Abstract
Staphylococcus aureus is an important pathogen in domestic ruminants. The main objective of this study was to determine the similarity of epidemiologically unrelated S. aureus isolates from bovine, ovine, and caprine mastitis. By pulsed-field gel electrophoresis, 160 different pulsotypes (PTs) were identified among 905 isolates recovered from 588 herds in 12 counties in Norway. Based on estimates of similarity, using an 80% cluster cutoff, the isolates were assigned to 47 clusters. One cluster included 62% of all the isolates and more than 45% of the isolates from each host species. Twenty-three PTs included isolates from more than one host species; these 23 PTs represented 72% of all the isolates. The six most prevalent PTs included isolates from all host species and contained 45% of the bovine isolates, 54% of the ovine isolates, and 37% of the caprine isolates. Antimicrobial susceptibility testing of 373 of the isolates revealed resistance to penicillin in 2.9% and to streptomycin in 2.4%; only 1.9% were resistant to 1 of the other 11 antimicrobials tested. The results of this study suggest that a small number of closely related genotypes are responsible for a great proportion of S. aureus mastitis cases in cows, ewes, and goats in Norway and that these genotypes exhibit little or no host preference among these species. Selection due to antimicrobial resistance appears not to have contributed to the predominance of these genotypes.
Staphylococcus aureus is the cause of a variety of infections in both humans and animals. It is a major pathogen of the mammary gland and a common cause of bovine, ovine, and caprine mastitis (5, 27). In Norway, two studies have found S. aureus to be responsible for 46 and 68% of cases of clinical mastitis in dairy heifers and ewes kept for meat production, respectively (20, 37). A study including more than 2,000 cows randomly selected from the Norwegian dairy cow population revealed that approximately 22% of the animals shed coagulase-positive staphylococci from at least one udder quarter at any time (30).
Host species specificity has been described for a variety of pathogenic bacterial species (29). Several studies have shown that different S. aureus types preferentially colonize or infect a single host species (9, 17, 18, 21, 28). Epidemiologic studies have suggested that a few S. aureus types are responsible for the majority of bovine intramammary infections and that these types have a broad geographic distribution (7, 10, 17, 33). A study of sheep dairy farms showed that most cases of S. aureus mastitis were caused by a few closely related genotypes, which were widely distributed (34).
Defined clonal lineages of methicillin-resistant S. aureus are spread among patients in hospitals and in communities (2, 26, 39). In domestic ruminants, widespread use of antimicrobial drugs for the treatment and prevention of mastitis may favor the selection of resistant strains within ruminant S. aureus populations. In many countries, a large proportion of S. aureus isolates recovered from bovine mastitis produce β-lactamases (35, 38), and multiresistant strains do occur (36, 38). In Norway, however, the proportion of antibiotic-resistant S. aureus isolates from ruminant mastitis is low (25). Bacterial populations with a low prevalence of antimicrobial-resistant strains are well suited for studying factors, other than antimicrobial usage, that may influence the dissemination of particular bacterial types. Improved knowledge of the epidemiology of S. aureus associated with ruminant mastitis, including factors affecting the spread and persistence of prevalent strains, would form a better basis for implementing control measures targeted at reservoirs and transmission routes.
In this study, typing of a large number of S. aureus isolates recovered from cases of ruminant mastitis was performed using pulsed-field gel electrophoresis (PFGE). The aim was to compare the distribution of pulsotypes (PTs) causing bovine, ovine, and caprine mastitis and to examine whether PTs were host species specific. Antimicrobial susceptibility testing of isolates was performed in order to assess whether selection due to antimicrobial resistance could have affected the observed proportions of PTs.
MATERIALS AND METHODS
Bacterial isolates.
A total of 905 S. aureus isolates were obtained from milk collected from cases of bovine, ovine, and caprine clinical and subclinical mastitis (23) in Norway between 1998 and 2004. Of these, 333 isolates were from 333 dairy cows, 384 isolates were from 332 ewes kept for meat production, and 188 isolates were from 180 dairy goats. The cows belonged to 287 herds located in 11 of the 19 counties in Norway, the ewes to 243 flocks located in 11 counties, and the goats to 64 herds located in 10 counties. Five farms that kept both sheep and dairy cows provided six ovine and six bovine S. aureus isolates, respectively. One farm that kept both sheep and dairy goats supplied 10 ovine and 12 caprine isolates.
Sample collection.
Milk samples were collected aseptically in 10-ml sterile plastic vials by veterinarians and dairy hygiene advisers according to procedures recommended by the International Dairy Federation (14). The samples were either sent fresh to the National Veterinary Institute in Oslo immediately after sampling or frozen and stored at −20°C until submission to the laboratory.
Identification of S. aureus.
Milk samples were examined and bacteria identified as recommended by the International Dairy Federation (14). Ten microliters of milk was plated on blood agar (Oxoid, Basingstoke, United Kingdom) containing 5% washed bovine erythrocytes and incubated for 24 h at 37°C. Cultures were read at 24 and 48 h. If growth was not detected after incubation for 24 h, the original sample was preincubated for 4 h at 37°C and 50-μl aliquots were subsequently plated and incubated for 24 h.
All suspected staphylococcal colonies were tested using the tube coagulase test (Becton Dickinson, Sparks, Md.). Coagulase-positive staphylococci were streaked onto peptone agar (p-agar) (Difco, Sparks, Md.) supplemented with 7 mg/liter of acriflavin (Sigma-Aldrich Chemie, Steinheim, Germany) (8) and incubated at 37°C for 24 h. Bacterial growth in the full length of the streak on p-agar was considered to be a positive reaction and confirmation of S. aureus. Isolates identified as S. aureus were stored at −70°C in heart infusion broth (Difco) with 15% glycerol.
Antimicrobial susceptibility testing.
Randomly selected subsets of S. aureus isolates from each of the host species (i.e., 231 bovine, 82 ovine, and 60 caprine isolates) were tested for susceptibility to 13 antimicrobial compounds. The 373 isolates were recovered from 332 different herds. For the bovine isolates, MICs were determined by using Etest (AB Biodisk, Solna, Sweden) as recommended by the manufacturer and as previously reported (24). The ovine and caprine isolates were tested using a commercially available microdilution system (VetMIC; Department of Antibiotics, National Veterinary Institute, Uppsala, Sweden), which is based on standard methods described by the National Committee for Clinical Laboratory Standards (22). For sulfadiazine, Etest was used for all 373 isolates. S. aureus ATCC 29213 was used for quality control.
When available, breakpoint values were applied according to National Committee for Clinical Laboratory Standards guidelines (22). Wild-type breakpoint values were used for fusidic acid, streptomycin, and trimethoprim (16). β-Lactamase activity of all isolates of S. aureus was tested by the cloverleaf method using S. aureus ATCC 25923 as the indicator strain (4, 15).
DNA isolation and digestion and PFGE.
Preparation of chromosomal DNA and enzymatic digestion were performed as described by Bannerman et al. (3), with some modifications. The bacteria were grown in 5 ml brain heart infusion broth (Difco) in a shaking incubator at 37°C overnight, harvested by centrifugation, and washed. The cells were redissolved in 250 μl EC buffer (0.01 M Tris Cl, 1 M NaCl, 0.2 M EDTA, 0.2% deoxycholate, 0.5% Sarkosyl), and 2 μl of a 1-mg/ml stock solution of lysostaphin (Sigma-Aldrich Chemie) and 250 μl 2% low melting point agarose (Invitrogen Life Technologies, San Diego, Calif.) were added to the suspension. The mixture was transferred to plug molds and allowed to solidify. The plugs were placed in 2 ml EC buffer and lysed at 37°C for 1 h, followed by washing in 3 ml TE buffer (10 mM Tris HCl, 1 mM EDTA) at 55°C for 1 h. The plugs were then washed twice in TE buffer for 1 h with gentle agitation at room temperature and finally stored in fresh TE buffer at 4°C until further analysis.
Digestion of DNA was performed with 20 U of the restriction enzyme SmaI (New England Biolabs, Beverly, Mass.). Digested DNA was electrophoresed in 1% SeaKem Gold Agarose (BioWhittaker Molecular Applications, Rockland, Maine) using CHEF-DR III (Bio-Rad Laboratories, Hercules, Calif.). Low Range PFGE Marker (New England Biolabs) was used as size standards in the first and last lanes of each gel. One strain of S. aureus (1685-4) isolated from a cow with clinical mastitis was used as a reference strain and run on every gel. Electrophoresis was performed as described by Bannerman et al. (3), with some modifications. The gel was run in 0.5× Tris-borate-EDTA buffer at 14°C with an initial switch time of 5 s to a final switch time of 40 s and a run time of 19 h. After electrophoresis, gels were stained for 20 min in ethidium bromide (1 μg/ml), destained for about 10 min in distilled water, and photographed under UV light using the GeneGenius gel documentation and analysis system (Syngene, Cambridge, United Kingdom).
Differentiation of PFGE profiles.
The number and sizes of electrophoretically separated DNA fragments were assessed both using the BioNumerics software (version 3.0; Applied Maths, Kortrijk, Belgium) and visual inspection. The software was used to calculate Dice coefficients of similarity and to cluster the strains and generate dendrograms by the unweighted-pair group method using average linkages. DNA fragments between approximately 45 kb and 650 kb were included in the analysis. A PT was defined as a unique electrophoretic banding pattern defined by a combination of computer-aided and visual interpretations. Banding patterns differing by less than one bandwidth for any band position were assigned to the same PT. Banding patterns were compared according to criteria defined by Tenover et al. (32). PTs with one to three band differences were considered closely related.
When discrepancies between visual and computer-aided interpretations were detected, the isolates in question and a subset of isolates with indistinguishable banding patterns recovered from more than one host species were run electrophoretically side by side on new gels in order to ease visual interpretation. One representative isolate from each PT was selected to create a dendrogram with optimization and position tolerance settings of 1%. The cluster cutoff was set at 80%, and all clusters were given arabic identification numbers.
A data subset was generated by the fingerprints of all S. aureus isolates belonging to PTs that included isolates from more than one of the three host species. One fingerprint from each of these PTs was selected for presentation.
A similarity of 98.2% was obtained by comparison (n = 83) of the internal control strain (1685-4) when analyzed in BioNumerics using the same settings as used for the cluster analysis.
RESULTS
PFGE profiles.
The 905 ruminant S. aureus isolates were assigned to 160 different PTs. The 333 bovine isolates belonged to 71 PTs, the 384 ovine isolates to 75 PTs, and the 188 caprine isolates to 48 PTs.
Cluster analysis.
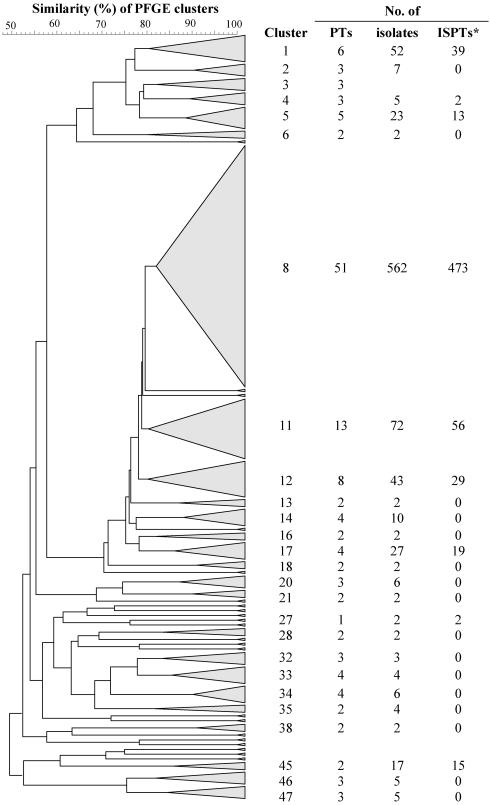
Forty-seven clusters were identified by computer analysis (Fig. 1). Cluster 8 was the dominant cluster and included 51 PTs and 62.1% of the isolates. Cluster 11 was the second largest cluster and included 13 PTs and 8.0% of the isolates. Twenty-two single-PT clusters were identified; 17 of these were represented by one isolate. The remaining five single-PT clusters contained two or three isolates.
FIG. 1.
Dendrogram generated by cluster analysis using the unweighted-pair group method using average linkages, based on the Dice coefficient. Band position tolerances and optimization were set to 1%. The 47 clusters were obtained from PFGE of SmaI-digested DNAs from 905 ruminant S. aureus mastitis isolates. The cluster cutoff was set at 80% similarity. For each cluster with more than one PT, the number of PTs, the number of isolates, and the number of isolates belonging to PTs found in more than one host species (interspecies PTs, ISPTs*) are presented next to the cluster triangle and cluster designation. The base of each cluster triangle is proportionate to the number of PTs within the cluster.
Distribution of PTs among host species.
Of the 905 isolates, 28.4% belonged to 137 PTs that contained isolates from only one host species, 8.5% to six PTs that contained isolates of bovine and ovine (BO) origin, 6.0% to six PTs that contained isolates of ovine and caprine (OC) origin, and 57.1% to 11 PTs that contained bovine, ovine, and caprine (BOC) isolates. Table 1 shows the corresponding distribution of isolates originating from each of the host species.
TABLE 1.
Distribution of S. aureus isolates recovered from bovine, ovine, or caprine mastitis according to the host spectrum of the PFGE PTs to which the isolates belong
| Host origin | No. of isolates | % of isolates in PTs grouped by host categorya |
||||||
|---|---|---|---|---|---|---|---|---|
| B | O | C | BO | BC | OC | BOC | ||
| Bovine | 333 | 26.4 | 14.1 | 0 | 59.5 | |||
| Ovine | 384 | 29.7 | 7.8 | 3.4 | 59.1 | |||
| Caprine | 188 | 29.3 | 0 | 21.8 | 48.9 | |||
Abbreviations: B, PTs with only bovine isolates; O, PTs with only ovine isolates; C, PTs with only caprine isolates; BO, PTs with bovine and ovine isolates; OC, PTs with ovine and caprine isolates; BOC, PTs with bovine, ovine, and caprine isolates.
Six hundred forty-eight isolates were assigned to the 23 PTs that included isolates from more than one host species. The distribution of these isolates by PT is shown in Table 2. Twelve of the 23 PTs, including 73.0% of the isolates assigned to these PTs, belonged to cluster 8, while 3 of the 23 PTs, including 8.6% of the isolates, belonged to cluster 11.
TABLE 2.
Distribution of 245 bovine, 270 ovine, and 133 caprine S. aureus mastitis isolates assigned to 23 PTs containing isolates from two or three domestic ruminant species
| PTa | Clusterb | No. (%) of isolates |
|||
|---|---|---|---|---|---|
| Bovine | Ovine | Caprine | All | ||
| BOC 1 | 8 | 34 (13.9) | 84 (31.1) | 10 (7.5) | 128 (19.8) |
| BOC 2 | 8 | 47 (19.2) | 58 (21.5) | 18 (13.5) | 123 (19.0) |
| BOC 3 | 8 | 33 (13.5) | 27 (10.0) | 6 (4.5) | 66 (10.2) |
| BOC 4 | 8 | 18 (7.3) | 16 (5.9) | 9 (6.8) | 43 (6.6) |
| BOC 5 | 1 | 2 (0.8) | 13 (4.8) | 24 (18.0) | 39 (6.0) |
| BOC 6 | 8 | 16 (6.5) | 10 (3.7) | 2 (1.5) | 28 (4.3) |
| OC 1 | 8 | 0 | 6 (2.2) | 20 (15.0) | 26 (4.0) |
| BOC 7 | 11 | 10 (4.1) | 7 (2.6) | 8 (6.0) | 25 (3.9) |
| BO 1 | 8 | 4 (1.6) | 21 (7.8) | 0 | 25 (3.9) |
| BOC 8 | 17 | 12 (4.9) | 5 (1.9) | 2 (1.5) | 19 (2.9) |
| BOC 9 | 11 | 4 (1.6) | 4 (1.5) | 9 (6.8) | 17 (2.6) |
| BOC 10 | 8 | 13 (5.3) | 2 (0.7) | 2 (1.5) | 17 (2.6) |
| BO 2 | 12 | 16 (6.5) | 1 (0.4) | 0 | 17 (2.6) |
| BO 3 | 45 | 14 (5.7) | 1 (0.4) | 0 | 15 (2.3) |
| BO 4 | 11 | 9 (3.7) | 5 (1.9) | 0 | 14 (2.2) |
| OC 2 | 5 | 0 | 1 (0.4) | 12 (9.0) | 13 (2.0) |
| BOC 11 | 12 | 9 (3.7) | 1 (0.4) | 2 (1.5) | 12 (1.9) |
| OC 3 | 8 | 0 | 3 (1.1) | 5 (3.8) | 8 (1.2) |
| BO 5 | 8 | 3 (1.2) | 1 (0.4) | 0 | 4 (0.6) |
| OC 4 | 8 | 0 | 1 (0.4) | 2 (1.5) | 3 (0.5) |
| OC 5 | 4 | 0 | 1 (0.4) | 1 (0.8) | 2 (0.3) |
| OC 6 | 8 | 0 | 1 (0.4) | 1 (0.8) | 2 (0.3) |
| BO 6 | 27 | 1 (0.4) | 1 (0.4) | 0 | 2 (0.3) |
PTs containing isolates from various combinations of host species. B. bovine; O, ovine; C, caprine.
The cluster to which each PT belonged is given (Fig. 1).
The six most frequently occurring PTs (BOC 1 through BOC 6) contained 47.2% of all isolates, including 45.0% of the bovine, 54.2% of the ovine, and 36.7% of the caprine isolates. Isolates belonging to each of these PTs were found in 8 to 11 of the 12 counties.
Of the PTs that included isolates from only one host species, 54 contained bovine, 52 ovine, and 31 caprine isolates. None of these PTs contained more than 13 isolates.
Of the PTs that contained more than four isolates, 2 of 9 single-species PTs and none of 18 PTs common to more than one species were found in a single county.
Band comparisons of PFGE profiles.
The most frequently occurring PT (BOC 1) included 128 (14.1%) isolates and had two bands that were different from the banding patterns of the second (BOC 2, 123 isolates), third (BOC 3, 66 isolates), and fourth (BOC 4, 43 isolates) most frequently occurring PTs (Table 2; Fig. 2). Seventeen of the PTs that included isolates from more than one host species had six bands or fewer that were different from BOC 1; these 18 PTs contained 89.0% of the 648 isolates assigned to the 23 PTs shared by isolates from more than one host species. PFGE profiles of isolates representing each of these 23 PTs are shown in Fig. 2.
FIG. 2.
PFGE of SmaI-digested DNAs of S. aureus isolates representing the 23 PTs that included isolates from more than one ruminant species; B, bovine; O, ovine; C, caprine. The band differences (BD*) between the most commonly occurring PT (BOC 1) and the 22 other PTs are indicated to the right of the PT designation. The banding pattern above BOC 1 represents lambda ladder size standards, with numbers indicating molecular sizes (kilobases).
In addition to BOC 1, cluster 8 contained 50 PTs; 35 of these PTs differed from BOC 1 by three bands or fewer, and the remaining 15 PTs differed from BOC 1 by four to six bands.
Antimicrobial susceptibility.
Results of the antimicrobial susceptibility testing are shown in Table 3. Three hundred fifty-one isolates (94.1%) were susceptible to all 13 antimicrobial agents, 17 isolates (4.6%) were resistant to 1 agent, and 5 isolates were resistant to 2 agents. The following resistance combinations were found in isolates resistant to two agents: penicillin and fusidic acid (one bovine isolate), tetracycline and trimethoprim (one bovine isolate), tetracycline and streptomycin (one ovine isolate), and penicillin and streptomycin (two caprine isolates). Eight of the 11 penicillin-resistant isolates belonged to PTs that each contained single isolates. The remaining 3 isolates were assigned to PTs that each contained fewer than 18 isolates, 2 of which were caprine isolates resistant to both penicillin and streptomycin. No penicillin-resistant isolates were found among the six most frequently occurring PTs. One of the nine streptomycin-resistant isolates belonged to a PT that contained a single isolate, and one isolate belonged to BOC 3. None of the remaining 7 isolates belonged to PTs containing more than 18 isolates.
TABLE 3.
Antimicrobials, breakpoint values for resistance, and frequency of resistance in 231 bovine, 82 ovine, and 60 caprine S. aureus mastitis isolates from Norway
| Antimicrobial | Breakpoint (μg/ml) | No. (%) of resistant isolates |
|||
|---|---|---|---|---|---|
| Bovine | Ovine | Caprine | All | ||
| Cephalothin | ≥32 | 0 | 0 | 0 | 0 |
| Chloramphenicol | ≥32 | 0 | 0 | 0 | 0 |
| Clindamycin | ≥4 | 0 | 0 | 0 | 0 |
| Enrofloxacin | ≥4 | 0 | 0 | 0 | 0 |
| Erythromycin | ≥8 | 0 | 0 | 0 | 0 |
| Fusidic acid | ≥1 | 2 (0.9) | 0 | 0 | 2 (0.5) |
| Gentamicin | ≥16 | 0 | 0 | 0 | 0 |
| Oxacillin | ≥4 | 0 | 0 | 0 | 0 |
| Penicillina | ≥0.25 | 6 (2.6) | 2 (2.4) | 3 (5.0) | 11 (2.9) |
| Streptomycin | ≥32 | 5 (2.2) | 1 (1.2) | 3 (5.0) | 9 (2.4) |
| Sulfadiazin | ≥512 | 1 (0.4) | 0 | 0 | 1 (0.3) |
| Oxytetracycline | ≥16 | 1 (0.4) | 1 (1.2) | 0 | 2 (0.5) |
| Trimethoprim | ≥8 | 1 (0.4) | 1 (1.2) | 0 | 2 (0.5) |
β-Lactamase production of all isolates was determined. All β-lactamase-positive isolates had a MIC of >0.125 μg/ml, and all β-lactamase-negative isolates had a MIC of ≤0.125 μg/ml.
DISCUSSION
S. aureus mastitis in domestic ruminants in Norway appear to a great extent to be caused by PTs shared by at least two host species. Of the 905 isolates, more than 70% belonged to PTs that included isolates from more than one species. Also, within each subset of bovine, ovine, and caprine isolates, respectively, between 70 and 74% of the isolates were assigned to PTs shared by one or two of the other host species.
Considerable genetic diversity was observed among the isolates. A total of 160 different PTs were identified, and with an 80% similarity cutoff they were assigned to 47 clusters. PTs including isolates from more than one ruminant species were found in all seven clusters that contained more than 10 isolates. However, a great proportion (40%) of the 905 isolates belonged to four PTs, BOC 1 through BOC 4, each found in all three ruminant species. The most common of these, BOC 1, was considered closely related to each of the others since it differed by only two bands from BOC 2 through BOC 4 (32).
When aiming to describe and compare strain distribution of bacterial populations in different host species, studies should be based on isolates that reflect the actual bacterial populations. The S. aureus isolates included in this study were obtained from cases of ruminant mastitis on 588 different farms in 12 of the 19 counties of Norway during a 7-year period. Except for six farms from which isolates were recovered from two host species, there was no known animal-to-animal contact between the cases of the different ruminant species. Sixty pairs of isolates from ewes and goats that were infected with S. aureus in both mammary glands and a limited number of isolates from different animals in the same herd or flock were included. However, the great majority of isolates within each of the host species were considered epidemiologically unrelated. Isolates from healthy carriers may not be representative when the purpose is to compare the occurrence of disease-causing organisms in different host species. In this study, all isolates were from infected mammary glands. Thus, it is not unreasonable to assume that the isolates included in this study were representative of S. aureus populations that cause ruminant mastitis in Norway. The frequent occurrence of a few closely related PTs in all ruminant species suggests that the S. aureus strains most commonly associated with mastitis in domestic ruminants exhibit little or no host specificity.
The predominance of a small number of genotypes among S. aureus isolates associated with bovine mastitis has also been reported in other countries (7, 10, 17, 33). The reason for the clustering of these genotypes is unclear, but it might be due to a tropism for mammary gland tissues of domestic ruminants. A DNA microarray analysis of S. aureus strains selected from a large collection of isolates found that the most prevalent bovine and ovine strains causing mastitis were closely related (11). The authors concluded that a similar gene complement may be required to cause bovine and ovine mastitis. Evolutionary anatomic-site adaptation and development of S. aureus isolates with a particular preference for certain organs have been proposed on the basis of associations between S. aureus PTs and the sites of isolation (mammary secretions and healthy bovine skin) (40). The present observation, that certain frequently occurring PTs caused mastitis in different ruminant species, may indicate the existence of an anatomic-site specificity independent of the host species in such strains. The possible existence of organ specificity is supported by a study of S. aureus isolates from different human diseases (6). The authors of that study found that PFGE profiles of S. aureus from respiratory tracts differed significantly from isolates from other infection sites.
Although domestic ruminants are generally not housed or grazed together today, about 10% of the sheep flocks and dairy cow and dairy goat herds are located on farms with more than one ruminant species (31). In times past, trade of ruminants and the keeping of two or three ruminant species on the same farm were common in many parts of the country (12, 19). Thus, cows, sheep, and goats may have shared an S. aureus-contaminated environment through a period of several hundred years. This exposure may have induced an adaptation to ruminant udders among a small number of S. aureus genotypes.
Interestingly, the distribution of isolates by PT as observed in dairy cows and dairy goats was quite similar to that in ewes, the latter being kept for meat production. In dairy ruminants, spread during milking is considered to be the most important mode of transfer of contagious mastitis pathogens, thus allowing certain S. aureus strains to become prevalent within herds. However, the present sampling strategy limited the number of isolates obtained from single herds and prevented the potential overrepresentation of certain PTs as a result of strain clustering within herds. In meat-producing sheep, the epidemiology of S. aureus mammary pathogens is largely unknown.
For several decades, penicillin has been the antimicrobial compound most commonly used for the treatment of staphylococcal infections in ruminants in Norway (13, 25). Until recently, sulfonamides administered orally have also been used frequently to treat mastitis and other infections in sheep. Less than 6% of the isolates tested were resistant to any of the 13 antimicrobials, which is considerably lower than that reported for ruminant S. aureus in other countries (1, 35, 38). Only 11 (2.9%) of the isolates were resistant to penicillin, all of which produced β-lactamase. Eight of these isolates belonged to single-isolate PTs, and the remaining three belonged to PTs with a low prevalence that also contained penicillin-sensitive isolates. One of the nine streptomycin-resistant isolates was assigned to a PT that also contained 65 streptomycin-sensitive isolates. The remaining streptomycin-resistant isolates and those resistant to sulfonamides (0.3%) or any of the other antimicrobials tested (1.6%) were assigned to PTs with a low prevalence. Thus, antimicrobial resistance does not appear to have been a factor determining the strain distribution observed in this study.
The present results suggest that a limited number of closely related PTs are responsible for a great proportion of the cases of S. aureus mastitis in domestic ruminants in Norway. The most prevalent S. aureus PTs were almost equally represented among mastitis isolates from cows, ewes, and goats. For reasons not fully understood, these genotypes are successful mammary pathogens in ruminants without discriminating among the three domestic ruminant species.
Acknowledgments
We thank Hanne Tharaldsen and Tone Berdal from the National Veterinary Institute for help with the antimicrobial resistance analyses. We also thank the veterinarians and the dairy hygiene advisers who collected the milk samples.
REFERENCES
- 1.Aarestrup, F. M., and N. E. Jensen. 1998. Development of penicillin resistance among Staphylococcus aureus isolated from bovine mastitis in Denmark and other countries. Microb. Drug Resist. 4:247-256. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergan, T., J. N. Bruun, A. Digranes, E. Lingaas, K. K. Melby, and J. Sander. 1997. Susceptibility testing of bacteria and fungi—report from “the Norwegian Working Group on Antibiotics”. Scand. J. Infect. Dis. Suppl. 103:1-36. [PubMed] [Google Scholar]
- 5.Bergonier, D., R. de Cremoux, R. Rupp, G. Lagriffoul, and X. Berthelot. 2003. Mastitis of dairy small ruminants. Vet. Res. 34:689-716. [DOI] [PubMed] [Google Scholar]
- 6.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzzola, F. R., L. Quelle, M. I. Gomez, M. Catalano, L. Steele-Moore, D. Berg, E. Gentilini, G. Denamiel, and D. O. Sordelli. 2001. Genotypic analysis of Staphylococcus aureus from milk of dairy cows with mastitis in Argentina. Epidemiol. Infect. 126:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capurro, A., C. Concha, L. Nilsson, and K. Östensson. 1999. Identification of coagulase-positive staphylococci isolated from bovine milk. Acta Vet. Scand. 40:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devriese, L. A. 1984. A simplified system for biotyping Staphylococcus aureus strains isolated from animal species. J. Appl. Bacteriol. 56:215-220. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., D. E. Sturdevant, S. M. Macki, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insight into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gjeråker, B. 2002. The agricultural history of Norway: III. 1814-1920. Det Norske Samlaget, Oslo, Norway. (In Norwegian.)
- 13.Grave, K., C. Greko, L. Nilsson, K. Odensvik, T. Mørk, and M. Rønning. 1999. The usage of veterinary antibacterial drugs for mastitis in cattle in Norway and Sweden during 1990-1997. Prev. Vet. Med. 42:45-55. [DOI] [PubMed] [Google Scholar]
- 14.International Dairy Federation. 1981. Laboratory methods for use in mastitis work. Document 132. International Dairy Federation, Brussels, Belgium.
- 15.Jarløv, J. O., and V. T. Rosdahl. 1986. Quantitative determination of beta-lactamase production in Staphylococcus aureus strains compared to qualitative testing by a microbiological clover leaf test, a chromogenic cephalosporin test and a iodometric test. Acta Pathol. Microbiol. Immunol. Scand. Sect. B 94:415-421. [DOI] [PubMed] [Google Scholar]
- 16.Kahlmeter, G., D. F. Brown, F. W. Goldstein, A. P. MacGowan, J. W. Mouton, A. Österlund, A. Rodloff, M. Steinbakk, P. Urbaskova, and A. Vatopoulos. 2003. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J. Antimicrob. Chemother. 52:145-148. [DOI] [PubMed] [Google Scholar]
- 17.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, H. D., K. H. Sloth, C. Elsberg, C. Enevoldsen, L. H. Pedersen, N. H. Eriksen, F. M. Aarestrup, and N. E. Jensen. 2000. The dynamics of Staphylococcus aureus intramammary infection in nine Danish dairy herds. Vet. Microbiol. 71:89-101. [DOI] [PubMed] [Google Scholar]
- 19.Lunden, K. 2002. The agricultural history of Norway: II. 1350-1814. Det Norske Samlaget, Oslo, Norway. (In Norwegian.)
- 20.Mørk, T., S. Waage, T. Tollersrud, G. Mosdøl, and S. Sviland. 2004. Bacteria causing clinical mastitis in ewes in Norway. Nor. Vet. Tidsskr. 116:819-826. (In Norwegian.) [Google Scholar]
- 21.Musser, J. M., and R. K. Selander. 1990. Genetic analysis of natural populations of Staphylococcus aureus, p. 59-67. In R. P. Novick and R. A. Skurray (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 22.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.National Mastitis Council. 1996. Mastitis can take on several forms. Udder Topics 19:1. [Google Scholar]
- 24.Ngui-Yen, J. H., E. A. Bryce, C. Porter, and J. A. Smith. 1992. Evaluation of the E test by using selected gram-positive bacteria. J. Clin. Microbiol. 30:2150-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norwegian Zoonosis Centre. 2003. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Zoonosis Centre, National Veterinary Institute, Oslo, Norway.
- 26.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radostitis, O. M., C. C. Gay, D. C. Blood, and K. W. Hinchcliff (ed.). 2000. Veterinary medicine, 9th ed., p. 603-700. W. B. Saunders Company Ltd., London, United Kingdom.
- 28.Schlegelova, J., M. Dendis, J. Benedik, V. Babak, and D. Rysanek. 2003. Staphylococcus aureus isolates from dairy cows and humans on a farm differ in coagulase genotype. Vet. Microbiol. 92:327-334. [DOI] [PubMed] [Google Scholar]
- 29.Selander, R. K., and J. M. Musser. 1990. Population genetics of bacterial pathogenesis, p. 11-36. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis. Academic Press Inc., San Diego, Calif.
- 30.Sølverød, L., and O. Østerås. 2001. The Norwegian survey of subclinical mastitis during 2000, p. 126-130. In Proceedings of the 2nd International Symposium on mastitis and milk quality. National Mastitis Council, Madison, Wis.
- 31.Statens Landbruksforvaltning. 2003. Register of production subsidies as of 31 July, 2003. [Online.] http://www.slf.dep.no.
- 32.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tollersrud, T., K. Kenny, D. A. Caugant, and A. Lund. 2000. Characterisation of isolates of Staphylococcus aureus from acute, chronic and subclinical mastitis in cows in Norway. APMIS 108:565-572. [DOI] [PubMed] [Google Scholar]
- 34.Vautor, E., G. Abadie, J. M. Guibert, C. Huard, and M. Pepin. 2003. Genotyping of Staphylococcus aureus isolated from various sites on farms with dairy sheep using pulsed-field gel electrophoresis. Vet. Microbiol. 96:69-79. [DOI] [PubMed] [Google Scholar]
- 35.Vintov, J., F. M. Aarestrup, C. E. Zinn, and J. E. Olsen. 2003. Association between phage types and antimicrobial resistance among bovine Staphylococcus aureus from 10 countries. Vet. Microbiol. 95:133-147. [DOI] [PubMed] [Google Scholar]
- 36.Waage, S., J. Bjorland, D. A. Caugant, H. Oppegaard, T. Tollersrud, T. Mørk, and F. M. Aarestrup. 2002. Spread of Staphylococcus aureus resistant to penicillin and tetracycline within and between dairy herds. Epidemiol. Infect. 129:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waage, S., T. Mørk, A. Røros, D. Aasland, A. Hunshamar, and S. A. Ødegaard. 1999. Bacteria associated with clinical mastitis in dairy heifers. J. Dairy Sci. 82:712-719. [DOI] [PubMed] [Google Scholar]
- 38.Werckenthin, C., M. Cardoso, J. L. Martel, and S. Schwarz. 2001. Antimicrobial resistance in staphylococci from animals with particular reference to bovine Staphylococcus aureus, porcine Staphylococcus hyicus, and canine Staphylococcus intermedius. Vet. Res. 32:341-362. [DOI] [PubMed] [Google Scholar]
- 39.Witte, W. 2004. International dissemination of antibiotic resistant strains of bacterial pathogens. Infect. Genet. Evol. 4:187-191. [DOI] [PubMed] [Google Scholar]
- 40.Zadoks, R. N., W. B. van Leeuwen, D. Kreft, L. K. Fox, H. W. Barkema, Y. H. Schukken, and A. van Belkum. 2002. Comparison of Staphylococcus aureus isolates from bovine and human skin, milking equipment, and bovine milk by phage typing, pulsed-field gel electrophoresis, and binary typing. J. Clin. Microbiol. 40:3894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]