Abstract
Most studies of the bacterial etiology of periodontitis have used either culture-based or targeted DNA approaches, and so it is likely that pathogens remain undiscovered. The purpose of this study was to use culture-independent, quantitative analysis of biofilms associated with chronic periodontitis and periodontal health to identify pathogens and beneficial species. Samples from subjects with periodontitis and controls were analyzed using ribosomal 16S cloning and sequencing. Several genera, many of them uncultivated, were associated with periodontitis, the most numerous of which were gram positive, including Peptostreptococcus and Filifactor. The genera Megasphaera and Desulfobulbus were elevated in periodontitis, and the levels of several species or phylotypes of Campylobacter, Selenomonas, Deferribacteres, Dialister, Catonella, Tannerella, Streptococcus, Atopobium, Eubacterium, and Treponema were elevated in disease. Streptococcus and Veillonella spp. were found in high numbers in all samples and accounted for a significantly greater fraction of the microbial community in healthy subjects than in those with periodontitis. The microbial profile of periodontal health also included the less-abundant genera Campylobacter, Abiotrophia, Gemella, Capnocytophaga, and Neisseria. These newly identified candidates outnumbered Porphyromonas gingivalis and other species previously implicated as periodontopathogens, and it is not clear if newly identified and more numerous species may play a more important role in pathogenesis. Finally, more differences were found in the bacterial profile between subjects with periodontitis and healthy subjects than between deep and shallow sites within the same subject. This suggests that chronic periodontitis is the result of a global perturbation of the oral bacterial ecology rather than a disease-site specific microbial shift.
There is considerable evidence to show that bacterial plaque is the etiologic agent in chronic periodontitis. No single species has been implicated as a primary pathogen, and the available evidence is consistent with a polymicrobial disease etiology. Nearly all studies on the bacterial etiology of periodontitis have used either culture-based or directed DNA approaches, targeting known species. The prevailing paradigm that implicates minor constituents of the subgingival community, the gram-negative bacteria Porphyromonas gingivalis, Tanerella forsythensis, and Treponema denticola (24), as periodontopathogens is based on such approaches. However, culturing is not representative of the composition of a microbial community, since it is often too selective, especially for fastidious and as-yet-uncultivable species. Even culture-independent targeted approaches are limited to detecting the presence and levels of known species. Obviously cultivation will not detect uncultivated species, but the limitations of closed-ended molecular approaches such as PCR or hybridization assays such as checkerboard and microarrays are not as widely appreciated. Using these approaches it is possible to detect uncultivated species, but only if they have been previously characterized to allow specific primers or probes to be constructed. Perhaps more importantly, quantitative information is incomplete with these methods since the total number of bacteria is not easily determined with a closed-ended approach. Thus, it is possible that pathogens remain undiscovered with such approaches. To advance our understanding of oral biofilm communities and disease processes, it is necessary to more comprehensively identify the microbiota in periodontal health and disease.
Open-ended molecular approaches capable of detecting all bacteria in a sample, including uncultivated and previously unsuspected ones, are the most powerful methods available for exploring the microbial profile of any community. Recently, cloning and sequencing of bacterial 16S rRNA genes have been used to investigate the composition of environmental samples, as well as samples from the human oral cavity. This culture-independent approach has revealed vastly greater diversity than was apparent with culturing (10, 28). Investigations of oral bacteria using these tools have used enrichment primers for rare taxa, e.g., Spirochaetaceae and Bacteroidetes, or subtraction systems to eliminate predominant taxa such as Streptococcus, enabling selective amplification and identification of rare species. Using this approach, more than 700 orally derived 16S sequences have been deposited in GenBank, less than half of which are from species that have been cultivated and characterized. To identify which of these many oral inhabitants are important in health and disease-associated biofilm communities, an adequately powered clinical study design and a quantitative, representational approach to ribosomal 16S cloning and sequencing that maintains the relative proportions of individual bacterial species is needed.
The purpose of the present study was to achieve a culture-independent representational analysis of biofilms associated with chronic periodontitis and periodontal health and to identify candidate pathogens and beneficial species or taxa. Since approximately half of oral bacteria are uncultivated, it seems likely that new associations would be revealed by this approach.
MATERIALS AND METHODS
Subject selection.
Subjects for this institutionally approved study were recruited from the dental clinics at the College of Dentistry of the Ohio State University, and informed consent was obtained. Fifteen subjects with moderate to severe generalized chronic periodontitis were identified after clinical and radiographic examination. The subjects ranged in age from 42 to 80 years. A control group of 15 age- and sex-matched periodontally healthy individuals was also selected. Exclusion criteria for both groups included diabetes, antibiotic therapy in the previous 3 months, oral prophylactic procedures within the last 3 months, fewer than 20 teeth in the dentition, and a history of smoking.
Sample collection and DNA isolation.
Subgingival plaque samples were collected on sterile endodontic paper points (Caulk-Dentsply) after isolation and supragingival plaque removal. Plaque was collected and pooled from the mesial sulcus of every tooth for the healthy subjects. For the periodontitis group, sites for microbial sampling were selected based on probe depth measurements. Plaque from four nonadjacent proximal sites with probe depths of 6 mm or more was collected and pooled (disease or deep-site samples). Samples were similarly acquired from four sites with probe depths of 3 mm or less and separately pooled (healthy or shallow-site samples). Samples were placed in 1.5-ml microcentrifuge tubes and frozen until further analysis. DNA was isolated by using a previously described methodology (20). Briefly, bacteria was removed from the paper points by adding 750 μl of sampling buffer, followed by vortexing for 1 min. The paper points were then removed, the sample pelleted, and the supernatant discarded. The pellet was suspended in 1% sodium dodecyl sulfate in Tris-EDTA (TE), proteinase K was added, and the samples were incubated overnight. DNA was isolated on glass beads and eluted in TE.
Amplification of 16S rRNA.
Bacterial 16S rRNA genes were amplified from the community DNA with universal eubacterial primers A17 (5′-GTT TGA TCC TGG CTC AG-3′) and 317 (5′-AAG GAG GTG ATC CAG GC-3′) (Biosynthesis, Lewisville, TX). PCR was performed by adding 1 μl of community DNA to a reaction mixture (50-μl final volume) containing 20 nmol of each primer, 40 nmol of deoxynucleotide triphosphates, and 1 U of Taq polymerase. The following cycling conditions were used: denaturation at 94°C for 1 min, annealing at 42°C for 2 min, and elongation at 72°C for 3 min. A final, 10-min elongation at 72°C followed 22 cycles of amplification. The PCR products were purified using the QiaQuik PCR purification kit (QIAGEN, Valencia, CA).
Cloning and sequencing.
The 16S amplicons generated by PCR were cloned into Escherichia coli by using a commercially available kit (TOPO TA cloning kit; Invitrogen, San Diego, CA). Competent TOP10 E. coli cells provided with the kit were transformed, plated onto Luria-Bertani agar plates supplemented with ampicillin, and incubated overnight. Colonies were further selected for the presence of an insert with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The presence of inserts of the correct molecular size (≅1,500 bp) was confirmed by PCR amplification of the white colonies with the same primers used for initial amplification and gel electrophoresis of the amplicons on 1% agarose. DNA was stained with ethidium bromide and visualized under UV light (wavelength, 320 nm). The products were then purified with a Millipore kit (Millipore, Billerica, MA) and sequenced with an ABI Prism cycle sequencing kit (BigDye terminator cycle sequencing kit) using an ABI 3700 instrument.
Sequence analysis.
Partial sequences of 500 to 800 bp were obtained from each amplicon. The sequences generated were compared to the GenBank database to identify the closest relatives by using a Time Logic DeCypher Tera BLAST server hosted by the Ohio Supercomputer Center. Sequences with low homology to GenBank entries were screened for chimeras by using the ChimeraCheck program of the Ribosomal Database Project II (http://rdp.cme.msu.edu/html/). Twenty-three clones were identified as chimeric sequences and excluded from further analysis. Sequences were aligned, and a similarity matrix was constructed from the alignments by the method of Jukes and Cantor. Phylogenetic trees were constructed by using the neighbor-joining method. MacVector software was used to generate alignments, similarity matrices, and in phylogenetic tree construction. A novel phylotype was defined as a sequence that differed from the closest GenBank entry by >2%. Sequence data for the whole 16S gene was obtained for novel sequences and submitted to GenBank.
Statistical analysis.
Statistical analysis was carried out with JMP (SAS Institute, Inc., Cary, NC). The microbial profile of periodontally healthy subjects was compared to that of healthy sites and deep pockets in subjects with periodontitis by using Kruskal-Wallis analysis of variance. Within-subject comparisons between deep and shallow sites for individual species were made by using the Wilcoxon signed-rank test. Chi-square analysis was used to test for the presence or absence of species in health and disease.
RESULTS
Plaque samples for the study were collected from 15 subjects with moderate to severe chronic periodontitis (separate deep and shallow site samples were collected from each subject) and from 15 age-matched, periodontally healthy control subjects (pooled samples from all teeth). The mean age of the experimental group was 63.6 years (standard deviation [SD] = 9.1), and the mean age of the control group was 60.2 years (SD = 11.3). The difference, as determined by a Student t test, was not significant. The healthy group was 71% male, whereas the periodontitis group was 73% male. No significant difference was found by chi-square analysis. The healthy group was 100% white, and the periodontitis group was 87% white and 13% African-American. The sample size did not permit statistical comparisons by race.
Sequence data of 500 to 800 bp was obtained for 100 clones from each sample for a total of 4,500 clones. The identification of 100 clones per sample provided a 95% probability of detecting species present at ≥3% of total bacteria, and a 60% probability at ≥2%, calculated by using the binomial probability distribution. A total of 42 clones were <98% identical to current GenBank entries, and these clones were grouped into six novel phylotypes (GenBank accession numbers AY947495 to AY947500). A total of 274 species or phylotypes were identified. Table 1 lists these species in order of their ranking by overall prevalence and shows the mean prevalences in the three groups of samples. A table showing the same data sorted by phylogeny (Table S1A in the supplemental material) is available online.
TABLE 1.
Species and phylotypes from three sample groups showing the percentage of total clones and the mean prevalence in each group arranged in order of decreasing overall prevalence
| Overall rank | Species/phylotype(s) | % Clones | Mean prevalence ± SD |
||
|---|---|---|---|---|---|
| Healthy subjects | Shallow sites | Deep sites | |||
| 1 | Veillonella sp. oral clone X042 | 7.38 | 13.1 ± 9.4 | 4.5 ± 3.3 | 4.6 ± 1.7 |
| 2 | Campylobacter gracilis | 6.71 | 8.1 ± 4.9 | 7.8 ± 3.9 | 4.2 ± 4.3 |
| 3 | Peptostreptococcus sp. oral clone FG014 | 4.27 | 2.2 ± 2.7 | 5.2 ± 5.0 | 5.4 ± 7.9 |
| 4 | Selenomonas sputigena/EW051a/DD020 | 4.02 | 3.4 ± 3.6 | 4.0 ± 3.3 | 4.7 ± 3.2 |
| 5 | Veillonella sp. oral clone BU083 | 3.64 | 3.3 ± 3.9 | 3.5 ± 3.4 | 4.1 ± 2.5 |
| 6 | Peptostreptococcus sp. oral clone BS044 | 3.11 | 0.9 ± 2.1 | 4.1 ± 6.9 | 4.4 ± 8.2 |
| 7 | Filifactor alocis | 3.07 | 0.9 ± 1.1 | 4.4 ± 3.9 | 3.9 ± 3.6 |
| 8 | Streptococcus mitis | 2.71 | 4.9 ± 5.2 | 1.8 ± 2.2 | 1.4 ± 1.8 |
| 9 | Selenomonas infelix | 2.51 | 2.0 ± 1.9 | 2.7 ± 2.1 | 2.9 ± 1.9 |
| 10 | Selenomonas noxia/EQ054 | 2.18 | 2.9 ± 2.3 | 1.7 ± 1.8 | 2 ± 2.2 |
| 11 | Dialister sp. strain E2_20 E1 oral isolate | 2.09 | 1.8 ± 1.8 | 1.9 ± 2.0 | 2.6 ± 2.5 |
| 12 | Streptococcus gordonii | 1.98 | 2.2 ± 2.7 | 1.9 ± 2.4 | 1.9 ± 3.6 |
| 13 | Selenomonas dianae/AJ036/DY027 | 1.62 | 1.3 ± 1.3 | 1.3 ± 1.4 | 2.3 ± 2.1 |
| 14 | Streptococcus oralis | 1.56 | 1.7 ± 2.1 | 1.7 ± 2.0 | 1.3 ± 1.3 |
| 15 | Peptostreptococcus oral clone CK035 | 1.51 | 0.5 ± 0.8 | 0.8 ± 1.3 | 3.3 ± 4.7 |
| 16 | Megasphaera oral clone BB166 | 1.42 | 0.2 ± 0.4 | 2.1 ± 3.7 | 1.9 ± 2.7 |
| 17 | Desulfobulbus oral clone CH031 | 1.40 | 0.3 ± 0.6 | 2.1 ± 3.3 | 1.9 ± 2.8 |
| 18 | Dialister sp. oral clone BS095 | 1.09 | 1.5 ± 1.9 | 0.8 ± 0.9 | 0.9 ± 1.2 |
| 19 | Dialister pneumosintes | 1.04 | 0.1 ± 0.4 | 1.7 ± 2.4 | 1.3 ± 1.3 |
| 20 | Campylobacter sputorum sputorum | 1.00 | 0.2 ± 0.4 | 1.9 ± 1.9 | 0.9 ± 1.1 |
| 21 | Abiotrophia adiacens | 0.96 | 2.1 ± 2.1 | 0.5 ± 1.0 | 0.3 ± 0.6 |
| 22 | Neisseria meningitidis | 0.93 | 1.4 ± 1.7 | 0.9 ± 1.2 | 0.5 ± 0.7 |
| 23 | Streptococcus intermedius | 0.91 | 1.3 ± 1.4 | 0.5 ± 1.1 | 0.9 ± 1.3 |
| 24 | Desulfobulbus sp. oral clone R004 | 0.89 | 0.0 ± 0.0 | 0.9 ± 1.5 | 1.7 ± 2.5 |
| 25 | Streptococcus pneumoniae | 0.87 | 1.3 ± 1.2 | 0.8 ± 1.3 | 0.5 ± 0.8 |
| 26 | Eubacterium sp. oral clone BP1-82 | 0.84 | 1.1 ± 1.5 | 1.1 ± 2.1 | 0.3 ± 0.5 |
| 27 | Campylobacter sp. oral clone BB120 | 0.82 | 0.0 ± 0.0 | 1.3 ± 3.2 | 1.1 ± 2.4 |
| 28 | Gemella morbillorum | 0.80 | 0.9 ± 1.5 | 0.9 ± 1.2 | 0.6 ± 0.6 |
| 29 | Firmicutes sp. oral clone A0068 | 0.73 | 0.6 ± 1.1 | 0.5 ± 0.7 | 1.1 ± 1.8 |
| 30 | Eikenella corrodens | 0.69 | 0.6 ± 1.3 | 0.8 ± 1.4 | 0.7 ± 1.1 |
| 31 | Eubacterium saburreum | 0.64 | 1.6 ± 1.5 | 0.1 ± 0.4 | 0.2 ± 0.4 |
| 32 | Veillonella sp. oral clone AA050 | 0.64 | 0.5 ± 1.8 | 1.1 ± 3.1 | 0.3 ± 0.6 |
| 33 | Campylobacter concisus | 0.64 | 1.0 ± 1.1 | 0.5 ± 0.7 | 0.5 ± 0.9 |
| 34 | Megasphaera (Anaerosphaera) micronuciformis | 0.60 | 0.3 ± 0.8 | 0.7 ± 1.1 | 0.7 ± 1.1 |
| 35 | Peptostreptococcus sp. oral clone AJ062 | 0.60 | 0.1 ± 0.3 | 0.7 ± 1.1 | 1.1 ± 1.8 |
| 36 | Selenomonas sp. oral clone D0042 | 0.58 | 0.1 ± 0.3 | 0.8 ± 1.3 | 0.9 ± 1.0 |
| 37 | Veillonella atypica | 0.58 | 0.7 ± 1.5 | 0.2 ± 0.4 | 0.9 ± 2.3 |
| 38 | Campylobacter showae | 0.58 | 1.1 ± 1.8 | 0.5 ± 0.8 | 0.1 ± 0.4 |
| 39 | Campylobacter rectus | 0.56 | 0.2 ± 0.4 | 0.9 ± 1.2 | 0.5 ± 0.8 |
| 40 | Streptococcus hyointestinalis | 0.53 | 0.6 ± 0.8 | 0.8 ± 1.6 | 0.2 ± 0.6 |
| 41 | Streptococcus sp. oral strain H3-M2 | 0.53 | 1.2 ± 2.0 | 0.1 ± 0.3 | 0.3 ± 0.7 |
| 42 | Gemella sp. strain 1754-94 | 0.51 | 1.1 ± 1.4 | 0.3 ± 0.7 | 0.1 ± 0.3 |
| 43 | Veillonella ratti | 0.51 | 0.3 ± 0.6 | 0.8 ± 1.1 | 0.4 ± 1.1 |
| 44 | Streptococcus sanguis | 0.49 | 1.3 ± 2.2 | 0.1 ± 0.4 | 0.0 ± 0.0 |
| 45 | Eubacterium yurii/A03MT | 0.47 | 0.4 ± 0.6 | 0.9 ± 1.4 | 0.1 ± 0.4 |
| 46 | Selenomonas sp. oral clone CS015 | 0.47 | 0.3 ± 0.9 | 0.5 ± 1.1 | 0.6 ± 0.9 |
| 47 | Kingella oralis | 0.47 | 0.7 ± 1.1 | 0.3 ± 0.6 | 0.4 ± 0.9 |
| 48 | Eubacterium sp. sp. oral clone EI074 | 0.44 | 0.7 ± 1.0 | 0.1 ± 0.3 | 0.5 ± 1.8 |
| 49 | Capnocytophaga gingivalis | 0.42 | 0.9 ± 1.3 | 0.0 ± 0.0 | 0.4 ± 1.1 |
| 50 | Deferribacteres sp. oral clone W090 | 0.42 | 0.0 ± 0.0 | 0.6 ± 1.0 | 0.7 ± 1.1 |
| 51 | Centipeda periodontii | 0.42 | 0.5 ± 0.8 | 0.3 ± 0.8 | 0.5 ± 0.9 |
| 52 | Eubacterium sp. oral clone EW049 | 0.42 | 0.5 ± 1.1 | 0.6 ± 1.5 | 0.2 ± 0.6 |
| 53 | Megasphaera sp. oral clone MCE3_141 | 0.42 | 0.0 ± 0.0 | 0.3 ± 0.6 | 1.0 ± 1.2 |
| 54 | Selenomonas sp. oral clone EY047 | 0.42 | 0.7 ± 1.2 | 0.0 ± 0.0 | 0.6 ± 0.8 |
| 55 | Neisseria elongata | 0.42 | 0.3 ± 1.0 | 0.7 ± 2.1 | 0.3 ± 1.0 |
| 56 | Gemella haemolysans | 0.38 | 0.6 ± 0.9 | 0.3 ± 0.6 | 0.3 ± 0.7 |
| 57 | Treponema socranskii subsp. socranskii | 0.38 | 0.4 ± 1.3 | 0.3 ± 0.8 | 0.4 ± 0.6 |
| 58 | Eubacteriaceae sp. oral clone MCE10_174 E2 | 0.36 | 0.1 ± 0.4 | 0.5 ± 0.9 | 0.4 ± 0.9 |
| 59 | Peptostreptococcus sp. clone FX38-1 | 0.36 | 0.4 ± 1.1 | 0.5 ± 0.9 | 0.1 ± 0.4 |
| 60 | Streptococcus sp. oral strain 7A | 0.33 | 0.2 ± 0.6 | 0.5 ± 0.9 | 0.3 ± 0.8 |
| 61 | Catonella sp. oral clone EZ006 | 0.33 | 0.3 ± 0.6 | 0.3 ± 0.7 | 0.4 ± 0.8 |
| 62 | Eubacterium brachy | 0.33 | 0.2 ± 0.6 | 0.7 ± 1.3 | 0.1 ± 0.3 |
| 63 | Johnsonella ignava | 0.33 | 0.7 ± 0.8 | 0.1 ± 0.4 | 0.2 ± 0.4 |
| 64 | Lachnospiraceae sp. oral clone MCE9_104 E2 | 0.33 | 0.4 ± 0.8 | 0.3 ± 0.6 | 0.3 ± 0.6 |
| 65 | Neisseria sp. oral clone AP132 | 0.33 | 0.2 ± 0.4 | 0.5 ± 0.6 | 0.3 ± 1.0 |
| 66 | Abiotrophia para adiacens | 0.31 | 0.5 ± 0.9 | 0.3 ± 0.6 | 0.1 ± 0.3 |
| 67 | Peptoniphilus ivorii (Peptostreptococcus ivoricus) | 0.31 | 0.1 ± 0.5 | 0.7 ± 1.8 | 0.1 ± 0.3 |
| 68 | Selenomonas sp. oral clone CS024 | 0.31 | 0.7 ± 1.3 | 0.1 ± 0.5 | 0.1 ± 0.4 |
| 69 | Selenomonas-like sp. oral clone DM071 | 0.31 | 0.0 ± 0.0 | 0.4 ± 0.7 | 0.5 ± 1.2 |
| 70 | Streptococcus pyogenes | 0.29 | 0.0 ± 0.0 | 0.7 ± 1.3 | 0.2 ± 0.4 |
| 71 | Selenomonas-like sp. oral strain FNA3 | 0.29 | 0.2 ± 0.4 | 0.3 ± 0.8 | 0.3 ± 0.7 |
| 72 | Neisseria sp. oral clone AP085 | 0.29 | 0.7 ± 1.9 | 0.2 ± 0.6 | 0.0 ± 0.0 |
| 73 | Streptococcus sp. oral clone BM 035 | 0.27 | 0.2 ± 0.6 | 0.1 ± 0.4 | 0.5 ± 1.3 |
| 74 | Streptococcus sp. oral strain 12F | 0.27 | 0.7 ± 1.4 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 75 | Eubacterium saphenum | 0.27 | 0.1 ± 0.4 | 0.3 ± 0.8 | 0.3 ± 0.8 |
| 76 | Mitsuokella jalaludinii | 0.27 | 0.1 ± 0.4 | 0.3 ± 0.8 | 0.4 ± 1.3 |
| 77 | Firmicutes sp. oral clone F058 | 0.27 | 0.3 ± 0.6 | 0.3 ± 0.5 | 0.2 ± 0.6 |
| 78 | Kingella denitrificans | 0.27 | 0.3 ± 0.6 | 0.3 ± 0.6 | 0.2 ± 0.6 |
| 79 | Veillonella sp. oral clone OH1A | 0.27 | 0.3 ± 0.7 | 0.2 ± 0.6 | 0.3 ± 0.6 |
| 80 | Streptococcus mutans | 0.24 | 0.7 ± 1.2 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 81 | Streptococcus suis | 0.24 | 0.1 ± 0.3 | 0.3 ± 0.6 | 0.4 ± 0.8 |
| 82 | Streptococcus sp. oral clone AA007 | 0.24 | 0.2 ± 0.8 | 0.1 ± 0.3 | 0.5 ± 1.1 |
| 83 | Eubacterium sp. oral clone EH006 | 0.24 | 0.1 ± 0.4 | 0.2 ± 0.4 | 0.4 ± 0.9 |
| 84 | Campylobacter sputorum | 0.24 | 0.1 ± 0.3 | 0.6 ± 1.1 | 0.1 ± 0.3 |
| 85 | Treponema socranskii subsp. buccale | 0.24 | 0.2 ± 0.8 | 0.4 ± 0.9 | 0.1 ± 0.4 |
| 86 | Porphyromonas gingivalis | 0.22 | 0.0 ± 0.0 | 0.2 ± 0.4 | 0.5 ± 1.8 |
| 87 | Capnocytophaga granulosa | 0.22 | 0.4 ± 0.9 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 88 | Capnocytophaga sp. oral clone AH015 | 0.22 | 0.6 ± 1.7 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 89 | Streptococcus sp. oral clone 2056B | 0.22 | 0.1 ± 0.4 | 0.1 ± 0.4 | 0.4 ± 1.3 |
| 90 | Dialister sp. oral clone MCE7_134 | 0.22 | 0.0 ± 0.0 | 0.1 ± 0.5 | 0.5 ± 0.9 |
| 91 | Eubacterium sp. oral clone E1-K17 | 0.22 | 0.2 ± 0.4 | 0.3 ± 0.6 | 0.2 ± 0.6 |
| 92 | Selenomonas sp. oral clone D027 | 0.22 | 0.1 ± 0.3 | 0.3 ± 0.8 | 0.3 ± 0.6 |
| 93 | Alysiella filiformis | 0.22 | 0.1 ± 0.3 | 0.3 ± 0.6 | 0.3 ± 0.7 |
| 94 | Neisseria denitrificans | 0.22 | 0.3 ± 0.9 | 0.1 ± 0.4 | 0.2 ± 0.6 |
| 95 | Capnocytophaga sp. oral strain S3 | 0.20 | 0.3 ± 0.5 | 0.0 ± 0.0 | 0.3 ± 1.3 |
| 96 | Deferribacteres sp. oral clone BH017 | 0.20 | 0.1 ± 0.3 | 0.2 ± 0.4 | 0.3 ± 1.3 |
| 97 | Streptococcus salivarius | 0.20 | 0.1 ± 0.5 | 0.3 ± 0.6 | 0.2 ± 0.6 |
| 98 | Streptococcus sinensis | 0.20 | 0.3 ± 0.6 | 0.3 ± 0.8 | 0.0 ± 0.0 |
| 99 | Catonella sp. oral clone BR063 | 0.20 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.5 ± 0.6 |
| 100 | Catonella morbi | 0.20 | 0.1 ± 0.4 | 0.3 ± 0.6 | 0.2 ± 0.4 |
| 101 | Eubacterium sp. oral clone EW053 | 0.20 | 0.1 ± 0.3 | 0.3 ± 0.8 | 0.2 ± 0.6 |
| 102 | Peptostreptococcus micros | 0.20 | 0.5 ± 1.2 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 103 | Selenomonas sp. oral clone EW076 | 0.20 | 0.2 ± 0.6 | 0.1 ± 0.4 | 0.3 ± 0.6 |
| 104 | Selenomonas sp. oral clone EW079 | 0.20 | 0.1 ± 0.3 | 0.2 ± 0.6 | 0.3 ± 0.9 |
| 105 | Selenomonas sp. oral clone CS023 | 0.20 | 0.1 ± 0.3 | 0.3 ± 1.0 | 0.3 ± 0.5 |
| 106 | Streptococcus sp. oral clone 4093B | 0.18 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.4 ± 1.1 |
| 107 | Eubacterium sp. oral clone CK047 | 0.18 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.3 ± 0.7 |
| 108 | Selenomonas sp. oral clone AA024 | 0.18 | 0.4 ± 0.9 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 109 | Neisseria sp. oral clone AP060 | 0.18 | 0.2 ± 0.8 | 0.3 ± 0.6 | 0.0 ± 0.0 |
| 110 | Selenomonas sp. oral clone OH4A | 0.18 | 0.1 ± 0.4 | 0.3 ± 0.6 | 0.1 ± 0.4 |
| 111 | Campylobacter sp. oral clone OH5A | 0.18 | 0.2 ± 0.6 | 0.2 ± 0.6 | 0.1 ± 0.4 |
| 112 | Prevotella sp. oral clone BR014 | 0.16 | 0.1 ± 0.4 | 0.3 ± 0.8 | 0.0 ± 0.0 |
| 113 | Eubacterium sp. oral clone DO016 | 0.16 | 0.3 ± 1.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 114 | Eubacterium sp. oral clone DA014 | 0.16 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.3 ± 0.8 |
| 115 | Selenomonas sp. oral clone CI002 | 0.16 | 0.2 ± 0.6 | 0.1 ± 0.3 | 0.2 ± 0.4 |
| 116 | Veillonella dispar | 0.16 | 0.3 ± 0.7 | 0.2 ± 0.6 | 0.0 ± 0.0 |
| 117 | Campylobacter curvus | 0.16 | 0.3 ± 0.6 | 0.1 ± 0.5 | 0.1 ± 0.3 |
| 118 | Tannerella forsythia (Bacteroides forsythus) | 0.13 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.3 ± 0.7 |
| 119 | Deferribacteres sp. oral clone BH007 | 0.13 | 0.0 ± 0.0 | 0.4 ± 0.7 | 0.0 ± 0.0 |
| 120 | Lactobacillus lactis subsp. lactis | 0.13 | 0.0 ± 0.0 | 0.3 ± 1.0 | 0.1 ± 0.3 |
| 121 | Streptococcus agalactiae | 0.13 | 0.3 ± 1.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 122 | Streptococcus anginosus | 0.13 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.3 ± 0.8 |
| 123 | Streptococcus infantis | 0.13 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.2 ± 0.8 |
| 124 | Streptococcus sp. oral clone 3097C | 0.13 | 0.2 ± 0.6 | 0.2 ± 0.4 | 0.0 ± 0.0 |
| 125 | Streptococcus sp. oral clone BW009 | 0.13 | 0.0 ± 0.0 | 0.3 ± 0.8 | 0.1 ± 0.4 |
| 126 | Streptococcus sp. oral strain 9F | 0.13 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.3 ± 0.6 |
| 127 | Selenomonas-like sp. oral clone GAA14 | 0.13 | 0.1 ± 0.3 | 0.1 ± 0.5 | 0.2 ± 0.4 |
| 128 | Abiotrophia sp. oral clone OH2A | 0.13 | 0.2 ± 0.4 | 0.2 ± 0.6 | 0.0 ± 0.0 |
| 129 | Atopobium sp. oral clone C019 | 0.11 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 1.0 |
| 130 | Corynebacterium sp. oral clone DS081 | 0.11 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 131 | Corynebacterium sp. oral clone AK143 | 0.11 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.2 ± 0.4 |
| 132 | Abiotrophia sp. oral clone P4PA_155 P1 | 0.11 | 0.3 ± 1.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 133 | Abiotrophia defectiva | 0.11 | 0.3 ± 0.5 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 134 | Gemella sanguinis | 0.11 | 0.3 ± 0.8 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 135 | Streptococcus oligofermentans | 0.11 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 136 | Streptococcus sp. oral clone 2061A | 0.11 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 137 | Eubacterium sp. oral clone DZ073 | 0.11 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 138 | Megasphaera sp. oral clone BU057 | 0.11 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.2 ± 0.8 |
| 139 | Peptostreptococcus anaerobius | 0.11 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.9 |
| 140 | Peptococcus sp. oral clone MCE10_265 E1 | 0.11 | 0.1 ± 0.4 | 0.1 ± 0.5 | 0.1 ± 0.3 |
| 141 | Firmicutes sp. oral clone CK051 | 0.11 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 142 | Haemophilus segnis | 0.11 | 0.2 ± 0.6 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 143 | Treponema sp. strain V:19:D36 | 0.11 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.2 ± 0.6 |
| 144 | Corynebacterium matruchotii | 0.09 | 0.2 ± 0.4 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 145 | Rothia dentocariosa | 0.09 | 0.3 ± 0.8 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 146 | Deferribacteres sp. oral clone D084 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.5 | 0.1 ± 0.5 |
| 147 | Streptococcus cristatus | 0.09 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 148 | Eubacterium clone vadinBB14 | 0.09 | 0.1 ± 0.5 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 149 | Eubacterium sp. oral strain A35MT | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.2 ± 0.4 |
| 150 | Lachnospiraceae sp. oral clone MCE9_173 E4 | 0.09 | 0.1 ± 0.5 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 151 | Megasphaera sp. oral clone BS073 | 0.09 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.5 |
| 152 | Selenomonas flueggei-like sp. clone AH132 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.2 ± 0.4 |
| 153 | Selenomonas sp. oral clone DS051 | 0.09 | 0.3 ± 0.8 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 154 | Selenomonas sp. oral clone DS071 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 155 | Anaeroglobus geminatus | 0.09 | 0.0 ± 0.0 | 0.2 ± 0.4 | 0.1 ± 0.3 |
| 156 | Lactobacillus cateneforme | 0.09 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.2 ± 0.4 |
| 157 | Firmicutes sp. oral clone MCE3_120E | 0.09 | 0.0 ± 0.0 | 0.3 ± 0.7 | 0.0 ± 0.0 |
| 158 | Fusobacterium nucleatum subsp. nucleatum | 0.09 | 0.1 ± 0.4 | 0.1 ± 0.4 | 0.0 ± 0.0 |
| 159 | Burkholderia cepacia | 0.09 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 160 | Neisseria weaveri | 0.09 | 0.1 ± 0.3 | 0.2 ± 0.6 | 0.0 ± 0.0 |
| 161 | Niesseria flava | 0.09 | 0.2 ± 0.6 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 162 | Cardiobacterium sp. strain B | 0.09 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 163 | Treponema sp. strain Smibert-5 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.1 ± 0.4 |
| 164 | Treponema sp. strain 6:H:D15A-4 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.2 ± 0.4 |
| 165 | Treponema sp. strain VI:G:G47 | 0.09 | 0.1 ± 0.3 | 0.2 ± 0.4 | 0.0 ± 0.0 |
| 166 | Spirochaeta sp. clone Nt17 | 0.09 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.1 ± 0.5 |
| 167 | Eubacterium sp. oral clone OH3A | 0.09 | 0.2 ± 0.4 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 168 | Olsenella profusa | 0.07 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 169 | Leptotrichia goodfellowii | 0.07 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 170 | Deferribacteres sp. oral clone W028 | 0.07 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.4 |
| 171 | Lactobacillus sp. MR-2 | 0.07 | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.0 ± 0.0 |
| 172 | Streptococcus sp. oral clone EK048 | 0.07 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 173 | Streptococcus sp. oral clone DP009 | 0.07 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.4 |
| 174 | Streptococcus sp. oral clone FP064 | 0.07 | 0.1 ± 0.5 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 175 | Streptococcus sp. oral clone KL-27-1-5 | 0.07 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.8 |
| 176 | Streptococcus sp. oral strain B5SC | 0.07 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 177 | Streptococcus genomosp. strain C7 | 0.07 | 0.2 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 178 | Dialister sp. oral strain GBA27 | 0.07 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 179 | Eubacterium sp. oral clone BS091 | 0.07 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.4 |
| 180 | Selenomonas lacticifix | 0.07 | 0.1 ± 0.4 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 181 | Selenomonas sp. oral clone CS002 | 0.07 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 182 | Firmicutes sp. oral clone BB124 | 0.07 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 183 | Fusobacterium sp. oral clone BS019 | 0.07 | 0.2 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 184 | Lautropia sp. oral clone AP009 | 0.07 | 0.0 ± 0.0 | 0.2 ± 0.6 | 0.0 ± 0.0 |
| 185 | Neisseria genomosp. P1 clone P4PC_20 | 0.07 | 0.2 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 186 | Neisseria sp. oral clone AK105 | 0.07 | 0.1 ± 0.3 | 0.1 ± 0.5 | 0.0 ± 0.0 |
| 187 | Cardiobacterium hominis HS-A | 0.07 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 188 | Haemophilus parainfluenzae | 0.07 | 0.1 ± 0.5 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 189 | Actinomyces naeslundii | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 190 | Bacteroides oral clone AU126 | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.4 | 0.0 ± 0.0 |
| 191 | Porphyromonas sp. oral clone DS033 | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.5 |
| 192 | Flexistipes sp. oral clone BB062 | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 193 | Deferribacteres sp. oral clone D006 | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 194 | Abiotrophia elegans | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 195 | Lactobacillus sp. oral clone CX036 | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 196 | Marinococcus halophilus | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 197 | Streptococcus equi | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 198 | Streptococcus sp. oral clone 3192A | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 199 | Streptococcus sp. oral clone CH016 | 0.04 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 200 | Streptococcus sp. oral clone FX003 | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 201 | Dialister sp. oral clone FY011 | 0.04 | 0.1 ± 0.4 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 202 | Eubacterium minutum | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 203 | Eubacterium sp. equine clone CL11 | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 204 | Eubacterium sp. oral clone DN050 | 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 205 | Megasphaera sp. oral clone CS025 | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 206 | Mogibacterium pumilum | 0.04 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 207 | Selenomonas ruminantium | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 208 | Selenomonas sp. oral clone EZ011 | 0.04 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 209 | Veillonella parvula | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 210 | Fusobacterium sp. oral clone CY024 | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 211 | Neisseria lactamica | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 212 | Neisseria perflava | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 213 | Neisseria subflava | 0.04 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 214 | Vogesella indigofera | 0.04 | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 215 | Campylobacter mucosalis | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 216 | Cardiobacterium hominis HS-B | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 217 | Haemophilus influenzae | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 218 | Spirochaeta sp. clone Nt25 | 0.04 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 219 | Uncultured bacterial clone UB611 | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 220 | Anaerosphaera sp. oral clone OH6A | 0.04 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 221 | Slackia heliotrinreducens | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 222 | Atopobium parvulum | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 223 | Atopobium rimae | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 224 | Actinobacillus actinomycetemcomitans | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 225 | Actinomyces odontolyticus | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 226 | Actinomyces oral strain C29KA | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 227 | Actinomyces sp. oral clone DR002 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 228 | Corynebacterium glutamicum | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 229 | Rothia sp. oral clone BP2-13 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 230 | Bacteroidales sp. oral cloneMCE7_120E3 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 231 | Capnocytophaga sp. strain ChDC OS44 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 232 | Capnocytophaga sputigena | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 233 | Capnocytophaga sp. oral clone DS022 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 234 | Capnocytophaga sp. oral clone X089 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 235 | Prevotella sp. oral clone AO009 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 236 | Prevotella intermedia | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 237 | Lactobacillus sp. strain CLE-4 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 238 | Lactobacillus sp. strain Y10 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 239 | Streptococcus constellatus | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 240 | Streptococcus ferus | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 241 | Streptococcus parasanguis | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 242 | Uncultured bacterium ECS55 | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 243 | Dialister sp. strain ADV 04.01 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 244 | Mogibacterium (Eubacterium) timidum | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 245 | Eubacterium sp. strain WFeA1-59 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 246 | Eubacterium sp. equine clone PL35 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 247 | Eubacterium sp. oral clone BE088 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 248 | Eubacterium sp. oral clone BB142 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 249 | Eubacterium sp. oral clone P2PC | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 250 | Lachnospiraceae sp. oral clone MCE7_60 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 251 | Lachnospiraceae sp. oral clone P4PC_12P1 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 252 | Mogibacterium diversum | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 253 | Peptoniphilus (Peptostreptococcus) lacrimalis | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 254 | Veillonella sp. strain ADV 281.99 | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 255 | Zymophilus paucivorans | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 256 | Acholeplasma palmae | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 257 | Erysipelothrix rhusiopathiae | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 258 | Firmicutes sp. oral clone CD4B11 | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 259 | Firmicutes sp. oral clone CH017 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 260 | Firmicutes sp. oral clone A0069 | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 261 | Ehrlichia muris | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 262 | Methylobacterium organophilum | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 263 | Burkholderia sp. strain PJ431 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 264 | Vitreoscilla stercoraria | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 265 | Simonsiella steedae | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 266 | Simonsiella muelleri | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 267 | Campylobacter lari | 0.02 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 268 | Campylobacter fecalis | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 269 | Brenneria (Erwinia) salicis | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 270 | Serratia liquefaciens | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 271 | Treponema sp. strain I:G:C1 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 272 | Treponema vincentii | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 273 | Treponema clone RFS18 | 0.02 | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 274 | Firmicutes sp. oral strain FTB41 | 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.3 |
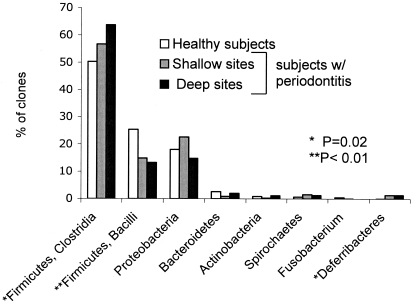
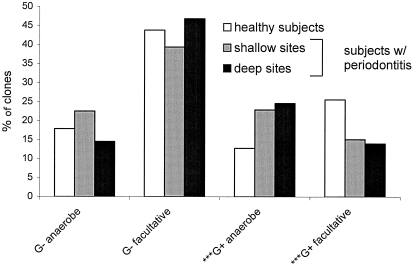
Figure 1 shows the distribution of bacterial phyla in health and disease. Bacilli and Clostridia, two classes in the phylum Firmicutes, are displayed separately due to their high numbers. Both Clostridia and Deferribacteres showed a significant (P < 0.05) association with periodontitis, and the bacilli were significantly associated with periodontal health. Figure 2 shows the distribution of gram-positive and gram-negative anaerobes and facultative bacteria in relation to health status. Gram-positive but not gram-negative bacteria showed significant differences in relation to periodontal health status: gram-positive facultative bacteria accounted for a greater fraction of total bacteria in healthy subjects than in subjects with periodontitis, and gram-positive anaerobic species were more common in subjects with periodontitis than in healthy subjects.
FIG. 1.
Distribution of bacterial phyla in health and disease. Two classes of Firmicutes (Bacilli and Clostridia) are displayed individually due to their high prevalence.
FIG. 2.
Distribution of gram-positive (G+) and gram-negative (G−) anaerobes and facultative species in relation to disease status. The gram status of uncharacterized phylotypes was inferred from that of their closest neighbor. ✽✽✽, P < 0.005.
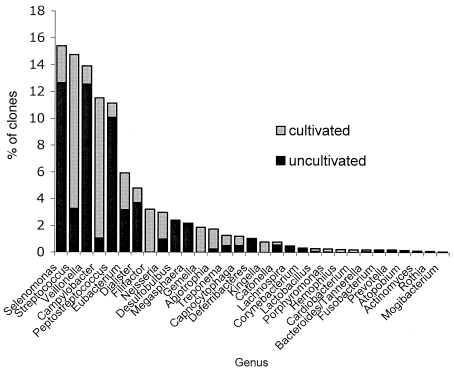
Overall, 59.9% of the clone population was made up of as-yet-uncultivable phylotypes. Figure 3 shows the relative prevalence of uncultivated phylotypes to cultivated species within each genus. The genera Deferribacteres, Megasphaera, Desulfobulbus, and Lachnospira were composed entirely of uncultivated phylotypes. Uncultivated phylotypes were predominant within the genera Selenomonas, Veillonella, and Peptostreptococcus. Other genera such as Campylobacter, Gemella, Streptococcus, and Neisseria were composed predominantly of named species.
FIG. 3.
Distribution of cultivated and uncultivated bacteria by genus for all samples.
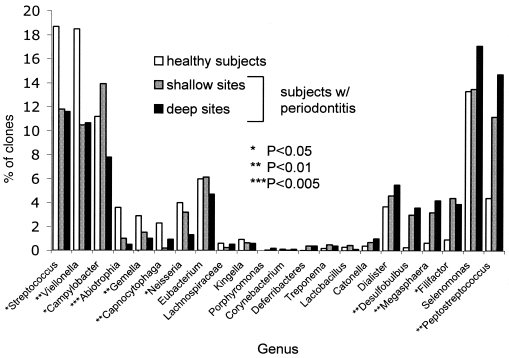
Figure 4 shows the distribution of the 22 most common bacterial genera in relation to disease status. Table 2 lists species or phylotypes that showed an association with disease or health (P < 0.1). The ranking of these species indicates their relative prevalence among all clones.
FIG. 4.
Distribution by health status for genera accounting for >0.025% of total bacteria are shown. The genera are arranged in a gradient from those predominant in health shown on the left to those predominant in periodontitis shown on the right.
TABLE 2.
Species and phylotypes significantly associated with disease and health (P < 0.1)
| Clinical status and overall rank | Species and/or phylotype |
P |
||
|---|---|---|---|---|
| Levels |
Presence (between subjectsc) | |||
| Between sitesa | Between subjectsb | |||
| Disease | ||||
| 6 | Peptostreptococcus sp. oral clone BS044 | 0.07 | ||
| 7 | Filifactor alocis | 0.04 | ||
| 15 | Peptostreptococcus sp. oral clone CK035 | 0.06 | 0.05 | 0.06 |
| 16 | Megasphaera sp. oral clone BB166 | 0.01 | 0.009 | |
| 17 | Desulfobulbus sp. oral clone CH031 | 0.03 | ||
| 19 | Dialister pneumosintes | 0.01 | 0.002 | |
| 20 | Campylobacter sputorum sputorum | 0.008 | 0.06 | |
| 24 | Desulfobulbus sp. oral clone R004 | 0.006 | 0.001 | |
| 27 | Campylobacter sp. oral clone BB120 | 0.03 | 0.008 | |
| 36 | Selenomonas sp. oral clone D0042 | 0.01 | 0.002 | |
| 50 | Deferribacteres sp. oral clone W090 | 0.03 | 0.008 | |
| 53 | Megasphaera sp. oral clone MCE3_141 | 0.003 | 0.0003 | |
| 54 | Selenomonas sp. oral clone EY047 | 0.031 | ||
| 90 | Dialister sp. oral clone MCE7_134 | 0.06 | 0.01 | |
| 100 | Catonella sp. oral clone BR063 | 0.01 | 0.002 | |
| 118 | Tannerella forsythia (Bacteroides forsythus) | 0.03 | ||
| 119 | Deferribacteres sp. oral clone BH007 | 0.01 | ||
| 123 | Streptococcus sp. oral strain 9F | 0.05 | 0.01 | |
| 129 | Atopobium sp. oral clone C019 | 0.03 | ||
| 140 | Peptostreptococcus anaerobius | 0.03 | ||
| 151 | Eubacterium sp. oral strain A35MT | 0.04 | ||
| 153 | Megasphaera sp. oral clone BS073 | 0.01 | 0.002 | |
| 154 | Selenomonas flueggei-like sp. clone AH132 | 0.03 | ||
| 165 | Treponema sp. strain 6:H:D15A-4 | 0.04 | ||
| 172 | Streptococcus sp. oral clone DP009 | 0.04 | 0.008 | |
| Health | ||||
| 1 | Veillonella sp. oral clone X042 | 0.0008 | ||
| 2 | Campylobacter gracilis | 0.04 | 0.02 | |
| 21 | Abiotrophia adiacens | 0.003 | 0.007 | |
| 31 | Eubacterium saburreum | 0.0009 | 0.005 | |
| 38 | Campylobacter showae | 0.05 | 0.02 | |
| 42 | Gemella sp. strain 1754-94 | 0.009 | 0.002 | |
| 44 | Streptococcus sanguis | 0.01 | 0.002 | |
| 49 | Capnocytophaga gingivalis | 0.02 | 0.05 | |
| 81 | Streptococcus mutans | 0.02 | 0.003 | |
| 133 | Abiotrophia sp. oral clone P4PA_155 P1 | 0.03 | ||
| 145 | Rothia dentocariosa | 0.03 | ||
| 150 | Eubacterium sp. oral clone OH3A | 0.04 | ||
| 155 | Selenomonas sp. oral clone DS051 | 0.03 | ||
Comparison of levels between deep and shallow sites in subjects with periodontitis by Wilcoxon signed-rank test.
Comparison of levels between healthy and subjects with periodontitis by Kruskal-Wallis analysis of variance.
Comparison of presence or absence of species by chi-square test.
DISCUSSION
The current paradigm of the microbial etiology of periodontitis implicates numerically minor gram-negative anaerobic components of the plaque biofilm, such as P. gingivalis, T. forsythensis, and T. denticola, as the primary etiologic agents. Although several lines of evidence are available to support an etiologic role for these species, the epidemiologic data linking these species to disease was obtained with closed-ended approaches that would not allow the detection and enumeration of previously unidentified species. The present study used 16S PCR amplification with universal 16S primers of dental plaque samples, followed by cloning and sequencing to allow an open-ended and quantitative exploration of the bacterial populations present in periodontal health and disease. Using this approach an unexpected profile of health and disease-associated bacteria populations was observed.
Molecular approach.
Subgingival bacterial populations have previously been explored by using 16S cloning and sequencing. These studies have been qualitative studies exploring the diversity of subgingival bacterial populations and have included the use of primers targeted to specific, previously suspected groups of bacteria such as the Bacteroidetes (3, 28), Eubacterium (32), and even Archaea (14), or subtraction systems to eliminate major species, such as streptococci (13), and have used high cycle numbers to enrich for minor species. In the present, quantitative study, in order to retain a representative set of amplicons, a low PCR cycle number was used to avoid plateau effects, and a set of broad, universal eubacterial primers were used. One hundred clones were sequenced and identified from every sample to allow statistical comparisons to be made. Disease-associated samples were collected from the four deepest sites in subjects with established periodontitis. Control samples were collected from shallow sites in these same subjects and also from a separate, age-matched healthy control group. Including samples from completely healthy individuals, as well as from sites that did not exhibit signs of disease in individuals with disease, allowed questions regarding site-specific versus global ecological perturbation to be addressed.
The most numerous species by 16S clonal analysis belonged to the genera Selenomonas, Streptococcus, Veillonella, Campylobacter, and Peptostreptococcus (Fig. 4). These genera were all detected in a previous culture-based study of periodontal bacteria (34), although all but Streptococcus appeared to account for a relatively smaller fraction of total bacteria. Other major groups of bacteria detected in previous studies by using DNA hybridization included Fusobacterium and, by using cultivation and DNA hybridization, Actinomyces (8, 34). Both were rare in the present study. The greater sensitivity of cultivation compared to molecular analyses for the detection of actinobacteria has been previously reported (22). To investigate this, the DNA isolation and amplification method was tested on Actinomyces viscosus in a mixture with other species, and A. viscosus was detected with comparable sensitivity (data not shown), suggesting that the bias might be attributed to over-representation with cultivation.
The genera Bacteroides and Porphyromonas were numerically minor, a finding also consistent with earlier studies (31, 34, 35), and spirochetes were also found in low numbers. Centrifugation, freezing, and long storage times before isolation of DNA have been suspected of contributing to loss of delicate, easily lysed organisms such as Spirochetes. However, the DNA isolation method was tested, both with and without centrifugation on both fresh and frozen samples, for recovery of DNA from Spirochetes, and no differences were detected (data not shown). The methodology used for DNA isolation in the present study may have been slightly biased toward gram-negative species, since the protocol did not include disruption of cell wall by vigorous agitation. Nevertheless, large numbers of gram-positive bacteria were detected. Undoubtedly some bias is present with 16S cloning and sequencing of bacterial populations due to differences in isolation of DNA from structurally varied bacteria, varied affinities for universal primers, and differences in the copy numbers of ribosomal genes. For the present study, efforts were made to minimize bias and, compared to cultivation, with less than half of species detectable and many inaccuracies inherent in phenotypic identification, molecular analysis offers a more comprehensive and accurate approach.
Overall, 274 species or phylotypes of bacteria including six novel phylotypes were detected (Table 1), and they belonged to six different phyla (Fig. 1). Consistent with earlier observations (13, 28), ca. 60% of these species were uncultivated. Several of the most numerous genera, including Selenomonas, Veillonella, and Peptostreptococcus, were composed primarily of uncultivated species (Fig. 3). Distributions of several uncultivated bacteria were found to differ between healthy subjects and subjects with periodontitis, and it appears that significant relationships may have been undetectable in previous studies using cultivation-based or closed-ended DNA approaches.
Only 0.5% chimeric sequences were detected in the present study. Studies using similar approaches have found 1 to 15% of clones to be chimeric sequences (28). For the current study, formation of chimeras was minimized by limiting the PCR cycle number (37). Colonies were also screened for inserts of the expected size by PCR and gel electrophoresis before sequencing, eliminating many potential chimeric sequences.
The large number of species observed necessitated grouping data into phyla and genera to obtain sufficient power for statistical analysis of all but the most numerous species. However, the data were analyzed at the level of species (alpha = 0.10) to identify candidate species for subsequent investigation. Because of the non-normal distributions typically observed with bacterial counts, nonparametric statistics were used for all analyses.
Phyla associated with periodontitis.
The subgingival flora in both health and periodontitis was dominated by the phylum Firmicutes. The classes Clostridia and Bacilli of the Firmicutes together accounted for 75% of all clones and were associated with opposite ends of the health spectrum: the class Bacilli (most numerous genera were Streptococcus and Gemella) accounted for a greater fraction of the bacteria in healthy subjects; in contrast, the class Clostridia (most numerous genera were Peptostreptococcus, Veillonella, and Selenomonas) was more common in subjects with periodontitis. Several additional opposing patterns of association within phyla were observed, suggesting that analysis at the level of the phylum is not informative for disease classification.
Analysis at the level of genera showed several statistically significant associations with periodontitis and health. Surprisingly, many of these occurred among the gram positives rather than the gram negatives usually thought to be important in disease.
Genera and species associated with periodontitis.
The taxonomy of the gram-positive anaerobic cocci (GPAC) commonly referred to as “peptostreptococci” is evolving, and some species previously classified as Peptostreptococcus have recently been reassigned to closely related genera (9, 23) such as Anaerococcus, Peptococcus, Micromonas, and Peptonephilus (9). In addition, several uncultivated peptostreptococci were detected in large numbers in the present study. Based on their phylogenetic similarity and evolving taxonomy, the peptostreptococci were grouped together for this analysis. The association of the peptostreptococci with periodontitis was particularly robust, and they were far more numerous than the gram-negative anaerobes commonly associated with periodontitis. At the species level, Peptostreptococcus strains BS044 and CK035 were very numerous and were associated with disease (Table 2). The selectivity of culturing and low specificity of chemical and phenotypic characterization may have prevented their identification as potential pathogens in previous studies.
GPAC have been isolated from a wide range of human infections, typically constituting one-fourth or more of anaerobic species from clinical specimens (23). Most infections involving GPAC are polymicrobial and appear to involve synergistic interactions with other bacteria (23). Previous epidemiologic evidence has linked peptostreptococci with dental infections, although investigations have been limited to Peptostreptococcus micros, a rare species in the current study (and not associated with disease). P. micros has been associated with odontogenic infections (4, 16) and is significantly higher in smokers, a population that has more extensive and severe periodontitis than nonsmokers (36). It is also more common around mobile teeth (7) and has been found at higher levels in epithelium-associated plaque compared to unattached plaque in the gingival sulcus (5). Targeted DNA approaches have also found P. micros to be elevated in advanced chronic periodontitis (26) and more common in subjects with periodontitis (15). Evidence regarding the mechanism of pathogenesis for GPAC-associated infections is limited. Peptostreptococci isolated from chronic skin ulcers have been shown to inhibit keratinocyte and fibroblast proliferation and wound repopulation in a tissue culture model system (33). P. micros demonstrates both adhesion to epithelium and coaggregation with other species such as P. gingivalis and F. nucleatum mediated by extracellular polysaccharides (11, 12). These data suggest that peptostreptococci may play a role in preventing wound healing in chronic disease and may be important in the physical structure of a disease-associated biofilm. Further exploration of the role these bacteria play in periodontitis is needed.
The gram-positive rod Filifactor alocis is related to the peptostreptococci, was also common in the samples (Table 2), and was significantly elevated in subjects with disease (Fig. 2). This organism has been previously associated with both chronic periodontitis (15) and endodontic lesions (29).
Several gram-negative bacteria were also associated with periodontitis, although they occurred in low numbers relative to the gram-positive, disease-associated species. The genus Megasphaera was elevated in cases of periodontitis, and at the species level Megasphaera oral clones BB166, MCE3_141, and BS073 were associated with disease. Megasphaera clone BB166 has been previously associated with chronic periodontitis (15). Megasphaera spp. have been reported as normal inhabitants of the gut and vagina (38), and M. elsdenii has been implicated in bacterial endocarditis in immunocompromised patients (1).
The genus Desulfobulbus was also associated with disease, and at the species level both Desulfobulbus CH031 and R004 were significantly associated with deep sites. Desulfobulbus species have been previously detected in the gingival sulcus (15, 28) and the human gut (6). Desulfobulbus are sulfate-reducing bacteria and have been frequently detected in aquatic environmental samples.
Campylobacter sputorum subsp. sputorum and Campylobacter strain BB120 were strongly associated with disease. Taken as a whole the genus Campylobacter was associated with health, but this association was accounted for by the highly prevalent species C. gracilis and C. showae.
Many clones of Selenomonas were detected, most from the cultivable species S. sputigena, S. infelix, and S. noxia. None of these were associated with disease, although S. noxia has been previously linked to active periodontitis (34). The less numerous and uncultivated Selenomonas phylotypes D0-042, EY047, and AH132 were associated with disease, and, in contrast, Selenomonas strain DS051 was detected more frequently in healthy subjects.
Dialister pneumosintes and Dialister phylotype ME_134 were associated with periodontitis. D. pneumosintes has been previously linked to periodontitis (2, 25) and to endodontic infections (30). Deferribacteres phylotypes W090 and BH007 were associated with periodontitis, and W090 has been previously linked to disease (15). In addition, uncultivated phylotypes of Catonella, Streptococci, Atopobium, Eubacterium, and Treponema were also significantly associated with disease (Table 2). However, because of the large number of species examined, some associations are likely to occur by random chance, and these candidates require further investigation.
P. gingivalis, T. denticola, and T. forsythia were rarely detected in the present study and, of these, only T. forsythia was associated with disease. Strong associations with disease have been observed for these species in many previous studies, but when quantitative results have been reported, they have comprised only a small fraction of the total bacteria. The sample size in the present study did not provide adequate power to detect association for minor species. More numerous bacteria did show strong associations with disease, however, indicating that potentially important bacteria have been overlooked in previous studies due to technical challenges. What remains unclear at the present time is whether these newly identified and more numerous species play a more important role in pathogenesis than the less numerous previously implicated species.
Genera and species associated with health.
Streptococcus and Veillonella spp. were found in high numbers in all samples and accounted for a significantly greater fraction of the microbial community in healthy subjects than in those with periodontitis. At the species level both S. sanguis and S. mutans were associated with periodontal health, as was the overall most abundant species, Veillonella sp. oral clone X042. Both Streptococcus and Veillonella have been previously associated with periodontal health (8, 34). Veillonella oral clone X042 is very closely related to V. parvula and V. dispar by 16S phylogeny and may be part of an indistinguishable cluster (18). The parallel relationship observed between levels of streptococci and Veillonella is not surprising in view of the fact that veillonellae utilize short-chain acids such as lactates that are secreted by gram-positive facultatives such as streptococci (21), and it has been shown that veillonellae will not colonize tooth surfaces without streptococci (19).
The microbial profile of periodontal health also included the less-abundant genera Campylobacter, Abiotrophia, Capnocytophaga, Gemella, and Neisseria. This confirms earlier studies linking Capnocytophaga (8, 27, 34) and Campylobacter gracilis (17) to health.
Levels of the genera Streptococcus and Veillonella were more similar between shallow and deep sites in individuals with periodontitis than between healthy individuals and those with periodontitis. A similar phenomenon was observed for many health- and disease-associated species (Table 2): many more differences were observed between healthy and diseased subjects than were found between shallow and deep sites in individuals with disease. It appears that disease may involve a disruption in the microbial ecology of the entire dentition rather than a disease site-specific shifts and that transitions between health and chronic periodontitis are associated with shifts in the relative proportions of major bacteria.
Several issues regarding molecular epidemiologic approaches to the study of chronic bacterial diseases deserve mention. First, these studies can demonstrate association but do not establish causation; subsequent studies are needed. Second, interactions with the host are likely to be important and are poorly understood at the present time. Finally, the diversity in bacterial communities is just beginning to be explored. We have little knowledge of the genetic heterogeneity in these communities beyond that occurring in ribosomal genes, so it is not clear whether explorations should be conducted at the level of genus, species, or even virulence genes. Polymicrobial bacterial communities are complex and undergo interactions within the community that could be critical determinants. Bacterial profiles also vary among individual hosts, suggesting that periodontitis has a heterogeneous etiology. Because of this complexity, much larger sample sizes than those achievable with current technology may be required for a full understanding of chronic polymicrobial diseases.
In summary, the largest differences between health-associated and periodontitis-associated biofilm communities were found among the gram-positive species. Peptostreptococcus and Filifactor were elevated in subjects with periodontitis, and Streptococcus, Abiotrophia, and Gemella were elevated in healthy subjects. Differences were also observed among the gram-negative bacteria: Veillonella, Campylobacter, and Capnocytophaga levels were higher in the plaque of healthy subjects, and Megasphaera and Desulfobulbus levels were increased in cases of periodontitis. Several species were also identified as candidates for further study, including many uncultivated phylotypes. These newly identified candidates outnumbered P. gingivalis and other species previously implicated as periodontopathogens, and it is not clear whether newly identified and more numerous species may play a more important role in pathogenesis. Finally, more differences were found in the bacterial profile of the two subject groups than between deep and shallow sites within the same mouth. This suggests that chronic periodontitis is the result of a global perturbation of the oral bacterial ecology rather than a disease-site specific microbial shift.
Supplementary Material
Acknowledgments
This study was supported by Public Health Service grant DE10467 from the National Institute of Dental and Craniofacial Research.
High-performance computing access was provided by the Ohio Supercomputer Center. We thank Ashley Beroski and Erin Gross for technical assistance.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Brancaccio, M., and G. G. Legendre. 1979. Megasphaera elsdenii endocarditis. J. Clin. Microbiol. 10:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Contreras, A., N. Doan, C. Chen, T. Rusitanonta, M. J. Flynn, and J. Slots. 2000. Importance of Dialister pneumosintes in human periodontitis. Oral Microbiol. Immunol. 15:269-272. [DOI] [PubMed] [Google Scholar]
- 3.de Lillo, A., V. Booth, L. Kyriacou, A. J. Weightman, and W. G. Wade. 2004. Culture-independent identification of periodontitis-associated Porphyromonas and Tannerella populations by targeted molecular analysis. J. Clin. Microbiol. 42:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dymock, D., A. J. Weightman, C. Scully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolar abscesses. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzink, J. L., R. J. Gibbons, W. C. d. Childs, and S. S. Socransky. 1989. The predominant cultivable microbiota of crevicular epithelial cells. Oral Microbiol. Immunol. 4:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Gibson, G. R., G. T. Macfarlane, and J. H. Cummings. 1988. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J. Appl. Bacteriol. 65:103-111. [DOI] [PubMed] [Google Scholar]
- 7.Grant, D. A., M. J. Flynn, and J. Slots. 1995. Periodontal microbiota of mobile and non-mobile teeth. J. Periodontol. 66:386-390. [DOI] [PubMed] [Google Scholar]
- 8.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 9.Hill, K. E., C. E. Davies, M. J. Wilson, P. Stephens, M. A. Lewis, V. Hall, J. Brazier, and D. W. Thomas. 2002. Heterogeneity within the gram-positive anaerobic cocci demonstrated by analysis of 16S-23S intergenic rRNA polymorphisms. J. Med. Microbiol. 51:949-957. [DOI] [PubMed] [Google Scholar]
- 10.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer, B. H., A. J. Herscheid, W. Papaioannou, M. Quirynen, and T. J. van Steenbergen. 1999. Adherence of Peptostreptococcus micros morphotypes to epithelial cells in vitro. Oral Microbiol. Immunol. 14:49-55. [DOI] [PubMed] [Google Scholar]
- 12.Kremer, B. H., and T. J. van Steenbergen. 2000. Peptostreptococcus micros coaggregates with Fusobacterium nucleatum and non-encapsulated Porphyromonas gingivalis. FEMS Microbiol. Lett. 182:57-62. [DOI] [PubMed] [Google Scholar]
- 13.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulik, E. M., H. Sandmeier, K. Hinni, and J. Meyer. 2001. Identification of archaeal rDNA from subgingival dental plaque by PCR amplification and sequence analysis. FEMS Microbiol. Lett. 196:129-133. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 16.Kuriyama, T., T. Karasawa, K. Nakagawa, E. Yamamoto, and S. Nakamura. 2002. Bacteriology and antimicrobial susceptibility of gram-positive cocci isolated from pus specimens of orofacial odontogenic infections. Oral Microbiol. Immunol. 17:132-135. [DOI] [PubMed] [Google Scholar]
- 17.Macuch, P. J., and A. C. Tanner. 2000. Campylobacter species in health, gingivitis, and periodontitis. J. Dent. Res. 79:785-792. [DOI] [PubMed] [Google Scholar]
- 18.Marchandin, H., C. Teyssier, M. Simeon De Buochberg, H. Jean-Pierre, C. Carriere, and E. Jumas-Bilak. 2003. Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiology 149:1493-1501. [DOI] [PubMed] [Google Scholar]
- 19.McBride, B. C., and J. S. Van der Hoeven. 1981. Role of interbacterial adherence in colonization of the oral cavities of gnotobiotic rats infected with Streptococcus mutans and Veillonella alcalescens. Infect. Immun. 33:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClellan, D. L., A. L. Griffen, and E. J. Leys. 1996. Age and prevalence of Porphyromonas gingivalis in children. J. Clin. Microbiol. 34:2017-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikx, F. H., and J. S. Van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20:407-410. [DOI] [PubMed] [Google Scholar]
- 22.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murdoch, D. A. 1998. Gram-positive anaerobic cocci. Clin. Microbiol. Rev. 11:81-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- 25.Nonnenmacher, C., A. Dalpke, R. Mutters, and K. Heeg. 2004. Quantitative detection of periodontopathogens by real-time PCR. J. Microbiol. Methods 59:117-125. [DOI] [PubMed] [Google Scholar]
- 26.Nonnenmacher, C., R. Mutters, and L. F. de Jacoby. 2001. Microbiological characteristics of subgingival microbiota in adult periodontitis, localized juvenile periodontitis and rapidly progressive periodontitis subjects. Clin. Microbiol. Infect. 7:213-217. [DOI] [PubMed] [Google Scholar]
- 27.Papapanou, P. N., P. N. Madianos, G. Dahlen, and J. Sandros. 1997. “Checkerboard” versus culture: a comparison between two methods for identification of subgingival microbiota. Eur. J. Oral Sci. 105:389-396. [DOI] [PubMed] [Google Scholar]
- 28.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siqueira, J. F., Jr., and I. N. Rocas. 2003. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol. Immunol. 18:263-265. [DOI] [PubMed] [Google Scholar]
- 30.Siqueira, J. F., Jr., and I. N. Rocas. 2002. Dialister pneumosintes can be a suspected endodontic pathogen. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 94:494-498. [DOI] [PubMed] [Google Scholar]
- 31.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 32.Spratt, D. A., A. J. Weightman, and W. G. Wade. 1999. Diversity of oral asaccharolytic Eubacterium species in periodontitis: identification of novel phylotypes representing uncultivated taxa. Oral Microbiol. Immunol. 14:56-59. [DOI] [PubMed] [Google Scholar]
- 33.Stephens, P., I. B. Wall, M. J. Wilson, K. E. Hill, C. E. Davies, C. M. Hill, K. G. Harding, and D. W. Thomas. 2003. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br. J. Dermatol. 148:456-466. [DOI] [PubMed] [Google Scholar]
- 34.Tanner, A., M. F. Maiden, P. J. Macuch, L. L. Murray, and R. L. Kent, Jr. 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85-98. [DOI] [PubMed] [Google Scholar]
- 35.Uematsu, H., and E. Hoshino. 1992. Predominant obligate anaerobes in human periodontal pockets. J. Periodontal Res. 27:15-19. [DOI] [PubMed] [Google Scholar]
- 36.van Winkelhoff, A. J., C. J. Bosch-Tijhof, E. G. Winkel, and W. A. van der Reijden. 2001. Smoking affects the subgingival microflora in periodontitis. J. Periodontol. 72:666-671. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G. C., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142(Pt. 5):1107-1114. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.