Abstract
Verocytotoxin-producing Escherichia coli causes zoonotic food- or waterborne infection that may be associated with massive outbreaks and with the serious complication of hemolytic uremic syndrome (HUS). Serotypes O157:H7 and O157:NM are more commonly associated with HUS and outbreaks than other serotypes, such as O26:H11. To determine whether a genetic basis exists for why serotype O157:H7/NM causes HUS and outbreaks more often than other serotypes, such as O26:H11, we conducted suppression subtractive hybridization (SSH) between the genomes of the sequenced O157:H7 strain EDL933 and CL1, a clinical serotype O26:H11 isolate. Genes from four EDL933 fimbria-encoding genomic O islands (OIs) (OI-1, -47, -141, and -154) were identified in the SSH library. OI-47 encodes several additional putative virulence factors, including secreted and signaling proteins, a hemolysin locus, a lipoprotein, an ABC transport system, and a lipid biosynthesis locus. The distribution of the OIs was investigated by PCR and Southern hybridization (when PCR was negative) with 69 VTEC strains belonging to 39 different serotypes corresponding to 5 seropathotypes that differ in their disease and epidemic potential. The four OIs described here were distributed almost exclusively in serotypes O157:H7 and O157:NM, which indicates that they may be associated with the ability of these strains to colonize human and/or animal intestinal tracts and to cause epidemic and serious disease more frequently than other serotypes. The occurrence of the four OIs in enteropathogenic E. coli O55:H7 strains is consistent with their vertical inheritance by VTEC O157:H7/NM from this clonally related ancestor.
Verocytotoxin-producing Escherichia coli (VTEC) (31), also referred to as Shiga toxin-producing E. coli (8), is a major cause of zoonotic food- or waterborne infection that may be associated with massive outbreaks and with the serious complication of hemolytic uremic syndrome (HUS) (15, 27, 29, 30, 39). More than 200 serotypes of VTEC have been isolated from humans (65), but their incidence and association with serious or epidemic disease are highly variable, suggesting differences in epidemicity and virulence between VTEC serotypes (28). To determine why some serotypes are more common and cause serious and epidemic disease more frequently than other serotypes, we have classified VTEC serotypes in the following five “seropathotypes” based on their relative incidence and their association with epidemic and/or serious disease, such as HUS (28). Serotypes O157:H7 and O157:NM, which are associated most commonly with outbreaks and HUS (3, 15, 16, 22, 26, 39), are classified as seropathotype A. Seropathotype B comprises serotypes O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM, which are associated with outbreaks and HUS but much less frequently than serotypes O157:H7 and O157:NM (6, 16, 22, 34, 65). Serotypes such as O91:H21 and O113:H21, which have an occasional association with sporadic HUS but not with outbreaks (6, 22, 39), are classified as seropathotype C. Seropathotype D consists of a large number of serotypes which have been isolated from patients with diarrhea but have not been associated with either outbreaks or HUS (6, 22, 39, 65). Seropathotype E comprises multiple VTEC serotypes from animals that have not been associated with human disease (64, 65).
Growing evidence suggests that major differences in virulence between groups of strains within bacterial species, such as E. coli, Salmonella enterica serovar enteritidis, and Helicobacter pylori, are related to the presence of specific genomic islands that encode virulence. Such structures may also be referred to as pathogenicity islands (PAIs) (18, 25). To determine whether the presence of specific PAIs might explain the apparent difference in virulence between seropathotypes, we investigated the distribution of the locus of enterocyte effacement (LEE) and O island 122 (OI-122) among 39 VTEC serotypes classified according to the 5 seropathotypes indicated above (28). LEE, a well-known PAI in enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli, is responsible for the characteristic attaching and effacing lesion on enterocytes (36, 39). OI-122, on the other hand, is one of several novel putative PAIs that were identified in the sequenced genome of E. coli serotype O157:H7 strain EDL933 (44) and whose possible pathogenic significance remains to be elucidated. Our findings (28) showed that both LEE and OI-122 were highly correlated with seropathotypes A and B, which are associated with both HUS and epidemic disease, in contrast to seropathotypes C, D, and E, which are not. However, neither LEE nor OI-122 could adequately distinguish between seropathotypes A and B. To determine if seropathotype A strains contain unique PAIs that are absent in seropathotype B and that might account for the higher frequency with which seropathotype A strains are associated with outbreaks and HUS, we conducted genome subtractive hybridization between EDL933, the reference seropathotype A strain (serotype O157:H7), and CL1, a serotype O26:H11 (seropathotype B) strain. This led to the identification of four fimbria-encoding genomic islands, OI-1, -47, -141, and -154 (44), that, as reported here, have a high specificity for seropathotype A strains and thus could explain the apparently greater epidemicity and HUS frequency associated with seropathotype A strains than with strains of seropathotype B and other seropathotypes, the vast majority of which lack the four genomic islands.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Subtractive hybridization was performed between O157:H7 strain EDL933 and O26:H11 strain CL1 (28). E. coli K-12 strain MG1655 (ATCC 700926) (7) was used as a negative control. The distribution of EDL933 OI-1, OI-47, OI-141, and OI-154 was investigated in 69 additional VTEC strains of 39 different serotypes corresponding to the five seropathotypes A to E (28). The presence of these OIs was also investigated with two strains of EPEC serotype O55:H7 strains DEC5B and DEC5D, which were kindly provided by T. Whittam, University of Michigan. EPEC O55:H7 strains have a close clonal relationship to VTEC O157:H7 strains, which are thought to originate ancestrally from EPEC O55:H7 (61-63). Strains were grown routinely at 37°C overnight in Luria-Bertani broth (1% tryptone, 0.5% yeast extract, 171 mM NaCl) (2).
DNA techniques.
Standard techniques were used for DNA extraction, purification, analysis, and PCR (2, 47). Genomic DNA was isolated using the QIAGEN Genomic-tip 100/G (QIAGEN GmbH, Hilden, Germany).
Subtractive hybridization.
The Clontech PCR-Select bacterial genome subtraction kit (Clontech Laboratories, Inc., Palo Alto, CA) was used to identify DNA sequences present in the genome of one strain but absent in the genome of another (52). Genomic DNA of strain EDL933 was used as the tester and subtracted by genomic DNA of CL1 (driver). Subtractive hybridization was performed according to the manufacturer's instructions. Briefly, the tester and driver DNA samples were digested with a four-base cutting restriction enzyme (RsaI) that yields blunt ends. The tester fragments were divided into two portions, each of which was ligated with a different adaptor that facilitates suppression PCR. The driver fragments and the adaptor-ligated tester fragments then underwent a two-step round of hybridization, followed by PCR to enrich tester-specific DNA fragments. The latter were then cloned into a pGEM T-Easy TA cloning vector (Promega, Madison, WI) and sequenced. BLASTN searches (1) were performed against the EDL933 genome on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST) to identify the genes from the subtractive library.
Investigation of strains for the presence of the OIs.
When putative EDL 933 virulence genes were identified in the subtraction library, their absence in CL1 was confirmed by PCR and Southern hybridization. The presence of OIs encoding these genes in the 69 VTEC strains of 39 different serotypes and 5 seropathotypes, as described above, was then investigated by screening for representative open reading frames (ORFs) by PCR as described previously (28). All strains that were negative for any of the OI genes by PCR were retested by Southern hybridization using digoxigenin-labeled probes following the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany), as reported previously (28).
Sequence homology and conserved-domain search.
The homology between two related genes or proteins was calculated by the CLUSTAL V alignment method (DNASTAR, Madison, WI). A search of public databases was conducted for the homology of nucleotide sequences of the genes in the putative PAIs and of amino acid sequences of their products with other genes and proteins using the BLASTN, BLASTP, and BLASTX programs. Structural similarities of proteins were studied by searching the Conserved Domain Database using the RPSBLAST program (1) on the NCBI website (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), as well as by searching the protein signature database InterPro (37) using the InterProScan program (66) on the European Bioinformatics Institute website (http://www.ebi.ac.uk/InterProScan).
G+C content analysis.
The G+C content and dinucleotide bias of each gene were analyzed using the IslandPath program (20) (http://www.pathogenomics.sfu.ca/islandpath).
Prediction of signal peptides, transmembrane helices, and lipoproteins.
The presence and location of signal peptide cleavage sites in amino acid sequences were predicted by SignalP V3.0 based on hidden Markov models (41) on the website http://www.cbs.dtu.dk/services/SignalP. Probable transmembrane helices in proteins were predicted by TMHMM2.0 (53) on the website http://www.cbs.dtu.dk/services/TMHMM. Prediction of lipoproteins was done using LipoP 1.0 (24) (http://www.cbs.dtu.dk/services/LipoP).
RESULTS
Identification of EDL933-specific genes in the subtractive library.
Preliminary analysis of sequenced clones from the subtractive library revealed the presence of 10 putative virulence-encoding ORFs (Z0021, Z1534, Z1536, Z1537, Z1538, Z1550, Z1556, Z4967, Z4968, and Z5220) that were present in the EDL933 (serotype O157:H7) genome but absent from CL1 (serotype O26:H11). These ORFs belong to four different fimbria-encoding genomic islands, OI-1, -47, -141, and -154. To confirm that these ORFs were truly positive for EDL933 and negative for CL1, specific PCR primers were designed (Table 1) Negative PCR results were confirmed by Southern hybridization. The results (Table 2) showed that all four OIs are distributed almost exclusively in seropathotype A strains. However, evidence for the presence of OI-1 and OI-141 is seen in all three serotype O145:NM strains and a strain of O113:NM (Table 2). The presence of OI-1 is also evident in a strain of O132:NM (Table 2). Strains that were negative for a particular gene by PCR were also usually negative by Southern blotting. However, in several cases in which PCR results were negative, Southern hybridization results were positive (Table 2). The significance of this is currently not known.
TABLE 1.
PCR primers used to amplify genes in O islands 1, 47, 141, and 154
| Island | Gene | Primer name | Primer sequence (5′-3′) | Island | Gene | Primer name | Primer sequence (5′-3′) | |
|---|---|---|---|---|---|---|---|---|
| OI-1 | Z0020-21 | Z0021-F1 | CGACCAACGCCATTAAAGTTCA | |||||
| Z0020-R1 | GCCCTGGATGCAGTCAAATC | |||||||
| Z0022 | Z0022-F1 | GGGCGAAGTGGAAATGCTAAAG | ||||||
| Z0022-R1 | CACCGGGCCAGACAACTAATC | |||||||
| Z0023 | Z0023-F1 | CTGTTCATGCCGGTGTGGTC | ||||||
| Z0023-R1 | GCGAAAAGGGAGGCAGTAAGATA | |||||||
| Z0024 | Z0024-F1 | GTCGATCAAGGAACACTAACCATT | ||||||
| Z0024-R1 | TTTTTAACCCTGGTCTCATTCTGG | |||||||
| Z0025 | Z0025-F1 | CTCGCCCAGTCGGTTAATAGAG | ||||||
| Z0025-R1 | CGGCCAGGCATAATATCTTCAC | |||||||
| OI-47 | Z1528 | Z1528-F1 | CTGCTATCACCCGCTTTTACTGG | |||||
| Z1528-R1 | TAGGGGGTCAATGCTCCGTTAG | |||||||
| Z1530 | Z1530-F1 | GAGCCATATTGCGTTTCATCAT | ||||||
| Z1530-R1 | GCGTGGTGTCCTGCCTGTAG | |||||||
| Z1531 | Z1531-F1 | AGCGATAGCAGTGCCGATGTC | ||||||
| Z1531-R1 | TTTTTATAGTGTGGGATTTGCTTTGA | |||||||
| Z1533 | Z1533-F1 | GCAGGGGCTAATCTATTGTTGGTTG | ||||||
| Z1533-R1 | GTTGAGCCGGTATACGCCCTTTTA | |||||||
| Z1534 | Z1534-F2 | CCGCCAGAAACTCCCCATCAA | ||||||
| Z1534-R1 | TCGCCGCAACCACACCACTAA | |||||||
| Z1535 | Z1535-F1 | TAATGTCCGCGTCGAGTGTAGTAAC | ||||||
| Z1535-R1 | TTCAACCGGGCGATAAAAGAGTAA | |||||||
| Z1536 | Z1536-F1 | GCCCGCTGCCCGCCATAAATA | ||||||
| Z1536-R1 | ATCCCGTGCTGCCAGGTAAGGTCA | |||||||
| Z1537 | Z1537-F3 | CCATCCAGTTGCGAAATACCAG | ||||||
| Z1537-R3 | GAATTCGCGAGGGGGACTTTTT | |||||||
| Z1538 | Z1538-F1 | CAGCTATTGCATTACCTTTTTCA | ||||||
| Z1538-R1 | CTTCGGTATCGCCAACTTTC | |||||||
| Z1539 | Z1539-F1 | CTTCTTGTCGCAGTGGATGA | ||||||
| Z1539-R1 | GTTTTTAAAACCGGTAAATCTCC | |||||||
| Z1540-41 | Z1540-F4 | TTGCCGAAAGATGATAAACAGAGT | ||||||
| Z1541-R4 | ATTACGGGCAGGCGACATTT | |||||||
| Z1542 | Z1542-F2 | GTGGAATGGTCGGCAAGGAAG | ||||||
| Z1542-R2 | GTCGCCGCCAGCAGAAATA | |||||||
| Z1543 | Z1543-F1 | CACACCGGGGGAAAAACTGG | ||||||
| Z1543-R1 | ATATCCCGCGCCTGTAATGTCA | |||||||
| Z1544 | Z1544-F2 | GATTATGAACGGGCCTCTGGAT | ||||||
| Z1544-R2 | CTGCCCGATAAAAATTTCTTGTAG | |||||||
| Z1545 | Z1545-F1 | GCGGGCATCGTTAAAGACAGC | ||||||
| Z1545-R1 | AGACGGCGTAACGGGATGGTA | |||||||
| Z1546 | Z1546-F1 | CCTCGAAATGGGCGGCTG | ||||||
| Z1546-R1 | CGGTGAATGCGTGCGTGAA | |||||||
| Z1547 | Z1547-F2 | ACCATGAATGACGCTATTGAACAG | ||||||
| Z1547-R2 | CGGGTCGTCACTTTCGCTGAT | |||||||
| Z1548 | Z1548-F1 | GGCGGAGCAGTGGGAAAAAC | ||||||
| Z1548-R1 | TGACGCGTCGATATAACTGTGCTC | |||||||
| Z1549 | Z1549-F1 | GCAATAGGCCACACGCTCAAC | ||||||
| Z1549-R1 | GCCGCCAACCAATACCTGC | |||||||
| Z1550 | Z1550-F1 | GATGCCGTATGCCGAAGACTG | ||||||
| Z1550-R1 | CAATCCCGCCCTGCTCAATG | |||||||
| Z1551 | Z1551-F2 | ACAACAAATGCCGACTAAAGAACG | ||||||
| Z1551-R2 | CACGCTGGCTGAGAATAAAACAAC | |||||||
| Z1552 | Z1552-F1 | TGGGGCAGGAAAGGTTGATG | ||||||
| Z1552-R1 | GCCCGATAGTTGCCCAGGTTTA | |||||||
| Z1553 | Z1553-F1 | TGACCGAGGCGTTAAAGAAAGAGT | ||||||
| Z1553-R1 | GGTCGCGGATCGTTGTCGT | |||||||
| Z1554 | Z1554-F1 | CTGGGGGTCGAGCCGTTAC | ||||||
| Z1554-R1 | GACTGCGCCAATCACACCAA | |||||||
| Z1555 | Z1555-F2 | CGCAACCCAGCCAGAGAATAAAC | ||||||
| Z1555-R2 | CGGCTGGAAATCCTGATACAACAC | |||||||
| Z1556 | Z1556-F1 | GTCACAACGGTTATCAGGAGGCG | ||||||
| Z1556-R1 | GTATGACACTCGCCGAAAACGC | |||||||
| OI-141 | Z4965 | Z4965-R1 | AGTGGCCTGAGTGTTGACGAC | |||||
| Z4965-F1 | GTTGCCTGCGGGGTGAATG | |||||||
| Z4966 | Z4966-R1 | GTGACACGCCCCTGAATAGATAC | ||||||
| Z4966-F1 | CGCGGATGGCACGACTTA | |||||||
| Z4968m | Z4968m-F1 | CAAAAGCAACGCCGAAGTCAG | ||||||
| Z4968m-R1 | GGCTATCGCGATCGTTATTCAG | |||||||
| Z4969 | Z4969-F1 | GTAAATTTGTTGCGTTGGCTCTC | ||||||
| Z4969-R1 | ATTTGGCGCGGTTTTTACATC | |||||||
| Z4971 | Z4971-F1 | TACAGGCGAGATCGTGGATTCAC | ||||||
| Z4971-R1 | GACCTGCGCATTGCCGTAAC | |||||||
| OI-154 | Z5220 | Z5220-F1 | CCTGCTCCCGGTATGGTTTAT | |||||
| Z5220-R1 | CTTGCCCGGCGGTAGATG | |||||||
| Z5221 | Z5221-F1 | TATGGTTTCGCCGCTAATGGTG | ||||||
| Z5221-R1 | GGCGACTTCCGTTGTGGGTAT | |||||||
| Z5222 | Z5222-F1 | GGGGCTACCTGGTTACTGGAC | ||||||
| Z5222-R1 | ATCGCCGCCGCTACTGGT | |||||||
| Z5223-24 | Z5224-F | CGCAGATGTCGATGGCTACG | ||||||
| Z5223-R1 | CGTCTTTATCATCCACCGTCACA | |||||||
| Z5225 | Z5225-F1 | CCTTTAGTGGCGTCGATGATGT | ||||||
| Z5225-R1 | CTGCGAGTCGGCGTTAGC |
TABLE 2.
Distribution of OI-1, -47, -141, and -154 in different seropathotypes and serotypes of VTEC
| Pathotype | Serotype | Strain | Hosta | Sourceb | OI-1, Z0024 | OI-47 |
OI-141 |
OI-154 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z1528 | Z1534 | Z1538 | Z1542 | Z1548 | Z1552 | Z1556 | Z4965 | Z4966 | Z4971 | Z5220 | Z5221 | Z5225 | ||||||
| Ctrl | O157:H7 | EDL933 | H | J. Kaper | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ctrl | O157:H7 | Sakai | H | T. Honda | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ctrl | (K12) | MG1655 | ATCC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| A | O157:H7 | 93111 | H | T. Whittam | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| OK1 | H | T. Whittam | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 278F1 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 279F1 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 237F1 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 235F1 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| D103F5 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 254 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| E48F9 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| 157F1 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| O157:NM | 158F2 | H | LFZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| E32511 | H | H. Smith | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| ER63-94 | H | F. Jamieson | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| B | O111:NM | R82F2 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − |
| C69F1 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | ||
| CL101 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | ||
| O121:H19 | CL106 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Z3F3 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 274F4 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| O103:H2 | N01-2454 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| N01-7015 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| N02-1626 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| O26:H11 | CL4 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| CL9 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | ||
| O145:NM | N01-2051 | H | J. Isaac-Renton | + | −c | + | − | − | − | − | − | + | + | + | − | − | − | |
| N00-6496 | H | J. Isaac-Renton | + | −c | + | − | − | − | − | − | + | + | + | − | − | − | ||
| N02-5149 | H | J. Isaac-Renton | + | −c | + | − | − | − | − | − | + | + | + | − | − | − | ||
| C | O113:H21 | CL3 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| N99-3504 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| N89-0541 | H | J. Preiksaitis | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| N90-0657 | H | P. VanCaeseele | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| O91:H21 | B2F1 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| EC7-181 | H | LFZ | − | − | − | − | − | − | − | − | − | − | − | −c | − | − | ||
| EC6-990 | H | S. Aleksic | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| EC6-936 | H | L. Beutin | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | ||
| O104:H21 | G5506 | H | T. Whittam | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O165:H25 | N00-4540 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O5:NM | N00-4067 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| N00-4541 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| O121:NM | N99-4390 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| N99-4389 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| D | O103:H25 | N00-4859 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | −c | − | − | − |
| N02-2616 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | −c | − | − | ||
| OR:H2 | N00-3105 | H | G. Horsman | − | − | − | − | − | − | − | − | − | − | − | −c | − | − | |
| O117:H7 | N02-4495 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| N02-0035 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | −c | − | − | ||
| O69:H11 | EC97-821 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O132:NM | EC2-051 | H | LFZ | + | + | + | −c | + | + | + | + | −c | + | + | − | − | − | |
| O119:H25 | EC92-267 | H | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O146:H21 | N02-1625 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O174:H8 | A2EV659 | H | J. Isaac-Renton | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O172:NM | EC96-484 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O113:H4 | EC96-371 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O7:H4 | EC3-480 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O171:H2 | EC2-032 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| E | O39:H49 | EC92-293 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| O98:H25 | EC93-377 | B | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O88:H25 | EC94-453 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O6:H34 | EC96-626 | B | J. Preiksaitis | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O46:H38 | EC97-451 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O76:H7 | EC99-333 | O | P. Desmarchelier | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O163:NM | EC2-459 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O136:H12 | EC3-208 | B | LFZ | − | − | − | − | − | − | − | − | − | −c | − | − | − | − | |
| O153:H31 | EC2-104 | B | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O113:NM | EC2-211 | B | LFZ | + | + | + | −c | + | + | + | + | + | + | + | − | − | − | |
| O136:NM | EC2-258 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| O8:H19 | EC96-448 | B | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O156:NM | EC2-020 | B | LFZ | − | − | − | − | − | − | − | − | − | − | −c | − | − | − | |
| O84:NM | EC2-044 | B | LFZ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
Host: H, human; B, bovine; O, ovine.
Source: LFZ, Laboratory for Foodborne Zoonoses.
Negative in PCR but positive in Southern blot hybridization.
Distribution of OI-1, OI-47, OI-141, and OI-154 in EPEC O55:H7 strains.
All four OIs were present consistently in the two EPEC O55:H7 strains DEC5B and DEC5D.
Characterization of OI-1, -47, -141, and -154.
OI-1, -141, and -154 contain fimbrial operons only, whereas OI-47 encodes several putative virulence genes in addition to fimbrial genes.
(i) OI-47.
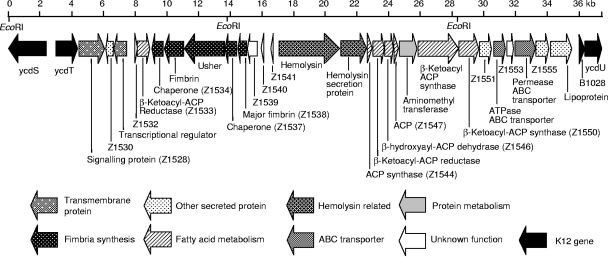
OI-47 is located at bp 1420969 to 1452695 in the EDL933 genome and has a size of 31,727 bp (44). It contains 28 ORFs flanked by E. coli K-12 genes ycdT upstream and B1028 downstream (Fig. 2). The average G+C content of this island is 46.24% (Table 3), compared to 50.36% ± 5.88% estimated for the entire EDL933 genome. Twelve ORFs have significantly lower G+C content than the genome, and seven ORFs have a dinucleotide bias above 1 standard deviation from the genome (Table 3). OI-47 is not inserted into a tRNA gene, does not contain genes encoding mobile genetic elements, and is not flanked by direct repeat sequences.
FIG. 2.
Physical and genetic map of O island 47 of E. coli O157 strain EDL933. The size of the genomic island and the presence of EcoRI restriction sites are marked. Gene clusters are indicated according to their predicted functions by different shadings. Arrows indicate the size, location, and transcription orientation of ORFs present in the island.
TABLE 3.
Nonfimbrial genes and their predicted proteins in OI-47
| Geneg | G+C contenta |
Dinucleotide biasc | No. of amino acids | Conserved domain | Signal peptided | Trans-helixe | Similar protein (reference) | % Identity/no. of aaf | |
|---|---|---|---|---|---|---|---|---|---|
| % G+C | STDb | ||||||||
| Z1528 | 36.69 | −2 | + | 536 | EAL | + | STY0376, predicted signal transduction protein in Salmonella enterica serovar Typhi (42) | 37/474 | |
| Z1530 | 33.54 | −2 | + | 163 | None | + | FidL, putative exported protein in serovar Typhi (42) | 29/145 | |
| Z1531 | 33.7 | −2 | + | 270 | Trans_reg_C | + | MarT, putative transcriptional regulatory protein in serovar Typhimurium LT2 (35) | 26/270 | |
| Z1532 | 17.28 | −2 | 26 | None | Not found | ||||
| Z1533 | 42.17 | −1 | FabG | YgcW, putative oxidoreductase in E. coli K12 (7) | 54/264 | ||||
| Z1540 | 30.63 | −2 | + | 36 | None | Not found | |||
| Z1541 | 36.67 | −2 | + | 39 | None | Not found | |||
| Z1542 | 49.28 | 1270 | Haemagg_act | + | RSp1073, probable hemagglutinin-related protein in the plant pathogen Ralstonia solanacearum (46) | 25/1260 | |||
| Z1543 | 42.47 | −1 | 539 | FhaC | + | SI and YPO2491, putative hemolysin-activator proteins in E. coli O113:H21 strain CL3 (50) and Yersinia pestis CO92 (43), respectively | 29/493 | ||
| Z1544 | 42.28 | −1 | 122 | AcpS | AcpS, holo-ACP synthase in Thermoanaerobacter tengcongensis (4) | 36/118 | |||
| Z1545 | 51.32 | 251 | FabG | FabG, 3-oxoacyl-ACP reductase in Clostridium perfringens (51) | 40/245 | ||||
| Z1546 | 48.09 | 182 | FabA | FabZ, (3R)-hydroxymyristoyl ACP dehydratase in Pasteurella multocida (33) | 40/92 | ||||
| Z1547 | 40.78 | −1 | 93 | AcpP | AcpC protein in Streptococcus agalactiae 2603V/R (57) | 38/86 | |||
| Z1548 | 49.01 | 386 | Gcv_T | GcvT, Glycine cleavage system T protein (aminomethyltransferase) in Thermoanaerobacter Tengcongensis (4) | 27/340 | ||||
| Z1549 | 53.54 | 852 | FabB | + | FabF, the 3-oxoacyl-ACP synthase II in Bacillus halodurans (56) | 27/805 | |||
| Z1550 | 52.78 | 407 | FabB | OlmA1, modular polyketide synthase in Streptomyces avermitilis (24) | 24/285 | ||||
| Z1551 | 48.95 | 317 | None | + | Not found | ||||
| Z1552 | 44.77 | 235 | ABC_ATPase | TM0705, ATPase component of the ABC transporter in Thermotoga maritima (40) | 50/222 | ||||
| Z1553 | 53.58 | 218 | None | Not found | |||||
| Z1554 | 47.52 | 436 | LolE | + | DUF214, permease component of the ABC transport system in Nitrosomonas europaea (9) | 36/405 | |||
| Z1555 | 47.78 | 262 | None | + | NE2245 in N. europaea ATCC 19718 (9) | 30/243 | |||
| Z1556 | 52.41 | 462 | None | + | Not found | ||||
G+C content: mean of whole genome EDL933 is 50.36%; standard deviation, 5.88%; mean of OI-47, 46.24% (42).
Standard deviation (STD): number of STD below genome mean.
Dinucleotide bias: “+” indicates the region has dinucleotide bias above 1 STD.
Signal peptide: “+” indicates the protein is predicted as a secreted protein.
Transmembrane helix: “+” indicates the protein is predicted as a transmembrane protein.
Number of amino acids (aa) in a contiguous stretch from which the identity was calculated.
Z1534 through Z1539 are fimbria-encoding genes. See Table 4 for details.
The deduced amino acid sequences of the OI-47 gene products show a variable degree of homology to other proteins (Table 3) (4, 7, 9, 21, 33, 35, 40, 42, 43, 46, 50, 51, 56, 57) in bioinformatics databases. Many proteins aligned very well with particular conserved domains, which allows the functions of OI-47 genes to be predicted (Table 3). From the predicted gene functions, OI-47 genes can be classified into seven groups (Fig. 2 and Tables 3 and 4): (i) genes encoding transmembrane proteins (Z1528, Z1531, and Z1554); (ii) genes encoding secreted proteins with signal peptides (Z1530, Z1551, Z1555, and Z1556); (iii) a fimbrial biogenesis locus (Z1534 through Z1539); (iv) hemagglutinin/hemolysin-related genes (Z1542 and ZZ1543); (v) genes involved in fatty acid synthesis (Z1533, Z1544 through Z1547, Z1549, and Z1550); (vi) ABC (ATP-binding cassette) transport system (Z1552 and Z1554); and (vii) an aminomethyltransferase gene (Z1548).
TABLE 4.
Fimbrial operons OI-1, -141, and -154 and the fimbrial locus in OI-47
| Island and gene | G+Ca |
Dinucleotide biasc | No. of amino acids | Conserved domain | Signal peptided | Similar protein (reference) | % Identity/no. of aae | |
|---|---|---|---|---|---|---|---|---|
| % G+C | STDb | |||||||
| OI-1 | ||||||||
| Z0020 | 37.34 | −2 | + | 207 | Adhes_bact | − | No significant match | |
| Z0021 | 37.00 | −2 | + | 108 | None | + | No significant match | |
| Z0022 | 41.00 | −1 | + | 816 | FimD | + | StcC, putative outer membrane usher protein, Salmonella enterica serovar Typhi CT18 (42) and serovar Typhimurium LT2 (35) | 60/810 |
| Z0023 | 36.55 | −2 | 227 | FimC | + | StcB, putative fimbrial chaperone protein, serovar Typhi, CT18 (42) and serovar Typhimurium LT2 (35) | 65/227 | |
| Z0024 | 39.14 | −1 | 177 | FimA Adhes_bact | + | StcA, putative fimbrial subunit protein, serovar Typhi CT18 (42) and serovar Typhimurium LT2 (35) | 34/177 | |
| Z0025 | 32.28 | −2 | 473 | None | − | No significant match | ||
| OI-47 (partial) | ||||||||
| Z1534 | 46.23 | 229 | FimC | + | Y2392, putative fimbrial chaperone, Yersinia pestis KIM (11) | 47/219 | ||
| Z1535 | 49.14 | 444 | Adhes_bact | + | Y2391, putative exported protein, Y. pestis KIM (11) | 35/420 | ||
| CupA4, fimbrial subunit, Pseudomonas aeruginosa PA01 (55) | 30/411 | |||||||
| Z1536 | 51.25 | 839 | FimD | + | Y2390, putative outer membrane usher protein, Y. pestis KIM (11) | 45/860 | ||
| Z1537 | 45.78 | 240 | FimC | + | Y0351, chaperone, Y. pestis KIM (11) | 40/206 | ||
| Z1538 | 51.16 | + | 186 | FimA Adhes_bact | + | Y2388, putative major fimbrial subunit, Y. pestis KIM (11) | 38/147 | |
| Z1539 | 31.89 | −2 | + | 207 | None | − | No significant match | |
| OI-141 | ||||||||
| Z4965 (lpfE) | 51.22 | 176 | FimA Adhes_bact | + | LpfE, long polar fimbrial minor protein, serovar Typhimurium LT2 (5,35) | 48/167 | ||
| Z4966 (lpfD) | 47.63 | 351 | FimA Adhes_bact | + | LpfD, long polar fimbrial protein, serovar Typhimurium LT2 (5,35) | 39/353 | ||
| Z4968m (lpfC) | 51.44 | 857 | FimD | + | LpfC, long polar fimbrial outer membrane usher protein, serovar Typhimurium LT2 (5,35) | 72/829 | ||
| Z4969 (lpfB) | 42.49 | −1 | 232 | FimC | + | LpfB, long polar fimbrial chaperone, serovar Typhimurium LT2 (5,35) | 68/233 | |
| Z4971 (lpfA) | 46.37 | 178 | FimA Adhes_bact | + | LpfA, long polar fimbrial major protein, serovar Typhimurium LT2 (5,35) | 72/178 | ||
| OI-154 | ||||||||
| Z5220 | 43.77 | −1 | 360 | FimA Adhes_bact | − | Z5221, probable fimbrial protein, E. coli O157:H7 EDL933 (44) | 46/356 | |
| StgD, probable fimbrial protein, serovar Typhi strain CT18 (42) | 41/341 | |||||||
| Z5221 | 47.43 | 356 | FimA Adhes_bact | + | StgD, probable fimbrial protein, serovar Typhi strain CT18 (42) | 35/362 | ||
| Z5222 | 48.52 | 844 | FimD | + | LpfC, putative fimbrial outer membrane usher protein, E. coli O113:H21 strain EH41 (13) | 65/844 | ||
| Z5223 | 41.36 | −1 | 53 | FimC (C terminus) | − | C terminus of StgB, fimbrial chaperone protein, serovar Typhi, strain CT18 (42) | 67/53 | |
| Z5224 | 38.28 | −2 | 127 | FimC (N terminus) | + | N terminus of StgB, fimbrial chaperone protein, serovar Typhi strain CT18 (42) | 69/116 | |
| Z5225 | 44.94 | + | 200 | FimA Adhes_bact | + | LpfA, putative major fimbrial subunit, E. coli O113:I121 strain EH41 (13) | 58/201 | |
| StgA, probable fimbrial subunit protein, serovar Typhi strain CT18 (13) | 61/202 | |||||||
| Z5226 | 27.78 | −2 | + | 29 | None | No match | ||
G+C content: mean of whole genome, 50.36%; STD, 5.88%. Mean of OI-1, -47, -141, and 154 are 36.81%, 46.24%, 47.46%, and 43.75%, respectively.
Standard deviation (STD): number of STD below genome mean.
Dinucleotide bias: “+” indicates the region has dinucleotide bias above 1 STD.
Signal peptide: “+” indicates the protein is predicted as a secreted protein.
Number of amino acids in a contiguous stretch from which the identity was calculated.
(ii) OI-1, -141, and -154.
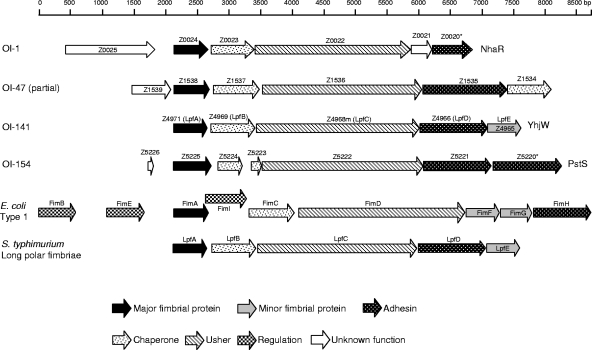
OI-1 (6,506 bp), OI-141 (5,904 bp), and OI-154 (6,943 bp) are located at bp 18464 to 24969, bp 4519710 to 4525613, and bp 4767320 to 4774262 in the EDL933 genome (44), respectively. The average G+C content of these islands (Table 4) is 36.8% (OI-1), 47.7% (OI-141), and 43.7% (OI-154), compared to 50.36% ± 5.88% estimated for the entire EDL933 genome. The G+C content and dinucleotide bias of individual genes in these O islands are shown in Table 4. None of these three islands is inserted into a tRNA gene, none contains genes encoding mobile elements, and none is flanked by repeat sequences. In each of these small O islands, the coding region carries only a fimbrial biosynthesis locus (Fig. 3).
FIG. 3.
Comparison of genetic organization of the four O islands that encode O157-specific fimbriae in EDL933 genome with that of type 1 fimbriae in E. coli and the long polar fimbriae in S. enterica serovar Typhimurium. Putative functions of genes are derived from domain search in the protein signature database InterPro and the Conserved Domain Database and from BLAST homology search (see Materials and Methods).
OI-1, -47, -141, and -154 encode fimbriae that resemble other fimbriae (Table 4) (5, 11, 13, 35, 42, 44, 55), including the prototype type 1 fimbriae (48, 54), which are exported by the chaperone/usher (C/U) pathway (48, 54). There are putative genes in each island encoding type 1-like fimbrial subunits, chaperone and usher proteins in highly conserved gene order of the E. coli type 1 fimbrial locus (49) and the long polar fimbrae in Salmonella enterica serovar Typhimurium (5) (Fig. 3 and Table 4).
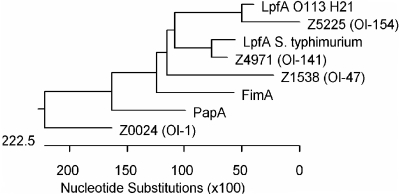
The phylogenetic relationships between the fimbriae reported here and the known P and type 1 fimbriae were analyzed based on the amino acid sequences of the putative major fimbrial proteins, as shown in Fig. 4. Results indicate that the OI-47, OI-141, and OI-154 fimbriae have a close phylogenetic relationship with the Salmonella long polar fimbriae, whereas the OI-1 fimbriae are phylogenetically distant from this cluster.
FIG. 4.
Phylogenetic relationships between the fimbriae encoded by OI-1, -47, -141, and -154 and P and type 1 fimbriae. The presumed evolutionary distance between any two members was calculated from an alignment using the CLUSTAL V method.
DISCUSSION
It is becoming increasingly recognized that unique or enhanced virulence properties of particular groups of strains within a species may be associated with the presence of specific horizontally acquired genetic elements, such as plasmids, bacteriophages, and PAIs (17, 18, 25). Consistent with this observation, we have previously shown (28) that the genomic islands LEE and OI-122 of the sequenced E. coli O157:H7 strain EDL 933 are specifically associated with VTEC seropathotypes (A and B) that are associated with severe and epidemic disease but not with seropathotypes C, D, and E, which are not associated with both severe and epidemic disease. However, LEE and OI-122 cannot adequately account for why seropathotype A strains (serotypes O157:H7 and O157:NM) cause serious and epidemic disease more commonly than seropathotype B strains (e.g., strains of serotypes O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM). To address this problem, in the present study, we used a subtractive hybridization approach to determine whether there are specific genomic islands in E. coli O157:H7 that are absent in seropathotype B strains that might explain why serotype O157:H7 is more often associated with epidemic and/or serious disease than seropathotype B strains. Our results show that four fimbria-encoding OIs (OI-1, OI-47, OI-141, and OI-154) are consistently present in seropathotype A strains (serotype O157:H7/NM strains) but are absent in seropathotype B strains as well as in virtually all other serotypes and seropathotypes (Table 2). This indicates that these OIs may confer unique virulence or adaptive traits to seropathotype A strains.
OI-1, -141, and -154 are solely fimbrial operons, whereas OI-47 contains additional putative virulence genes (Fig. 2). The major fimbrial proteins in OI-47, -141, and -154 are related phylogenetically to the Salmonella Lpf (long polar fimbriae), whereas that encoded by OI-1 is distant from this cluster (Fig. 4).
Gram-negative bacteria express a wide diversity of fimbriae of the C/U pathway that may be involved in colonization and/or pathological events on mucosal surfaces (48). The prototype type 1 fimbriae of the C/U pathway are the most common adhesin produced by E. coli and mediate adherence to mannose-containing glycoproteins found on the surfaces of many eukaryotic cells (48). The closely related type 1-like Pap fimbriae have been found to play an important role in the colonization and infection of the human urinary tract by E. coli (10, 12, 38), but type 1 or related C/U pathway fimbriae have not been proven to be associated with colonization of the human gastrointestinal tract by pathogenic E. coli, including strains of serotype O157:H7 (39, 48). However, type 1-like fimbriae of the C/U pathway have been implicated in diarrhea associated with enteropathogenic and enterotoxigenic E. coli in cattle and pigs (32, 48, 54).
E.
coli O157:H7 strains carry the fimA genes for the type 1 fimbrial subunit but do not appear to be able to express fimA (14). A 16-bp deletion in the fim switch locks the expression phase in the “off” orientation only in VTEC O157 (45). In contrast, other VTEC serotypes, such as O26 and O118, can express this adhesin (45). Since VTEC O157:H7 strains do not express the Fim fimbriae, it is possible that these strains synthesize novel type 1 or type 1-like fimbriae that are involved in the colonization of human and/or bovine gastrointestinal tracts. E. coli O157:H7 strains EDL 933 and Sakai have 14 loci for fimbrial biogenesis (19, 44). Ten of them are completely or partially conserved in E. coli K-12. The remaining four loci reported in our study appear to be unique to serotype O157 VTEC strains and may thus be associated with the ability of these strains to colonize human and/or animal intestinal tracts and to cause epidemic and serious disease more frequently than other serotypes.
Torres and colleagues (23, 59, 60) have investigated the pathogenic role of the long polar fimbrial operons encoded by OI-141 and OI-154, which they refer to as lpf1 and lpf2, respectively. lpf1 was thought to participate in the interaction of O157:H7 with eukaryotic cells by assisting in microcolony formation (59), whereas lpf2 was found to contribute to the early stages of enterohemorrhagic E. coli infection (60). Jordan et al. (23) challenged sheep, pigs, and gnotobiotic piglets with lpf1 and lpf2 mutants and wild-type parent strains. The mutant strains were recovered in significantly lower numbers and caused fewer attachment-and-effacement lesions than parent strains, providing in vivo evidence that long polar fimbriae contribute to intestinal adherence.
The presence of lpf1 and lpf2 was also investigated by Torres et al. (60) with a variety of pathogenic E. coli strains, including several EPEC, ETEC, and VTEC strains of different serotypes, and by Toma et al. with different VTEC seropathotypes (58). Both lpf1 and lpf2 were absent in serotype O26:H11 strains and in strains of other serotypes (58, 60), which is consistent with our findings. lpf1 was absent in five VTEC O111:H8 strains, but lpf2 was detected by colony blot hybridization in three of five strains of the latter serotype only at low stringency, suggesting the presence of a homolog (59, 60). Both lpf1 and lpf2 were detected in various EPEC and ETEC serotypes, notably EPEC serotype O55:H7 (60), which is considered to be the evolutionary ancestor of VTEC serotype O157:H7 strains (61).
While lpf1 and lpf2 are absent from the vast majority of other VTEC serotypes, Toma et al. (58) reported the presence of lpf1 homolog lpfO26 in all tested strains of seropathotype B and in 35%, 41%, and 42% of strains, respectively, in seropathotypes C, D, and E. Doughty et al. (13) reported the presence of a lpf2 homolog, lpfO113, in a VTEC strain of serotype O113:H21 and showed it was widely distributed in LEE-positive and LEE-negative non-O157 VTEC serotypes. Deletion of the putative major fimbrial subunit, LpfA, from a serotype O113:H21 strain resulted in decreased adherence to epithelial cells, suggesting it may play a role as an adhesin in non-O157 VTEC (13).
In contrast to progress in knowledge about OI-141 and OI-154, very little is known about the function and pathogenic significance of OI-1 and OI-47. The present study, using domain and motif searches in public bioinformatics databases, has provided significant new information on fimbriae and other potential virulence factors encoded by OI-1 and OI-47. Their role in pathogenesis remains to be investigated experimentally.
Typically, PAIs are inserted into tRNA genes, are often flanked by direct repeat sequences, contain mobile genetic elements, and have G+C content and codon usage patterns significantly different from those of the host genome (18). These features, which are characteristic of horizontally acquired genetic elements, are present only to a limited extent in the four OIs reported here (Tables 3 and 4). All four OIs have an average G+C content that is lower than the average G+C content of EDL 933, with that of OI-1 (G+C content, 36.1%) being markedly lower than that of EDL 933 (50.36%). The G+C content of several individual genes is also strikingly lower than the average G+C content of EDL 933, and several genes have a significant dinucleotide bias (Tables 3 and 4). On the other hand, none of the OIs are inserted into tRNA genes, contain genes encoding mobile genetic elements, or are flanked by direct repeat sequences. The partial evidence for their horizontal acquisition suggests that they may have been acquired very early in their evolution. The presence of OI-1, OI-47, OI-141, and OI-154 in the clonally related EPEC O55:H7 strains, which are considered to be the ancestors of E. coli O157:H7 strains (61), is consistent with this supposition.
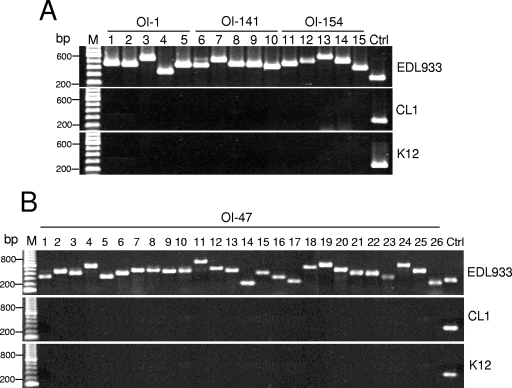
FIG. 1.
Ethidium bromide-stained agarose gel showing the results of PCR investigation of the presence of OI-1, -47, -141, and -154 in E. coli strain CL1 (serotype O26:H11). E. coli O157:H7 strain EDL933 was used as a positive control, and the E. coli K-12 strain MG1655 was used as a negative control. ORFs are designated as follows. Panel A, OI-1: 1, Z0020 and -21; 2, Z0022; 3, Z0023; 4, Z0024; 5, Z0025. OI-141: 6, Z4965; 7, Z4966; 8, Z4968m; 9, Z4969; 10, Z4971. OI-154: 11, Z5220; 12, Z5221; 13, Z5222; 14, Z5223 and -24; 15, Z5225. Panel B, ORFs in OI-47: 1, Z1528; 2, Z1530; 3, Z1531; 4, Z1533; 5, Z1534; 6, Z1535; 7, Z1536; 8, Z1537; 9, Z1538; 10, Z1539; 11, Z1540 and -41; 12, Z1542; 13, Z1543; 14, Z1544; 15, Z1545; 16, Z1546; 17, Z1547; 18, Z1548; 19, Z1549; 20, Z1550; 21, Z1551; 22, Z1552; 23, Z1553; 24, Z1554; 25, Z1555; 26, Z1556. M, DNA size marker; Ctrl, 23S rRNA gene.
Acknowledgments
This work was supported by Health Canada's postdoctoral fellowship, awarded to Songhai Shen, Health Canada Genomics R&D Fund award HCG10, and Canadian Institute for Health Research award 046163.
We thank S. Aleksic, L. Beutin, P. Desmarchellier, T. Honda, G. Horsman, J. Isaac-Renton, F. Jamieson, J. Kaper, J. Preiksaitis, H. Smith, P. VanCaeseele, and T. Whittam for generously providing strains used in this study. We also thank C. Clark for providing access to some of the above strains from culture collections at the National Microbiology Laboratory.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith. 1991. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Banatvala, N., P. M. Griffin, K. D. Greene, T. J. Barrett, W. F. Bibb, J. H. Green, and J. G. Wells. 2001. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J. Infect. Dis. 183:1063-1070. [DOI] [PubMed] [Google Scholar]
- 4.Bao, Q., Y. Tian, W. Li, Z. Xu, Z. Xuan, S. Hu, W. Dong, J. Yang, Y. Chen, Y. Xue, Y. Xu, X. Lai, L. Huang, X. Dong, Y. Ma, L. Ling, H. Tan, R. Chen, J. Wang, J. Yu, and H. Yang. 2002. A complete sequence of the T. tengcongensis genome. Genome Res. 12:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., S. Zimmermann, and K. Gleier. 1998. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg. Infect. Dis. 4:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Calderwood, S., D. W. K. Acheson, G. T. Keusch, T. J. Barrett, P. M. Griffin, N. A. Strockbine, B. Swaminathan, J. B. Kaper, M. M. Levine, B. S. Kaplan, H. Karch, A. D. O'Brien, T. G. Obrig, Y. Takeda, P. I. Tarr, and I. K. Wachsmuth. 1996. Proposed new nomenclature for SLT (VT) family. ASM News 62:118-119. [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell, I., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 93:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and R. A. Welch. 1996. Virulence determinants of uropathogenic Escherichia coli. American Society for Microbiology, Washington, D.C.
- 13.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami, M., N. Nakasone, Y. Honma, S. Kakinohana, J. Kudaka, and M. Iwanaga. 1999. Expression of type I pili is abolished in verotoxin-producing Escherichia coli O157. FEMS Microbiol. Lett. 179:467-472. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, P. M. 1995. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli, p. 739-761. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, Ltd., New York, N.Y.
- 16.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 19.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao, W., I. Wan, S. J. Jones, and F. S. Brinkman. 2003. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19:418-420. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, R., R. C. Clarke, J. B. Wilson, S. C. Read, K. Rahn, S. A. Renwick, K. A. Sandhu, D. Alves, M. A. Karmali, H. Lior, S. A. McEwen, J. S. Spika, and C. L. Gyles. 1996. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J. Food Prot. 59:1112-1122. [DOI] [PubMed] [Google Scholar]
- 23.Jordan, D. M., N. Cornick, A. G. Torres, E. A. Dean-Nystrom, J. B. Kaper, and H. W. Moon. 2004. Long polar fimbriae contribute to colonization by Escherichia coli O157:H7 in vivo. Infect. Immun. 72:6168-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juncker, A. S., H. Willenbrock, G. von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaper, J. B., J. L. Mellies, and J. Nataro. 1999. Pathogenicity islands and other mobile genetic elements of diarrheagenic Escherichia coli, p. 33-58. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 26.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 27.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 30.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, and B. T. Steele. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299-1300. [DOI] [PubMed] [Google Scholar]
- 31.Konowalchuk, J., J. I. Speirs, and S. Stavric. 1977. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 18:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lintermans, P. F., P. Pohl, A. Bertels, G. Charlier, J. Vandekerckhove, J. Van Damme, J. Schoup, C. Schlicker, T. Korhonen, and H. De Greve. 1988. Characterization and purification of the F17 adhesin on the surface of bovine enteropathogenic and septicemic Escherichia coli. Am. J. Vet. Res. 49:1794-1799. [PubMed] [Google Scholar]
- 33.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy, T. A., N. L. Barrett, J. L. Hadler, B. Salsbury, R. T. Howard, D. W. Dingman, C. D. Brinkman, W. F. Bibb, and M. L. Cartter. 2001. Hemolytic-uremic syndrome and Escherichia coli O121 at a lake in Connecticut, 1999. Pediatrics 108:E59. [DOI] [PubMed] [Google Scholar]
- 35.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 39.Nataro, J. P., and J. B. Kaper. Diarrheagenic Escherichia coli. 1998. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 42.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 43.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 44.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 45.Roe, A. J., C. Currie, D. G. Smith, and D. L. Gally. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145-152. [DOI] [PubMed] [Google Scholar]
- 46.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 49.Schilling, J. D., M. A. Mulvey, and S. J. Hultgren. 2001. Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J. Infect. Dis. 183(Suppl. 1):S36—S40. [DOI] [PubMed] [Google Scholar]
- 50.Shen, S., M. Mascarenhas, K. Rahn, J. B. Kaper, and M. A. Karmali. 2004. Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72:1496-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 54.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 56.Takami, H., K. Nakasone, C. Hirama, Y. Takaki, N. Masui, F. Fuji, Y. Nakamura, and A. Inoue. 1999. An improved physical and genetic map of the genome of alkaliphilic Bacillus sp. C-125. Extremophiles 3:21-28. [DOI] [PubMed] [Google Scholar]
- 57.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toma, C., E. E. Martinez, T. Song, E. Miliwebsky, I. Chinen, S. Iyoda, M. Iwanaga, and M. Rivas. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:4937-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333-344. [DOI] [PubMed] [Google Scholar]
- 61.Whittam, T. S. 1998. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 195-209. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 62.Whittam, T. S., I. K. Wachsmuth, and R. A. Wilson. 1988. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 157:1124-1133. [DOI] [PubMed] [Google Scholar]
- 63.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson, J. B., R. C. Clarke, S. A. Renwick, K. Rahn, R. P. Johnson, M. A. Karmali, H. Lior, D. Alves, C. L. Gyles, K. S. Sandhu, S. A. McEwen, and J. S. Spika. 1996. Vero cytotoxigenic Escherichia coli infection in dairy farm families. J. Infect. Dis. 174:1021-1027. [DOI] [PubMed] [Google Scholar]
- 65.World Health Organization. 1999. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a W.H.O. scientific working group meeting, Berlin, Germany, 22 to 23 June 1998, p. 1-30. World Health Organization, Geneva, Switzerland.
- 66.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]