Abstract
A molecular typing method based on the 16S rRNA sequence diversity was developed for Haemophilus influenzae isolates. A total of 330 H. influenzae isolates were analyzed, representing a diverse collection of U.S. isolates. We found a high level of 16S rRNA sequence heterogeneity (up to 2.73%) and observed an exclusive correlation between 16S types and serotypes (a to f); no 16S type was found in more than one serotype. Similarly, no multilocus sequence typing (MLST) sequence type (ST) was found in more than one serotype. Our 16S typing and MLST results are in agreement with those of previous studies showing that serotypable H. influenzae isolates behave as highly clonal populations and emphasize the lack of clonality of nontypable (NT) H. influenzae isolates. There was not a 1:1 correlation between 16S types and STs, but all H. influenzae serotypable isolates clustered similarly. This correlation was not observed for NT H. influenzae; the two methods clustered NT H. influenzae isolates differently. 16S rRNA gene sequencing alone provides a level of discrimination similar to that obtained with the analysis of seven genes for MLST. We demonstrated that 16S typing is an additional and complementary approach to MLST, particularly for NT H. influenzae isolates, and is potentially useful for outbreak investigation.
Haemophilus influenzae is an important cause of childhood invasive diseases, including meningitis and septicemia, in many parts of the world (30). In the United States, before the introduction of H. influenzae serotype b conjugate vaccines, between 10,000 and 20,000 cases of serotype b meningitis and other serious manifestations occurred each year (39). After the introduction of the serotype b conjugate vaccine in December 1987, invasive serotype b disease in the United States was reduced by more than 95% (2, 39). Since the dramatic decline in invasive serotype b disease, the relative contribution to disease burden has shifted to other serotypes and nontypable (NT) H. influenzae isolates. Estimates of the total number of cases of invasive H. influenzae disease have not changed significantly in the United States since 1997 (3,400 to 3,900 cases per year), with NT H. influenzae disease contributing ∼70% of the total number of cases (Centers for Disease Control and Prevention; Active Bacterial Core Surveillance reports, Emerging Infectious Program Network, Neisseria meningitidis; available at http://www.cdc.gov/ncidod/dbmd/abcs/survreports.htm, accessed 8 November 2004). Currently, rare cases of invasive serotype b disease continue to occur, most often identified in unvaccinated or undervaccinated populations with high carriage rates (12, 13, 22).
Methods for molecular characterization of bacterial pathogens are commonly used to establish genetic relatedness or similarities between individual strains, which is useful for understanding disease transmission, tracking the spread of virulent or antibiotic-resistant strains, and monitoring the evolution of bacterial populations. H. influenzae isolates have been characterized by different molecular methods, including pulsed-field gel electrophoresis and multilocus enzyme electrophoresis (MEE) (25, 26). To date, MEE has been the most discriminatory method, showing that H. influenzae serotypes c, d, e, and f form monophyletic clusters, unlike H. influenzae serotype a or serotype b isolates, which are more diverse (24, 26, 31). MEE also showed that NT H. influenzae isolates are more heterogeneous than serotypable isolates due to frequent genetic recombination (24, 31). In the last few years, though, multilocus sequence typing (MLST) has been replacing MEE as a molecular subtyping tool for a number of bacterial pathogens, including H. influenzae (7, 19, 23, 37), since it allows much better intra- and interlaboratory comparison and portability of data.
Recently, 16S rRNA gene sequencing (16S typing) has been used to characterize Neisseria meningitidis isolates (33), and the epidemiological benefit of this approach became apparent in investigations of disease outbreaks caused by N. meningitidis serogroup C and W135 isolates. 16S typing provided the highest sensitivity, specificity, and positive and negative predictive values for defining outbreak-related versus sporadic isolates when compared with pulsed-field gel electrophoresis, MEE, and MLST (21, 33).
Because of the increased contribution of non-serotype b H. influenzae in invasive disease in the United States after the introduction of the serotype b vaccine, the lack of an ideal molecular subtyping method, especially for NT H. influenzae isolates, and the successes of 16S typing for men in outbreak settings, we analyzed a representative and diverse collection of H. influenzae isolates by 16S typing and MLST to do the following: (i) investigate the diversity of H. influenzae 16S rRNA genes in H. influenzae isolates of different serotypes and in nonserotypable isolates, (ii) examine any association of 16S type with MLST sequence types (ST) or H. influenzae serotype, and (iii) evaluate the abilities of both methods to differentiate NT H. influenzae isolates.
MATERIALS AND METHODS
Bacterial isolates.
A total of 330 H. influenzae isolates (268 clinical isolates defined as causative agents of invasive disease and 62 isolates from asymptomatic carriers or reference strains) were selected for this study, 239 isolates of different serotypes and 91 nonserotypable isolates (13 serotype a isolates, 135 serotype b isolates, 3 serotype c isolates, 5 serotype d isolates, 45 serotype e isolates, and 38 serotype f isolates).
H. influenzae surveillance isolates.
Of the 330 H. influenzae isolates in this study, 293 were surveillance isolates. Two hundred twenty-seven H. influenzae case isolates recovered from 2000 to 2003 by the Active Bacterial Core Surveillance/Emerging Infectious Program for H. influenzae invasive disease (part of the ongoing multistate active laboratory-based, population-based surveillance program coordinated by Centers for Disease Control and Prevention [CDC]). They were selected for this study to provide representation for the U.S. H. influenzae isolates as follows: all isolates of serotypes a, b, c, and d (n = 12, 42, 1, and 3, respectively) and a random selection from serotypes e (43/80, 54%) and f (36/267, 13%) and NT H. influenzae (90/894, 10%).
H. influenzae community survey isolates.
Twenty-six H. influenzae isolates (25 serotype b isolates and 1 NT isolate) were recovered from 1999 to 2000 during an investigation of serotype b transmission among Amish children in Pennsylvania (12). Eight of these isolates were from patients with invasive disease; the other 18 isolates were from asymptomatic carriers in two of the communities in which there were cases of serotype b disease. These isolates were selected to assess usefulness of 16S typing in an outbreak-like setting. An additional 66 serotype b isolates were collected from a study in Alaska during 1992 to 1997; 24 isolates were from cases of invasive serotype b disease, and the remaining 42 isolates were cultured from the throats of healthy Alaska Native children. They were selected for this study due to the slightly different epidemiology of H. influenzae disease in Alaska and its remote geographic location (18).
H. influenzae reference strains.
Eleven H. influenzae strains were used as controls for serotype-specific PCR, including six CDC control isolates (serotype a, M4741; serotype b, M5216; serotype c, M6542; serotype d, M6548; serotype e, M5153; and serotype f, M6297) and five ATCC strains (serotype b, ATCC 9795; serotype c, ATCC 9007; serotype d, ATCC 9332; serotype e, ATCC 8142; and serotype f, ATCC 9833).
Identification, standard slide agglutination serotyping, and serotype-specific PCR.
Identification of all H. influenzae isolates was performed in the state public health laboratories according to standard microbiological methods. H. influenzae serotypes were confirmed at the CDC by standard slide agglutination for all 330 isolates. PCR specific for the capsule transport gene (bexA) and serotype-specific PCRs for molecular characterization of H. influenzae capsular types (a to f) were performed on all 330 isolates as previously described (9), except that the final MgCl2 concentration was increased to 2.1 mM and the annealing temperature was 55°C when serotype e-specific primers were used.
Whole-cell suspensions.
Whole-cell suspensions were used as templates for all PCRs. Cells were harvested from overnight growth on chocolate II agar plates (BD Bioscience, Baltimore, MD), transferred into 1.0 ml of 10 mM Tris buffer (pH 8), and vortexed to homogeneity. Suspensions were boiled at 100°C for 10 min and then stored at −20°C.
16S rRNA gene sequence.
The 16S rRNA genes of all H. influenzae isolates were amplified by PCR using external primers to the 16S rRNA gene, primers F15 and R1594, as previously described (33). The amplified product of 1,595 bp was sequenced using 16 different primers: primers 357, 530, 790, and 1390 in forward and reverse orientation; primers 15, 17, 24, 981, and 1230 in forward orientation; and primers 1492, 1583, and 1585 in reverse orientation. A total of six novel primers were used in this study: F15 (5′TAAGCAGTTTATTGAGCGAT 3′), F17 (5′ AGCAGTTTATTGAGCGATTG 3′), F24 (5′ TATTGAGCGATTGAACTTGA 3′), R1583 (5′ CTCGCTGTCTCTCGTCTTCA 3′), R1585 (5′ CTCGCTGTCTCTCGTCTT 3′), and R1594 (5′ GTGAGCACTCGCTGTC 3′). Primers were designed based on the 16S rRNA gene consensus sequence of the six operons of the serotype d strain Rd W20 (GenBank L42023) (11), using Oligo V6 software (Molecular Biology Insights, Inc., Cascade, CO). Primers 357, 530, 790, 981, 1230, 1390, and 1492 were described previously (8, 32, 35). Sequencing was performed using the BigDye terminator cycle sequencing kit version 2.0 (Applied BioSystems, Foster City, CA). Sequencing products were purified using Centri-Sep (Princeton Separations, Adelphia, NJ) and were resolved in an Applied BioSystems model 3100 automated DNA sequencing system (Applied BioSystems).
16S rRNA type determination.
The 16S rRNA gene sequence (1,538 bp) was obtained and used for analysis and comparison by the GCG (Wisconsin) package, version 10.1, (Accellrys, San Diego, CA). A type number was assigned for each different 16S sequence; a single base difference, including a mixed base (more than one nucleotide identified at a single position), was considered a novel 16S type. Because H. influenzae has six rRNA operons (11), the PCR products could have different nucleotides at any single position for a given operon, which could result in a heterologous base call or “mixed base” in the consensus sequence. When a unique 16S type was obtained, the 16S rRNA gene amplification and sequencing of the entire gene or parts containing the novel region were repeated.
MLST.
MLST was performed by sequencing gene fragments of adk, atpG, frdB, fucK, mdh, pgi, and recA as previously described (23) (http://www.mlst.net). MLST was performed on 304/330 H. influenzae isolates in this study. MLST testing of the 26 isolates from Pennsylvania was conducted previously (23). The detailed MLST results and sources of the isolates determined in this study have been deposited in the H. influenzae public database (http://www.mlst.net). The eBurst algorithm and the H. influenzae database at http://eburst.mlst.net were used to examine relationships among isolates (10).
Dendrogram.
Two dendrograms were generated: one for 16S and one for MLST data. Each dendrogram contained a single representative sequence for each of the 65 different 16S types or 89 different STs. For the MLST dendrogram, DNA sequences for each of the seven genes for each ST were concatenated and used as an input file. Evolutionary distance correlation was predicted by the method of Jukes and Cantor, and the phylogenetic dendrograms were generated using the unweighted pair group method with arithmetic mean (UPGMA) (16).
Nucleotide sequence accession numbers.
The 330 16S rRNA gene sequences determined in this study have been deposited in the GenBank database under the following accession numbers: AY613445 to AY613775.
RESULTS
bexA and serotype-specific PCR.
The bexA product was obtained for all 239 serotypable H. influenzae isolates; no bexA PCR product was obtained from any of the 91 NT H. influenzae isolates. Serotype-specific PCR was in agreement with the slide agglutination serotyping results for all 239 serotypable isolates.
16S rRNA gene sequencing. (i) Sequence diversity.
In total, 65 different 16S types were identified (Table 1). Sequence differences among these 65 16S types ranged from 0.06% to 2.73%, i.e., from a single base to small regions of up to seven consecutive base changes. The 16S rRNA gene sequence for serotype f isolates (16S types 13, 51, and 54) is shorter by one base due to a base missing at position 211 (1,537-bp length). When the nucleotide sequence of the 16S rRNA gene fragments (1,538 bp) from all 330 isolates were aligned and compared, 78 positions of difference throughout the gene were found. In 32 of these 78 positions (41%), mixed bases were detected. There were two hypervariable areas (46 bp and 34 bp) in the 16S rRNA gene responsible for 33 (42%) of the 78 positions of differences. These areas are between positions 170 and 216 (18 positions) and 1002 and 1036 (15 positions) in the 16S rRNA gene; these positions correspond to variable regions V2 and V6 (27). The remaining 45 positions were distributed across the entire length of the 16S rRNA gene sequence.
TABLE 1.
Distribution of 65 16S types and 89 STs and their correlation
| Serotype (na) | ST within 16S type |
16S type within ST |
||||
|---|---|---|---|---|---|---|
| 16S type | No. (%)b of isolates | ST | ST | No. (%)b of isolates | 16S type | |
| a (13) | 22 | 3 (23) | 23 | 56 | 8 (62) | 34, 35, 36 |
| 34 | 3 (23) | 56 | 23 | 4 (31) | 22, 35 | |
| 36 | 3 (23) | 56 | 4 | 1 (8) | 47 | |
| 35 | 2 (15) | 23, 56 | ||||
| 33 | 1 (8) | 56 | ||||
| 47 | 1 (8) | 4 | ||||
| b (135) | 6 | 67 (50) | 6, 24, 55, 95, 114, 118, 119, 128, 130 | 6 | 53 (39) | 6, 44 |
| 66 | 35 (26) | 54 | 54 | 39 (29) | 41, 44, 66 | |
| 45 | 20 (15) | 44, 157 | 44 | 16 (12) | 45 | |
| 44 | 4 (3) | 6, 54, 120 | 55 | 5 (4) | 6 | |
| 48 | 4 (3) | 45, 115 | 157 | 4 (3) | 45 | |
| 41 | 2 (1) | 54 | 45 | 3 (2) | 48 | |
| 27 | 1 (1) | 117 | 95 | 3 (2) | 6 | |
| 42 | 1 (1) | 115 | 115 | 2 (1) | 42, 48 | |
| 43 | 1 (1) | 116 | 119 | 2 (1) | 6 | |
| 24 | 1 (1) | 6 | ||||
| 114 | 1 (1) | 6 | ||||
| 116 | 1 (1) | 43 | ||||
| 117 | 1 (1) | 27 | ||||
| 118 | 1 (1) | 6 | ||||
| 120 | 1 (1) | 44 | ||||
| 128 | 1 (1) | 6 | ||||
| 130 | 1 (1) | 6 | ||||
| c (3) | 65 | 2 (67) | 9 | 9 | 3 (100) | 40, 65 |
| 40 | 1 (33) | 9 | ||||
| d (5) | 1 | 5 (100) | 10, 47 | 47 | 3 (60) | 1 |
| 10 | 2 (40) | 1 | ||||
| e (45) | 17 | 25 (56) | 18, 68, 122, 127 | 18 | 24 (53) | 17, 37 |
| 37 | 15 (33) | 18, 66, 69 | 66 | 8 (18) | 37 | |
| 38 | 3 (7) | 121 | 122 | 3 (7) | 17 | |
| 49 | 2 (4) | 129 | 121 | 3 (7) | 38 | |
| 129 | 2 (4) | 49 | ||||
| 127 | 1 (2) | 17 | ||||
| 69 | 1 (2) | 37 | ||||
| 68 | 1 (2) | 17 | ||||
| f (38) | 13 | 36 (95) | 123, 124, 126 | 124 | 35 (92) | 13, 51 |
| 51 | 1 (3) | 124 | 123 | 1 (3) | 13 | |
| 54 | 1 (3) | 125 | 125 | 1 (3) | 54 | |
| 126 | 1 (3) | 13 | ||||
| NT (91) | 3 | 16 (18) | 12, 14, 46, 135, 136, 137, 144, 165, 181, 183 | 3 | 4 (4) | 23, 26 |
| 4 | 10 (11) | 34, 140, 149, 153, 156, 159, 163, 186 | 14 | 4 (4) | 3, 59 | |
| 39 | 7 (8) | 11, 103, 145 | 159 | 4 (4) | 4, 7, 31 | |
| 7 | 4 (4) | 159, 165 | 165 | 4 (4) | 3, 7 | |
| 11 | 4 (4) | 57, 84 | 12 | 3 (3) | 3 | |
| 28 | 4 (4) | 150, 185 | 57 | 3 (3) | 11 | |
| 26 | 3 (3) | 3 | 103 | 3 (3) | 39 | |
| 31 | 3 (3) | 107, 159 | 11 | 2 (2) | 39 | |
| 2 | 2 (2) | 143 | 34 | 2 (2) | 4, 56 | |
| 5 | 2 (2) | 142, 155 | 41 | 2 (2) | 24, 30 | |
| 12 | 2 (2) | 162 | 46 | 2 (2) | 3 | |
| 20 | 2 (2) | 146 | 142 | 2 (2) | 5, 21 | |
| 32 | 2 (2) | 147, 148 | 143 | 2 (2) | 2 | |
| 46 | 2 (2) | 105, 149 | 145 | 2 (2) | 39 | |
| 53 | 2 (2) | 169 | 146 | 2 (2) | 20 | |
| 60 | 2 (2) | 167 | 147 | 2 (2) | 14, 32 | |
| 8 | 1 (1) | 139 | 149 | 2 (2) | 4, 46 | |
| 9 | 1 (1) | 160 | 150 | 2 (2) | 28 | |
| 10 | 1 (1) | 112 | 153 | 2 (2) | 4 | |
| 14 | 1 (1) | 147 | 156 | 2 (2) | 4 | |
| 15 | 1 (1) | 152 | 162 | 2 (2) | 12 | |
| 16 | 1 (1) | 134 | 167 | 2 (2) | 60 | |
| 19 | 1 (1) | 180 | 169 | 2 (2) | 53 | |
| 21 | 1 (1) | 142 | 180 | 2 (2) | 19, 61 | |
| 23 | 1 (1) | 3 | 183 | 2 (2) | 3 | |
| 24 | 1 (1) | 41 | 185 | 2 (2) | 28 | |
| 25 | 1 (1) | 166 | 84 | 1 (1) | 11 | |
| 30 | 1 (1) | 41 | 105 | 1 (1) | 46 | |
| 50 | 1 (1) | 158 | 107 | 1 (1) | 31 | |
| 52 | 1 (1) | 161 | 112 | 1 (1) | 10 | |
| 55 | 1 (1) | 141 | 134 | 1 (1) | 16 | |
| 56 | 1 (1) | 34 | 135 | 1 (1) | 3 | |
| 57 | 1 (1) | 182 | 136 | 1 (1) | 3 | |
| 58 | 1 (1) | 164 | 137 | 1 (1) | 3 | |
| 59 | 1 (1) | 14 | 138 | 1 (1) | 64 | |
| 61 | 1 (1) | 180 | 139 | 1 (1) | 8 | |
| 62 | 1 (1) | 151 | 140 | 1 (1) | 4 | |
| 63 | 1 (1) | 168 | 141 | 1 (1) | 55 | |
| 64 | 1 (1) | 138 | 144 | 1 (1) | 3 | |
| 67 | 1 (1) | 184 | 148 | 1 (1) | 32 | |
| 151 | 1 (1) | 62 | ||||
| 152 | 1 (1) | 15 | ||||
| 155 | 1 (1) | 5 | ||||
| 158 | 1 (1) | 50 | ||||
| 160 | 1 (1) | 9 | ||||
| 161 | 1 (1) | 52 | ||||
| 163 | 1 (1) | 4 | ||||
| 164 | 1 (1) | 58 | ||||
| 166 | 1 (1) | 25 | ||||
| 168 | 1 (1) | 63 | ||||
| 181 | 1 (1) | 3 | ||||
| 182 | 1 (1) | 57 | ||||
| 184 | 1 (1) | 67 | ||||
| 186 | 1 (1) | 4 | ||||
n, no. of isolates.
The percentage of 16S type or ST for that serotype. The percentages may not add up to 100% due to rounding.
(ii) 16S rRNA types among serotypable isolates.
There was exclusive correlation between 16S types and serotypes: 25 16S types were found among the 239 serotypable isolates (Table 1), and none of these 25 16S types was found among NT H. influenzae isolates.
(iii) 16S rRNA types among NT H. influenzae isolates.
Forty 16S types were found among the 91 NT H. influenzae isolates, and none of these 16S types was found among serotypable isolates (Table 1). Thirty-seven (92.5%) of these 40 16S types are identified in only 1 to 4 isolates each. Only three 16S types, types 3, 4, and 39, are represented by more than five isolates each. These account for 33 (36%) of the NT H. influenzae isolates; 16S type 3 (n = 16; 18%), type 4 (n = 10; 11%), and type 39 (n = 7; 8%).
MLST. (i) MLST diversity.
The nucleotide sequences of the seven MLST genes were aligned and compared with those on the MLST website. We found between 22 and 43 different alleles for each gene: 29 for adK, 22 for atpG, 30 for frdB, 26 for fucK, 43 for mdh, 36 for pgi, and 29 for recA. In total, 62 novel alleles were found and added to the MLST database: 8 for adK, 5 for atpG, 10 for frdB, 6 for fucK, 12 for mdh, 14 for pgi, and 7 for recA. When the alleles for the 7 genes for each of the 330 isolates were combined, we obtained 89 different STs, of which 47 were novel, and 9, 4, 4, and 30 new STs among serotype b, serotype e, serotype f, and NT H. influenzae, respectively, were found. The novel STs also were added to the MLST database. For each of the 89 different STs, a concatenated DNA sequence of 3,057 bp was generated using the individual sequences for each of the 7 genes. The concatenated sequences were aligned and used to generate a dendrogram (see Fig. 2).
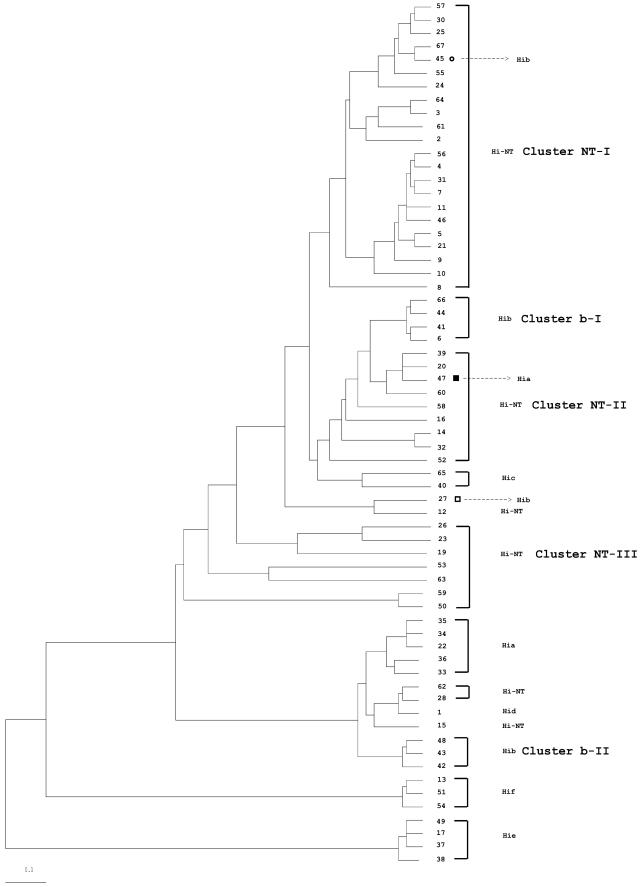
FIG. 2.
Consensus UPGMA phylogenetic tree constructed with 89 concatenated DNA sequences obtained from seven housekeeping genes of H. influenzae. The number in the branches indicates the ST, and the respective serotypes are represented. The symbols by STs 44 and 157 (circle), 117 (open square), and 4 (filled square) indicate STs of H. influenzae serotypes that correlate with 16S types in Fig. 1; they are indicated by the same symbols. The scale bar represents an expected substitution rate of 0.1 nucleotide substitution per base position. See the legend to Fig. 1 for abbreviations.
(ii) STs among serotypable H. influenzae isolates.
Thirty-five STs were found among the 239 serotypable isolates (Table 1). There was an exclusive correlation between STs and serotypes. None of these 35 STs was found among the NT H. influenzae isolates.
(iii) STs among NT H. influenzae isolates.
Fifty-four STs were found among and were exclusive to the 91 NT H. influenzae isolates (Table 1). Forty-seven (87%) of these STs are represented by 1 (n = 29; 54%) or 2 (n = 18; 33%) isolates only. The remaining 13% are STs represented by 3 (n = 3; 6%) or 4 (n = 4; 7%) isolates.
Correlation among H. influenzae 16S types, STs, and serotypes.
The correlation between 16S types and STs is presented in Table 1. Thirty-five of the 65 (54%) 16S types correlated exclusively with a particular ST; 27 of the 35 (77%) were among NT H. influenzae. For the remaining 30 16S types (46%) that did not correlate exclusively with a particular ST, 13 (43%) were also NT H. influenzae. Twenty-nine of the 89 (33%) STs correlated exclusively with a particular 16S type; 24 of the 29 (83%) were among NT H. influenzae. For the remaining 60 STs (67%) that did not correlate exclusively with a particular 16S type, 30 (50%) were also NT H. influenzae. There was a specific correlation between 16S types and STs with serotypes; not a single 16S type or ST was found in two or more different H. influenzae serotypes. Six major clones defined by a combination of ST/16S types represent 69% of serotypable isolates (Table 2).
TABLE 2.
The six most prevalent combinations of MLST and 16S rRNA types among serotypable H. influenzae isolatesa
| MLST cluster | MLST | No. (%) of isolates with MLST and 16S rRNA type combination |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serotype b (n = 135) and 16S rRNA type: |
Serotype e (n = 45) and 16S rRNA type: |
Serotype f (n = 38) and 16S rRNA type: |
||||||||
| 6 | 45 | 66 | Other | 17 | 37 | Other | 13 | Other | ||
| b-I | ST6 | 52 (39)b | 1 (<1) | |||||||
| ST44c | 16 (12) | |||||||||
| ST54c | 35 (26) | 4 (3) | ||||||||
| e | ST18 | 20 (44) | 4 (9) | |||||||
| ST66d | 10 (22) | |||||||||
| f | ST124 | 33 (87) | 2 (5) | |||||||
| Other STs | 15 (11) | 4 (3) | 12 (9) | 5 (11) | 1 (2) | 5 (11) | 3 (8) | |||
Sixty-nine percent of serotypeable isolates have one of these six combinations (in bold): ST6/16S type 6, ST44/16S type 45, ST54/16S type 66, ST18/16S type 17, ST66/16S type 37, and ST124/16S type 13. Note that in some cases additional 16S types or STs occur in less-frequent combinations.
Number of isolates (percentage for that serotype). Percentages do not add to 100% due to rounding.
ST44 and 54 differ from ST6 by two alleles.
ST66 differs from ST18 by one allele.
Pennsylvania and Alaska community isolates.
For the 25 Pennsylvania serotype b isolates, three different 16S types (16S types 6, 45, and 48) were obtained, and one NT H. influenzae was of 16S type 3. The 16S sequencing results were compared with previously published MLST results for the 26 isolates (23). The carriage and disease isolates from each of these two communities were shown to have identical, community-specific molecular markers by both 16S and MLST. Similar results were obtained for the 66 Alaska isolates, with the exception of 7 16S type 45 isolates discussed below.
Relationships among 16S rRNA or MLST sequences.
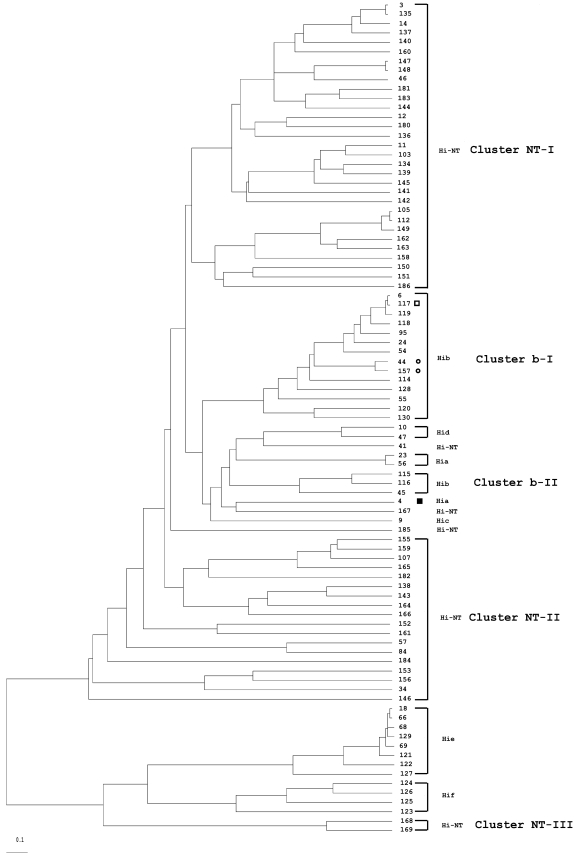
Dendrograms generated with 16S rRNA or MLST gene sequences show that isolates of a particular serotype are grouped as one unique, serotype-specific cluster, with the exception of serotype b isolates, which were grouped into two serotype b clusters (b-I and b-II), and that NT H. influenzae isolates are mainly grouped into three unclearly delineated clusters (NT-I, NT-II, and NT-III) (Fig. 1 and 2). However, the 16S rRNA dendrogram shows three exceptions: first, a single serotype a isolate (16S type 47) was grouped into cluster NT-II. The 16S type 47 sequence is similar to 16S type 34 (serotype a), with differences at four positions (positions 263, 468, 1131, and 1133) and a region of 19 bp (between positions 831 to 850). These differences in the sequence are identical to the sequence of 16S types 20 and 39, both part of the cluster NT-II. This suggests that 16S type 47 arose by recombination between 16S type 34 (serotype a) with 16S type 20 or 39 (NT H. influenzae). Second, serotype b of 16S types 27 (1 isolate) and 45 (20 isolates) were not in cluster b-I or b-II but clustered with NT H. influenzae isolates. Additional sequence examination showed that 16S types 27 and 45 are highly related to 16S type 6 (cluster b-II); 16S type 27 sequence is the same as 16S type 6, with 12 bp (between positions 180 and 191) identical to 16S type 12. The 16S type 45 sequence is also like 16S type 6, with a substitution of three consecutive bases (between positions 1131 and 1133) identical to those of 16S types 55 and 67. It appears that 16S types 27 and 45 arose by recombination between 16S types 6 and 12 to produce type 27 and between 16S types 6 and either 55 or 67 to produce type 45. MLST confirmed that these unusual serotype b isolates are within the MLST b-I cluster including the ST6 complex (Fig. 2). According to eBurst analysis, ST117 (found in isolates of 16S type 27), ST44, and ST157 (both 16S type 45) are all related to ST6 with one, two, or three locus variations from ST6, showing these isolates to be members of the ST6 clonal complex.
FIG.1.
Consensus UPGMA phylogenetic tree constructed with 65 identified H. influenzae 16S types. The numbers on the branches indicate the 16S type, and the respective serotypes are represented. The symbols denoting 16S types 45 (circle), 27 (open square), and 47 (filled square) indicate H. influenzae isolates with 16S types and serotypes that do not cluster with other 16S types in each respective serotype. These 16S types correlate with STs in Fig. 2, as indicated by the same symbols. The scale bar represents an expected substitution rate of 0.1 nucleotide substitution per base position. Hi-NT, nontypeable H. influenzae; Hib, H. influenzae serotype b; Hia, H. influenzae serotype a; Hic, H. influenzae serotype c; Hid, H. influenzae serotype d; Hie, H. influenzae serotype e; Hif, H. influenzae serotype f.
For the NT H. influenzae isolates, both 16S and MLST dendrograms show that NT H. influenzae isolates are distributed throughout the tree. More than 90% of the NT H. influenzae isolates are within one of the three major clusters defined by 16S typing or MLST. However, unlike the near-perfect correlation between 16S types and STs found for H. influenzae serotypable isolates, clustering of the NT H. influenzae isolates differs by these two methods. For example, 16S cluster NT-I contains 10 STs from MLST cluster NT-I and 12 STs from MLST cluster NT-II, plus ST41. In fact, 16S types 3, 4, and 5 are seen in isolates from both MLST clusters NT-I and NT-II. 16S cluster NT-II shows similar heterogeneity, and 16S cluster NT-III is a mixture of MLST clusters NT-I and NT-III.
DISCUSSION
16S rRNA gene sequencing has been extensively used for identification and classification of microorganisms. Its use in microbial systematics is based on two assumptions: (i) that multiple 16S operons within one genome are essentially identical, and (ii) that horizontal transfer of 16S rRNA genes does not occur. However, it has become apparent that these assumptions may not be correct for all prokaryotes (6, 36, 38, 40). We found in this study that the 16S rRNA gene diversity among H. influenzae isolates ranges from 0.06% to 2.73%, a much higher level of diversity than originally found in N. meningitidis isolates (0.06% to 0.95%) (33) or the range obtained using our entire N. meningitidis collection (0.06% to 1.44%; data not published). Differences of 16S rRNA gene sequences among the serotype f isolates (types 13, 51, and 54) and the serotype e isolates (types 17, 37, 38, and 49) are very small (0.06% to 0.13%) inside each group. However, these sequences are divergent from all other H. influenzae 16S rRNA gene sequences. The range of differences between serotype e and serotype f 16S types and all the other 16S types was 1.63% to 2.73% and 1.76% to 2.73%, respectively. These values could easily have excluded them from belonging to the H. influenzae species according to the ∼1.0% to 2.0% 16S rRNA gene sequence differences cutoff threshold generally used for species identification (17). It is also interesting that the 16S rRNA gene sequences of all 38 serotype f isolates are one base shorter than those of all other H. influenzae 16S rRNA genes analyzed so far. Greater divergence in 16S rRNA genes has been found previously in other bacteria, e.g., bacteria having multiple operons within one genome, differing from each other from 6.7% to 11.6% (1, 4, 6, 20). Therefore, recombination between 16S rRNA genes of different strains or species can occur at a much higher frequency than originally suspected. In contrast to the high level of diversity of 16S rRNA genes among N. meningitidis and H. influenzae isolates, we have previously found that for some species, the diversity of 16S rRNA genes can be limited or even absent (14, 15, 32), suggesting that the level of 16S rRNA gene diversity is also species related. Some of the reasons that would affect the level of diversity among different bacteria would be natural competency, differences in ecology, the presence of DNA uptake sequences, and restriction modification systems (3, 5, 28, 29, 34).
The 16S rRNA gene sequence diversity of H. influenzae isolates described here is greater than in any other human pathogenic bacterial species described so far. However, we still observed a direct correlation between 16S types and serotypes (a to f); no 16S type was found in 2 or more different serotypes. An identical correlation was also observed between STs and serotypes.
Previous studies using a variety of subtyping methods including MEE and MLST have shown that though serotypable H. influenzae behave as highly clonal populations, NT H. influenzae are not closely related to serotypable isolates, and they also appear to undergo more frequent recombination (24, 26, 31). Our 16S typing and MLST results are in agreement with these studies and even emphasize the lack of clonality of NT H. influenzae in that the 2 methods cluster isolates differently.
The results of this study demonstrate that 16S typing and MLST allow for defining relationships among isolates when used on a predominantly clonally structured bacteria like serotypable H. influenzae. This was further confirmed in the analysis of serotype b isolates collected in Pennsylvania, as well as clinical and carrier serotype b isolates from Alaska, both areas with persistent disease.
This is the first study using 16S typing to characterize an extensive collection of H. influenzae isolates demonstrating an unexpectedly high level of DNA sequence heterogeneity (up to 2.73%), and emphasizing that species identification using 16S rRNA gene sequence cutoffs of ∼1% - 2% are not universally applicable to all bacteria species. Although there is not a 1:1 correlation between 16S types and STs, both methods similarly clustered nearly all H. influenzae serotypable isolates, but not NT H. influenzae. Despite the diversity found by 16S typing among these isolates, the evidence of clonal structure and association of a particular 16S type with a specific serotype remain. Because 16S rRNA gene sequence alone provides a similar level of discrimination to that obtained with the analysis of 7 genes for MLST, 16S rRNA sequencing is potentially useful molecular typing method for characterizing H. influenzae isolates.
Acknowledgments
We acknowledge the contributions of the ABCs sites for collecting isolates used in this study, the technical contribution of Susanna Schmink and Jordan Hughes for maintaining the H. influenzae collection and serotyping the isolates used in this study; the submission of isolates from Pennsylvania by Lind Allen, Jean Bennetch, Richard Berman, Padget Burkey, Geoffrey Cantor, Nancy Caruso, Vicki Gordon, Stephanie Laubach, Perrianne Lurie, Susan Miller, Janey Schear, and Suzanne Yeager; and the submission of isolates from Alaska by microbiology laboratories in hospitals and other medical facilities in collaboration with Lynne Lucher, Thomas Hennessy, and Alan Parkinson of the Arctic Investigations Program, NCID, CDC.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children—United States, 1998-2000. Morb. Mortal. Wkly. Rep. 51:234-237. [PubMed] [Google Scholar]
- 3.Bakkali, M., T. Y. Chen, H. C. Lee, and R. J. Redfield. 2004. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc. Natl. Acad. Sci. USA 101:4513-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, Y., C. J. Douady, A. K. Sharma, M. Kamekura, and W. F. Doolittle. 2004. Intragenomic heterogeneity and intergenomic recombination among Haloarchaeal rRNA genes. J. Bacteriol. 186:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M. 2004. Molecular phylogenetics of restriction endonucleases, p. 63-93. In A. Pingoud (ed.), Restriction endonucleases, vol. 14. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 6.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eden, P. A., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 9.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 12.Fry, A. M., P. Lurie, M. Gidley, S. Schmink, J. Lingappa, M. Fischer, and N. E. Rosenstein. 2001. Haemophilus influenzae type b disease among Amish children in Pennsylvania: reasons for persistent disease. Pediatrics 108:E60. [DOI] [PubMed] [Google Scholar]
- 13.Galil, K., R. Singleton, O. S. Levine, M. A. Fitzgerald, L. Bulkow, M. Getty, B. A. Perkins, and A. Parkinson. 1999. Reemergence of invasive Haemophilus influenzae type b disease in a well-vaccinated population in remote Alaska. J. Infect. Dis. 179:101-106. [DOI] [PubMed] [Google Scholar]
- 14.Gee, J. E., C. T. Sacchi, M. B. Glass, B. K. De, R. S. Weyant, P. N. Levett, A. M. Whitney, A. R. Hoffmaster, and T. Popovic. 2003. Use of 16S rRNA gene sequencing for rapid identification and differentiation of Burkholderia pseudomallei and B. mallei. J. Clin. Microbiol. 41:4647-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gee, J. M., M. E. Kovach, V. K. Grippe, S. Hagius, J. V. Walker, P. H. Elzer, and R. M. Roop II. 2004. Role of catalase in the virulence of Brucella melitensis in pregnant goats. Vet. Microbiol. 102:111-115. [DOI] [PubMed] [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 17.Kolbert, C. P., P. N. Rys, M. Hopkins, D. T. Lynch, J. J. Germer, C. E. O'Sullivan, A. Trampuz, and R. Patel. 2004. 16S ribosomal DNA sequence analysis for identification of bacteria in a clinical microbiology laboratory, p. 361-377. In D. H. Persing, F. C. Tenover, J. Versalovic, Y.-W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology: diagnostic principals and practice. American Society for Microbiology, Washington, D.C.
- 18.Lucher, L. A., M. Reeves, T. Hennessy, O. S. Levine, T. Popovic, N. Rosenstein, and A. J. Parkinson. 2002. Reemergence, in southwestern Alaska, of invasive Haemophilus influenzae type b disease due to strains indistinguishable from those isolated from vaccinated children. J. Infect. Dis. 186:958-965. [DOI] [PubMed] [Google Scholar]
- 19.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchandin, H., C. Teyssier, M. Simeon De Buochberg, H. Jean-Pierre, C. Carriere, and E. Jumas-Bilak. 2003. Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiology 149:1493-1501. [DOI] [PubMed] [Google Scholar]
- 21.Mayer, L. W., M. W. Reeves, N. Al-Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schmink, C. A. Noble, M. L. Tondella, A. M. Whitney, Y. Al-Mazrou, M. Al-Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. E. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electrophoretic type-37 complex. J. Infect. Dis. 185:1596-1605. [DOI] [PubMed] [Google Scholar]
- 22.McVernon, J., C. L. Trotter, M. P. Slack, and M. E. Ramsay. 2004. Trends in Haemophilus influenzae type b infections in adults in England and Wales: surveillance study. BMJ 329:655-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser, J. M., D. M. Granoff, P. E. Pattison, and R. K. Selander. 1985. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 82:5078-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, and G. Hammond. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 27.Neefs, J. M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman, H. 2001. Lateral and oblique gene transfer. Curr. Opin. Genet. Dev. 11:616-619. [DOI] [PubMed] [Google Scholar]
- 29.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Peltola, H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porras, O., D. A. Caugant, B. Gray, T. Lagergard, B. R. Levin, and C. Svanborg-Eden. 1986. Difference in structure between type b and nontypable Haemophilus influenzae populations. Infect. Immun. 53:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sacchi, C. T., A. M. Whitney, L. W. Mayer, R. Morey, A. Steigerwalt, A. Boras, R. S. Weyant, and T. Popovic. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacchi, C. T., A. M. Whitney, M. W. Reeves, L. W. Mayer, and T. Popovic. 2002. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J. Clin. Microbiol. 40:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 35.Stackebrandt, E., and O. Charfreitag. 1990. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. 136:37-43. [DOI] [PubMed] [Google Scholar]
- 36.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Y., and Z. Zhang. 2000. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of nonrandom base variations in bacterial rRNA genes. Microbiology 146:2845-2854. [DOI] [PubMed] [Google Scholar]
- 39.Wenger, J. D. 1998. Epidemiology of Haemophilus influenzae type b disease and impact of Haemophilus influenzae type b conjugate vaccines in the United States and Canada. Pediatr. Infect. Dis. J. 17:S132-S136. [DOI] [PubMed] [Google Scholar]
- 40.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the Actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]