Abstract
Lacazia loboi is an uncultivated fungal pathogen of humans and dolphins that causes cutaneous and subcutaneous infections only in the tropical areas of the Americas. It was recently found by phylogenetic analysis that this unusual pathogen is closely related to Paracoccidioides brasiliensis and to the other fungal dimorphic members of the order Onygenales. That original phylogenetic study used universal primers to amplify well-known genes. However, this approach cannot be applied to the study of other proteins. We have developed a strategy for studying the gene encoding the gp43 homologous protein of P. brasiliensis in L. loboi. The gp43 protein was selected because it has been found that this P. brasiliensis antigen strongly reacts when it is used to test sera from patients with lacaziosis. The principle behind this idea was to obtain the gp43 amino acid sequence of P. brasiliensis and other homologous fungal sequences from GenBank and design primers from their aligned conserved regions. These sets of primers were used to amplify the selected regions with genomic DNA extracted from the yeast-like cells of L. loboi from experimentally infected mice. Using this approach, we amplified 483 bp of the L. loboi gp43-like gene. These sequences had 85% identity at the nucleotide level and 75% identity with the deduced amino acid sequences of the P. brasiliensis gp43 protein. The identity of the 483-bp DNA fragment was confirmed by phylogenetic analysis. This analysis revealed that the L. loboi gp43-like deduced amino acid sequence formed a strongly supported (100%) sister group with several P. brasiliensis gp43 sequences and that this taxon in turn was linked to the other fungal sequences used in this analysis. This study shows that the use of a molecular model for investigation of the genes encoding important proteins in L. loboi is feasible.
Lacaziosis (Jorge Lobo's disease or lobomycosis) is a chronic cutaneous and subcutaneous disease of humans and dolphins characterized by the development of nodular parakeloidal lesions, particularly on the ears, face, arms, and legs of the infected individuals (7, 13, 21). The disease is caused by the yet to be cultured fungal pathogen Lacazia loboi (26). The disease is limited to some Latin American countries. However, it has been also reported in dolphins inhabiting the coasts of Florida, the Gulf of Mexico, and South America (6, 18, 23). Anomalous cases have also been reported in individuals from Canada and the United States who had visited or who had been working in the areas of endemicity and in a European aquarium worker (3, 8, 21). Little or nothing is known about the epidemiology of the disease. It has been suggested that L. loboi may be a hydrophilic pathogen (1), but this hypothesis has yet to be confirmed.
Lacazia loboi develops abundant branched chains of thick-walled yeast-like cells 5 to 10 μm in diameter characteristically connected by short tubules in the tissues of the infected hosts. All attempts to isolate this pathogen in culture have failed, a fact that has contributed to the many colloquial names under which this pathogen has been known (5, 21, 26). Despite several decades of traditional taxonomic investigation, the relationship of L. loboi to other organisms remained elusive. This ambiguity ended when Herr et al. (11), using universal primers, amplified the 18S small-subunit rRNA gene and the 600-bp chitin synthase 2 gene of L. loboi. Those investigators reported that this unique pathogen was the phylogenetic sister taxon to the dimorphic onygenal pathogen Paracoccidioides brasiliensis, which is also restricted to Latin America, a result later confirmed by others (17).
These early studies suggested that a molecular approach might be a good strategy for the study of this uncultivated fungal pathogen. Its limitation, however, was the use of universal primers that amplify only well-known sequences. The uniqueness of our study is that we took advantage of the phylogenetic relationship of P. brasiliensis to L. loboi to design primers from their conserved regions and target homologous genes in L. loboi. This is a new approach to the study of L. loboi, and it can be used to study any homologous gene of P. brasiliensis in L. loboi, a feature not anticipated following previous protocols. This study deals with the development of a molecular model for the partial characterization of the immunodominant homologous antigen gp43 (1-3-β-glucanase, cellulase) of P. brasiliensis in L. loboi. This study showed that a molecular approach to the study of the genes encoding proteins in L. loboi is possible.
MATERIALS AND METHODS
Collection and isolation of DNA from tissue containing L. loboi yeast-like cells.
Since L. loboi cannot be isolated in culture, the isolates used in this study were maintained in mice, as described by Belone et al. (2). Essentially, several mice were inoculated with fresh yeast-like cells of this pathogen. The yeast-like cells were obtained from confirmed Brazilian cases of human lacaziosis, and the experimental infection was obtained as described by Madeira et al. (14) and Opromolla et al. (19). To confirm the presence of L. loboi yeast-like cells in mice, the tissues were fixed, sectioned, and stained with hematoxylin-eosin and silver stains. Its genomic DNA was extracted directly from skin lesions of the experimentally infected mice as described previously (11). Briefly, two aseptically collected tissue samples infected with two different isolates was transported to the laboratory and ground under liquid nitrogen. The DNA from the ground tissues was placed in microcentrifuge tubes, treated with sodium dodecyl sulfate, subjected to proteinase K digestion, and then extracted with phenol and chloroform. Genomic DNA from P. brasiliensis was obtained from previous studies (11). The genomic DNAs of L. loboi and P. brasiliensis were stored at −80°C until use. To rule out the presence of P. brasiliensis or other contaminant fungi in the mouse tissues, the samples were cultured in 2% dextrose Sabouraud agar before and after DNA extraction.
Molecular procedure for investigating the gene encoding the gp43 homologous protein of P. brasiliensis in L. loboi.
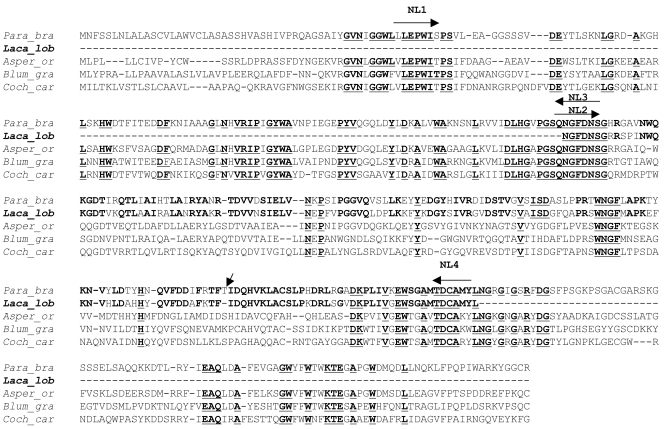
The strategy behind the selection of the antigenic protein gp43 for this study was the finding that the sera from patients with lacaziosis cross-reacted in serological assays with the purified gp43 protein obtained from isolates of P. brasiliensis (22). The fact that the gp43 of P. brasiliensis has been well characterized, the fact that its nucleotide sequences have been deposited in GenBank, and the finding that L. loboi is the sister taxon to P. brasiliensis (11) suggested that the gp43 from this pathogen may have several features in common with its homolog in L. loboi. Thus, this immunodominant antigen was considered the ideal target for the development and testing of a molecular model for L. loboi. The strategy was to obtain and align the gp43 amino acid sequence of P. brasiliensis with other homologous fungal sequences by Clustal analysis. The GenBank accession numbers of the amino acid sequences used in the Clustal analysis were as follows: Aspergillus oryzae, CAD97460; Blumeria graminis, AAL26905; Cochliobolus carbonum, AAF65310; and P. brasiliensis, AAG36681. Four sets of primers were designed from three of the highly conserved regions found within these sequences, as follows: primer NL1 (5′-TGCTGGAGCCATGGATC-3′), primer NL2 (5′-AACGGCTTCGACAACAGC-3′), primer NL3 (5′-GCTGTTGTCGAAGCCGTT-3′), and primer NL4 (5′-TAGATACATGGCGCAGTC-3′) (Fig. 1).
FIG. 1.
Clustal alignment of Paracoccidioides brasiliensis (Para_bra) gp43 amino acid sequence (GenBank accession no. AAG36681) and three homologous fungal amino acid sequences (Aspergillus oryzae [Asper_or], GenBank accession no.CAD97460; Blumeria graminis [Blum_gra], GenBank accession no. AAL26905; and Cochliobolus carbonum[Coch_car], GenBank accession no. AAF65310). The highly conserved motifs are shown in boldface and underlining. The regions were the primers NL1, NL2, NL3, and NL4 were originally designed (arrows) are shown above the selected sequences. The conserved gp43-like and the gp43 amino acid sequences of Lacazia loboi (Laca_lob) and P. brasiliensis, respectively, are shown in boldface. Note that the L. loboi gp43-like fragment lacks the amino acid threonine (inclined arrow).
PCR protocol and sequencing.
The genomic DNA extracted from two isolates of L. loboi was used to partially amplify the gp43-like gene by PCR with the primers described above, as follows: NL1-NL4 was designed to amplify 917 bp of the 1,329-bp gp43 molecule of P. brasiliensis, whereas NL1-NL3 and NL2-NL4 were designed to amplify the 917-bp amplicon in two fragments of 431 bp and 486 bp, respectively. The PCR protocol consisted of an initial activation at 95°C for 10 min (Taq Gold polymerase; Applied Biosystems, Foster City, CA) and 40 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 70°C, followed by an extension at 72°C for 7 min. Negative and positive (P. brasiliensis) controls were always included, and aseptic PCR conditions were strictly observed to ensure that foreign contamination was not a factor in the final results. The amplicons were run on 0.8% agarose gels stained with ethidium bromide and visualized on Bio-Rad Gel Doc 1,000 with Multi-Analysis version 1.0.2 (Bio-Rad, Hercules, CA).
The PCR amplicons were then cloned into a pCR 2.1-TOPO plasmid (Invitrogen, Calsbad, CA) and purified by using a SNAP miniprep kit protocol (Invitrogen), and sequencing of both strands was done by using BigDye terminator chemistry in an ABI Prism 310 genetic analyzer apparatus (Perkin-Elmer, Norwalk, CT). Ten different clones bearing the selected amplicons were edited and aligned by using Sequence Analysis and Sequencing Navigator software (Applied Biosystems/Perkin-Elmer).
Phylogenetic analysis.
The deduced amino acid sequence of the L. loboi gp43-like fragment was translated by using the standard eukaryotic codons and aligned by using Clustal analysis with 12 gp43 amino acid sequences of P. brasiliensis and 7 homologous sequences from other fungi, and their aligned sequences were visually inspected. To confirm the identities of the sequenced fragments, phylogenetic and evolutionary analyses were conducted with the computer software programs PAUP (Phylogenetic Analysis Using Parsimony, version 3.1; D. L. Swofford, Illinois Natural History Survey, Champaign), and MEGA, version 2.1 (Sudhir Kumar, Koichiro Tamura, Ingrid B. Jakobsen, and Masatoshi Nei, MEGA2: Molecular Evolutionary Genetics Analysis software, Arizona State University, Tempe). Neighbor-joining and parsimony analyses (heuristic) of the deduced amino acid sequences with maximum-likelihood multiple-hit correction and 1,000 bootstrap-resampled data set were used to assess branch support.
Nucleotide sequence accession number.
The edited gp43-like sequence of L. loboi was deposited in GenBank under accession number AY697436.
RESULTS
Clinical, histological, and laboratory findings.
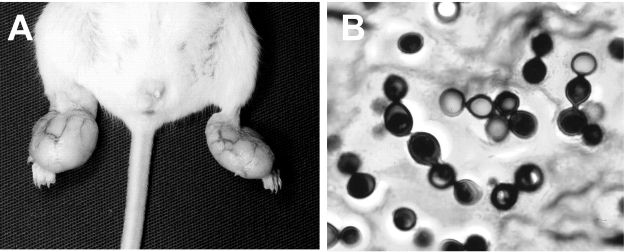
The clinical samples used for DNA isolation were collected from infected mice 3 months after experimental inoculation with L. loboi yeast-like cells. The injected mice developed parakeloidal tumoral-like lesions in the injection sites (Fig. 2A). Samples taken for histopathological and microbiological evaluation showed short chains of yeast-like cells connected by slender tubes, characteristic of L. loboi (Fig. 2B). All attempts to isolate the etiologic agent from the experimentally infected mice in 2% Sabouraud agar failed, ruling out the presence of P. brasiliensis in the tissue samples used for DNA analysis.
FIG. 2.
(A) Typical parakeloidal lesions on an experimental mouse injected with the yeast-like cells of Lacazia loboi; (B) chains of yeast-like cells of L. loboi connected by slender tubes obtained from the experimentally infected mouse. Gomori methenamine silver. Magnification, ×100.
Clustal and PCR analyses.
Clustal analysis of the gp43 amino acid sequence of P. brasiliensis and three other homologous fungal sequences (A. oryzae, B. graminis, and A. bisporus) showed several highly conserved amino acid regions (Fig. 1). Based on this result, the three most conserved motifs were selected for use in primer design. When the designed sets of primers were used with genomic DNA from a Brazilian isolate of P. brasiliensis, all the predicted 917-bp-, 431-bp-, and 486-bp-molecular mass fragments were always amplified by PCR (data not shown). Use of genomic DNA from L. loboi and primer sets NL1-NL4 and NL1-NL3 failed to amplify the predicted 917-bp and 431-bp fragments, respectively. However, when primer set NL2 and NL4 was used, a 483-bp fragment was consistently amplified from the two isolates of L. loboi used in this study (data not shown). Contamination during the PCR protocols was ruled out by the inclusion of the appropriate negative and positive controls in each experiment.
Phylogenetic analysis of L. loboi gp43-like sequence.
Sequence analysis showed that the 483-bp DNA fragments obtained from two different L. loboi isolates possessed identical sequences in this particular region of the gene. The edited 483-bp fragment from one of the L. loboi isolates was broadly compared in a BLAST (Basic Local Alignment Search Tool) search of GenBank and show that the closest matches were all the gp43 sequences of P. brasiliensis. The 483-bp DNA fragment encoded an open reading frame of 161 amino acids. The identity between the 483-bp nucleotide sequence of L. loboi and the gp43 sequences of P. brasiliensis was 85%, with several mismatches and three gaps, whereas the deduced amino acid sequence showed 75% identity with one gap. The three nucleotides encoding the amino acid threonine of P. brasiliensis at position 122 were missing from the gp43-like gene of L. loboi (Fig. 1).
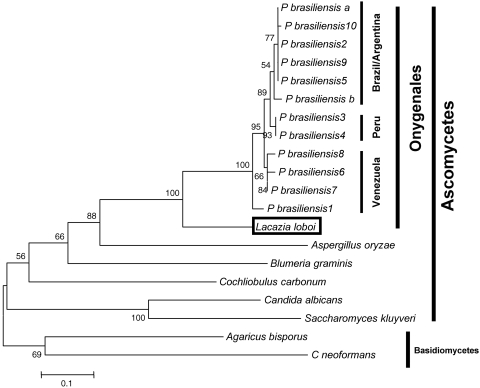
Phylogenetic analysis by the neighbor-joining and parsimony methods with the deduced amino acid sequence of the L. loboi gp43-like fragment and 12 other gp43 sequences of P. brasiliensis plus seven homologous fungal sequences showed very similar phylogenetic trees (Fig. 3). In these trees, the deduced amino acid sequence of the L. loboi 483-bp fragment was the sister group to all 12 gp43 sequences of P. brasiliensis used in this analysis. The position of the sister group was strongly supported (100%) with bootstrap searches by both parsimony and distance analyses. In addition, the 12 P. brasiliensis sequences clustered according to their geographical origins, but this distribution was not well supported. The gp43-like gene of L. loboi was also linked to the seven other homologous fungal sequences used in the study. These homologous fungal sequences clustered according to their phylogenetic preferences; the ascomycetes and the basidiomycetes both formed two poorly supported sister groups.
FIG. 3.
Phylogenetic analysis by neighbor joining in PAUP of 12 aligned gp43 amino acid sequences of Paracoccidioides brasiliensis, the Lacazia loboi gp43-like deduced amino acid sequence (GenBank accession no. AY697436), and seven other homologous fungal sequences (1-3-β-glucanases, cellulases). Multiple-hit-correction and 1,000 bootstrap-resampled data sets were used to assess branch support. In this tree the deduced amino acid sequence of the L. loboi gp43-like gene is the sister group (100% supported) to all P. brasiliensis gp43 sequences. The GenBank accession numbers for the gp43 sequences of P. brasiliensis used in this study are 2208385A, AAG36671, AAG36673, AAG36674, AAG36682, AAG36683, AAG36691, AAG36692, AAG36694, AAG36695, AAG36696, and AAG36697. The GenBank accession numbers of the homologous fungal sequences are as follows: Agaricus bisporus, CAA63536; Aspergillus oryzae, CAD97460; Blumeria graminis, AAL26905; Candida albicans, A47702; Cochliobolus carbonum, AAF65310; Cryptococcus neoformans, CAD31110; and Saccharomyces kluyveri, AAO32563. The scale bar represents the evolutionary distance, in substitutions per nucleotide.
DISCUSSION
Currently, several molecular approaches have been used to study uncultivated environmental (24) and microbial (9) pathogens, but these strategies are difficult to implement with L. loboi because >50% of its yeast-like cells within the infected tissues are dead (16, 17, 27). Dead L. loboi yeast-like cells harbor large quantities of lytic enzymes that directly interfere with RNA molecular analyses, such as reverse transcription-PCR. Moreover, genomic DNA extraction so far has yielded few L. loboi DNA molecules, and successful RNA isolation protocols are still under development for this pathogen (16, 17). This drawback in part explains why earlier molecular studies for the investigation of L. loboi met with failure (10, 16, 17).
The primer sets NL1-NL4 and NL1-NL3 were not able to amplify the predicted amplicons. This was more likely due to nucleotide mismatches in the designed primer NL1 with the nucleotide sequences of L. loboi. No introns were found in the fragment amplified from L. loboi, a feature also in common, in this particular region, with the homologous gene of P. brasiliensis. Our phylogenetic analysis confirmed that the continuous open reading frame of 161 amino acids of L. loboi was homologous to the gp43 of P. brasiliensis. However, this finding needs to be verified by mRNA analysis, an extraction protocol so far unsuccessful with this pathogen. This new approach can be used to study any homologous gene of P. brasiliensis in L. loboi. Thus, the main contribution of this study is not the amplification of the gp43-like DNA fragment of L. loboi but the molecular model that we have introduced for the study of this uncultivated fungus and that is based on the similarities between P. brasiliensis and L. loboi at the molecular level.
The L. loboi gp43-like sequence showed striking similarities with the gp43 of P. brasiliensis. For instance, the presence of numerous motifs with 100% identity between the gp43 of P. brasiliensis and the amplified fragment of L. loboi were evident. These data could explain why the P. brasiliensis gp43 antigen strongly reacted when it was tested in serological assays with sera from patients with lacaziosis (12, 15, 22). Because the gp43 antigen of P. brasiliensis has recently been linked to a protective vaccine that has successfully been used to prevent paracoccidioidomycosis in experimental animals (20, 25), we strongly believe that the gp43-like molecule of L. loboi might possess similar characteristics. This finding is of paramount importance because it predicts that this L. loboi gp43-like molecule could be used to develop strategies for the treatment and/or prevention of lacaziosis in South America. In addition, our phylogenetic analysis with the L. loboi gp43-like partial gene strongly supports previous molecular studies with the CHS2 partial gene, the 18S small-subunit rRNA gene, and the internal transcribed spacer sequences of this anomalous pathogen (11, 16, 17).
Among the organisms studied in medical mycology, L. loboi was the last of the taxonomic mysteries to be unveiled (11, 16). The recent discovery of an animal host was instrumental to maintaining in the laboratory several isolates of L. loboi for DNA extraction (14, 19). We took advantage of this to maintain a permanent supply of L. loboi genomic DNA. This study showed that the use of primers constructed from highly conserved regions of P. brasiliensis immunodominant antigens can be successful in the characterization of important homologous proteins of L. loboi. This initial step is of fundamental importance, since other P. brasiliensis immunological proteins showed cross-reactivity with sera from patients with lacaziosis as well (4, 12, 15, 22), and thus, these antigenic molecules could also be investigated by this methodology.
During this study, the name lacaziosis has been introduced to identify the diseases caused by Lacazia loboi in humans and dolphins. This designation is based on the genus name of the etiologic agent, a common practice in medical mycology. We hope that this new term ends decades of disagreement on terminology, such as lobomycosis, now considered synonymous with lacaziosis.
Acknowledgments
This study was supported by the Medical Technology Program, Michigan State University.
We thank Cristiane N. Pereira for her help with some of the molecular procedures. During this study our friend and colleague, Diltor Vladimir Araújo Opromolla, passed away. Without his support and collaboration, the studies on the molecular aspects of Lacazia loboi would not be possible.
REFERENCES
- 1.Ajello, L. 1998. Ecology and epidemiology of hydrophilic infectious fungi and parafungi of medical mycological importance: a new category of pathogens, p. 67-73. In L. Ajello and R. J. Hay (ed.), Topley and Wilson's microbiology and microbial infections, medical mycology, vol. 4, 9th ed. Arnold, London, England. [Google Scholar]
- 2.Belone, A. F. S. Madeira, P. S. Rosa, and D. V. A. Opromolla. 2002. Experimental reproduction of the Jorge Lobo's disease in Balb/c mice inoculated with Lacazia loboi obtained from a previous mouse. Mycopathologia 155:191-194. [DOI] [PubMed] [Google Scholar]
- 3.Burns, R. A., J. S. Roy, C. Woods, A. A Padhye, and D. W. Warnock. 2000. Report of the first case of lobomycosis in the United States. J. Clin. Microbiol. 38:1283-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camargo, Z. P., R. G. Baruzzi, S. M. Maeda, and M. C. Florino. 1998. Antigenic relationship between Loboa loboi and Paracoccidioides brasiliensis by serological methods. Med. Mycol. 36:413-417. [PubMed] [Google Scholar]
- 5.Ciferi, R., P. C. Acevedo, S. Campos, and L. S. Carneiro. 1956. Taxonomy of Jorge Lobo's disease fungus. Inst. Micol. Univ. Recife 53:1-21. [Google Scholar]
- 6.Cowan, D. F. 1993. Lobo's disease in a bottlenose dolphin (Tursiops truncatus) from Matagorda Bay, Texas. J. Wildl. Dis. 29:488-489. [DOI] [PubMed] [Google Scholar]
- 7.da Lacaz, C., S., R. G. Baruzzi, and M. D. B. Rosa. 1986. Doença de Jorge Lôbo, p. 1-92. IPSIS Gráfica e Editoral S.A., Sao Paulo, Brazil.
- 8.Elsayed, S., S. M. Kuhn, D. Barber, D. L. Church, S. Adams, and R. Kasper. 2004. Human case of lobomycosis. Emerg. Infect. Dis. 10:715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredericks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 9:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haubold, E. M., J. F. Aronson, D. F. Cowan, M. R. McGinnis, and C. R. Cooper, Jr. 1998. Isolation of fungal rDNA from bottlenose dolphin skin infected with Loboa loboi. Med. Mycol. 36:263-267. [PubMed] [Google Scholar]
- 11.Herr, R. A., E. J. Tarcha, P. R. Taborda, J. W. Taylor, L. Ajello, and L. Mendoza. 2001. Phylogenetic analysis of Lacazia loboi places this previously uncharacterized pathogen within the dimorphic Onygenales. J. Clin. Microbiol. 39:309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landman, G., M. A. Velludo, J. A. Lopes, and E. Mendes. 1988. Crossed-antigenicity between the etiologic agent of lobomycosis and paracoccidioidomycosis evidence by an immunoenzymatic method (PAP). Allergol. Immunopathol. (Madrid) 16:215-218. [PubMed] [Google Scholar]
- 13.Lobo, J. O. 1930. Nova especie de blastomycose. Brasil. Med. 44:1227. [Google Scholar]
- 14.Madeira, S., D. V. A. Opromolla, and A. D. F. Belones. 2000. Inoculation of Balb/C mice with Lacazia loboi. Rev. Inst. Med. Trop. Sao Paulo 42:239-243. [DOI] [PubMed] [Google Scholar]
- 15.Mendes-Giannini, M. J., M. E. Camargo, C. S. Lacaz, and A. W. Ferreira. 1984. Immunoenzymatic absorption test for serodiagnosis of paracoccidioidomycosis. J. Clin. Microbiol. 20:103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza, L., L. Ajello, and J. W. Taylor. 2001. The taxonomic status of Lacazia loboi and Rhinosporidium seeberi has been finally resolved with the use of molecular tools. Rev. Iberoam. Micol. 18:95-98. [PubMed] [Google Scholar]
- 17.Mendoza, L., and V. Silva. 2004. The use of phylogenetic analysis to investigate uncultivated microbes in medical mycology, p. 276-298. In G. San-Blas and R. A. Calderone (ed.), Pathogenic fungi. Structural biology and taxonomy. Caister Academic Press, Norfolk, England.
- 18.Migaki, G., M. G. Valerio, B. Irvine, and F. M. Garner. 1971. Lobo's disease in an Atlantic bottle-nose dolphin. J. Am. Vet. Med. Assoc. 159:578-582. [PubMed] [Google Scholar]
- 19.Opromolla, D. V. A., S. Madeira, A. D. F. Belones, and F. R. Vilani-Moreno. 1999. Jorge Lobo's disease: experimental inoculation in Swiss mice. Rev. Inst. Med. Sao Paulo 41:359-364. [DOI] [PubMed] [Google Scholar]
- 20.Pinto, A. R., R. Puccia, S. N. Diniz, M. F. Franco, and L. R. Travassos. 2000. DNA based vaccination against murine paracoccidioidomycosis using the gp43 gene from Paracoccidioides brasiliensis. Vaccine 18:3050-3058. [DOI] [PubMed] [Google Scholar]
- 21.Pradinaud, R. 1998. Loboa loboi, p. 67-73. In L. Ajello and R. J. Hay (ed.), Topley and Wilson's microbiology and microbial infections, vol. 4, 9th ed. Arnold, London, England. [Google Scholar]
- 22.Puccia, R., and L. R. Travassos. 1991. 43-kilodalton glycoprotein from Paracoccidioides brasiliensis: immunochemical reactions with sera from patients with paracoccidioidomycosis, histoplasmosis, or Jorge Lobo's disease. J. Clin. Microbiol. 29:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simose-Lopes, P. C., G. S. Paula, M. C. Both, F. M. Xavier, and A. C. Scaramello. 1993. First case of lobomycosis in a bottlenose dolphin from southern Brazil. Mar. Mammal Sci. 9:329-331. [Google Scholar]
- 24.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taborda, C. P., M. A. Juliano, R. Puccia, M. Franco, and L. R. Travassos. 1998. Mapping of the T-cell epitope in the major 43-kilodalton glycoprotein of Paracoccidioides brasiliensis which induces a Th-1 response protective against fungal infection in BALB/c mice. Infect. Immun. 66:786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taborda, P. R., V. A. Taborda, and M. R. McGinnis. 1999. Lacazia loboi gen nov., comb. nov., the etiologic agent of lobomycosis. J. Clin. Microbiol. 37:2031-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilani-Moreno, P. O., and D. V. A. Opromolla. 1997. Determinacão da viabilidade do Paracoccioides loboi em biópsias de pacientes portadores de doença de Jorge Lobo. Ann. Bras. Dermatol. (Rio de Janeiro) 72:433-437. [Google Scholar]