]
- 3.Baker, C. 2000. Group B Streptococcal infections, p. 222-237. In D. Stevens and E. Kaplan. (ed.), Streptococcal infections. Oxford University Press, New York, N.Y.
- 4.Berg, S., B. Trollfors, T. Lagergard, G. Zackrisson, and B. A. Claesson. 2000. Serotypes and clinical manifestations of group B streptococcal infections in western Sweden. Clin. Microbiol. Infect. 6:9-13. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack, J. F., A. A. Whiting, G. Martinez, N. Jones, E. E. Adderson, S. Detrick, A. J. Blaschke-Bonkowsky, N. Bisharat, and M. Gottschalk. 2004. Serotype III Streptococcus agalactiae from bovine milk and human neonatal infections. Emerg. Infect. Dis. 10:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 9.Coffey, T. J., M. C. Enright, M. Daniels, P. Wilkinson, S. Berron, A. Fenoll, and B. G. Spratt. 1998. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb. Drug Resist. 4:51-55. [DOI] [PubMed] [Google Scholar]
- 10.Davies, H. D., N. Jones, T. S. Whittam, S. Elsayed, N. Bisharat, and C. J. Baker. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097-1102. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franken, C., G. Haase, C. Brandt, J. Weber-Heynemann, S. Martin, C. Lammler, A. Podbielski, R. Lutticken, and B. Spellerberg. 2001. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 41:925-935. [DOI] [PubMed] [Google Scholar]
- 14.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 15.Granlund, M., F. Michel, and M. Norgren. 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 183:2560-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granlund, M., L. Oberg, M. Sellin, and M. Norgren. 1998. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J. Infect. Dis. 177:967-976. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 18.Hauge, M., C. Jespersgaard, K. Poulsen, and M. Kilian. 1996. Population structure of Streptococcus agalactiae reveals an association between specific evolutionary lineages and putative virulence factors but not disease. Infect. Immun. 64:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-209. [DOI] [PubMed] [Google Scholar]
- 20.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalia, A., B. G. Spratt, M. C. Enright, and D. E. Bessen. 2002. Influence of recombination and niche separation on the population genetic structure of the pathogen Streptococcus pyogenes. Infect. Immun. 70:1971-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieran, E., M. Matheson, A. G. Mann, A. A. Efstratiou, K. Butler, and W. Gorman. 1998. Group B streptococcus (GBS) colonisation among expectant Irish mothers. Ir. Med. J. 91:21-22. [PubMed] [Google Scholar]
- 23.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 24.Luan, S. L., M. Granlund, and M. Norgren. 2003. An inserted DNA fragment with plasmid features is uniquely associated with the presence of the GBSi1 group II intron in Streptococcus agalactiae. Gene 312:305-312. [DOI] [PubMed] [Google Scholar]
- 25.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, S., E. E. Adderson, Y. Nagano, N. Nagano, M. R. Briesacher, and J. F. Bohnsack. 1998. Identification of a highly encapsulated, genetically related group of invasive type III group B streptococci. J. Infect. Dis. 177:1116-1119. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi, S., S. Detrick, A. A. Whiting, A. J. Blaschke-Bonkowksy, Y. Aoyagi, E. E. Adderson, and J. F. Bohnsack. 2002. Correlation of phylogenetic lineages of group B streptococci, identified by analysis of restriction-digestion patterns of genomic DNA, with infB alleles and mobile genetic elements. J. Infect. Dis. 186:1034-1038. [DOI] [PubMed] [Google Scholar]
- 32.van Loo, I. H., K. J. Heuvelman, A. J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]