Abstract
The survival and microevolution of Helicobacter pylori strains in the niches of the stomach after eradication therapy have largely been unexplored. We analyzed genomic signatures for two successive isolates obtained 9 years apart from a duodenal ulcer patient who underwent eradication therapy for H. pylori. These isolates were genotyped based on 50 different parameters involving three different fingerprinting approaches and several evolutionarily significant and virulence-associated landmarks in the genome, including nine informative gene loci, the cag pathogenicity island and its right junction, members of the plasticity region cluster, and vacA and iceA alleles. Our observations reveal that the two isolates were derived from the same strain that colonized the patient for almost a decade and were almost identical. Microevolution, however, was observed in the cagA gene and its right junction, the vacA m1 allele, and a member of the plasticity region cluster (JHP926). These results suggest that H. pylori has a great ability to survive and reemerge as a microevolved strain posteradication, thereby hinting at the requirement for follow-up of patients after therapy.
Since the successful isolation of Helicobacter pylori in 1984 by Marshall and Warren (19), H. pylori infection has been recognized to have a causal role in peptic ulcer disease and a strong association with gastric carcinoma (1, 8). H. pylori characteristically displays a great genetic variability due to a high rate of spontaneous mutations and genetic recombination (6, 9, 23). The occurrence of spontaneous mutations after serial passages in gnotobiotic piglets revealed that H. pylori may undergo host-specific adaptation (2). This suggests that a primary colonizing strain of H. pylori may need to rearrange its genome in order to adapt to a new environment. It has also been shown that during mixed infections, recombinant strains could emerge with different combinations of parental genotypes (16). Various DNA fingerprinting methods applied to two or more H. pylori isolates taken from the same patient have shown that the fingerprint profiles of such strains are highly similar with only minor band differences (7). This implies that two or more isolates recovered from one patient may be derived from an ancestral parent strain but have undergone independent genomic changes. This phenomenon has been termed microevolution (7). However, there are few examples where H. pylori sequential evolution could be documented after many years in the same host (9, 11, 14, 18). Such cases are therefore of the utmost interest in order to appreciate the process of microevolution with respect to virulence potential, drug resistance, dissemination dynamics, persistence, and quasi species development. In this study, we examined the molecular clock of the H. pylori genome by analyzing 50 different genetic characteristics (PCR genotyping- and nucleotide sequencing-based parameters) in two isolates recovered 9 years apart from the same patient.
Patient 1, a black African male living in France, suffered from epigastric pain in March 1994. An endoscopic examination showed a duodenal ulcer, and H. pylori was grown from gastric biopsy specimens. The patient received a 10-day triple therapy comprised of lansoprazole, amoxicillin, and clarithromycin at standard doses. An endoscopic follow-up carried out 2 months later showed a healed ulcer, and the biopsy specimens were negative for H. pylori by culture and histology. The patient had no follow-up for 9 years. In January 2003, he suffered again from epigastric pain, motivating an endoscopy, and an H. pylori-positive ulcer was found. H. pylori strains isolated in years 1994 (maintained frozen at −70°C) and 2003 were first compared by using random amplified polymorphic DNA (RAPD)-PCR with two different primers: 1254 and 1290 (3). H. pylori reference strains J99 and 26695 were used as controls. Then, fingerprinting techniques based on enterobacterial repetitive intergenic consensus (ERIC) and repetitive extragenic palindrome sequences (REP) were employed as described earlier (12). The REP-based typing procedure involved primers used for amplification of unique DNA sequences between the two REP signatures (25). All the gel images corresponding to ERIC and REP PCRs were analyzed by using the Quantity 1.0 software in a gel documentation system (Bio-Rad). The images were then uploaded into the Diversity 2.2.0 database (Bio-Rad). Band sizes, band attributes, and standard molecular weights were assigned alongside the molecular weight markers. Cluster analysis of DNA profiles was conducted on the basis of fingerprint characteristics. All the data obtained through molecular genotyping and DNA profiling were deposited in the genoBASE pylori database (http://www.cdfd.org.in/amplibase/HP). The genoBASE pylori server was queried for genome-wide comparisons. Then, five housekeeping genes spread throughout the genome (atpA, efp, ureI, ppa, and mutY) were partially amplified and sequenced for the two isolates (9). Sequencing was performed with both forward and reverse primers by using an ABI Prism 3100 DNA sequencer (Applied Biosystems). PCR and direct sequencing were performed at least twice to determine and confirm the DNA sequences for each isolate. Consensus sequence for each of the samples was generated by using Genedoc (version 2.6.002). Multiple alignments of sequenced nucleotides were carried out by using Clustal X (version 1.81). The cag pathogenicity island (cag PAI) status was evaluated by PCR using eight sets of primers spanning the cagA gene, its promoter region, the cagE and cagT genes (13, 15), and the left end of the PAI as mentioned elsewhere (17). The presence and status of the cagA gene (24) and rearrangement analysis on the right end of the cag PAI (17), vacA genotyping (4), iceA allele status (22), babB gene status (21), and sequencing were carried out by molecular genotyping methods as described previously. A fragment containing the middle and final part of the vacA gene was amplified with VAG-F (4) and R-total-VacA (CCTAAATTGGAAGCGAAATGG) and sequenced with these primers plus R-vacA-3114 (AGCGTTAGAGCCGCTATTCA), F-vacA-3043 (TGTTGTATCAATTTGCCCCTAA), F-vacA-2008 (TCAACAATCAAGGCACTATCAA), and F-vacA-2592 (AGCGCCACTCCTAATTTAGTC). The (cag PAI) rearrangement profiles and cagA-glr motif types were also compared to existing records in the genoBASE pylori database. The presence or absence of the plasticity region open reading frames (ORFs) JHP947, HP986, JHP912, JHP926, JHP931, JHP933, JHP944, and JHP945 was detected based on PCR amplifications employing target sequences and reaction parameters as described by Occhialini et al. (20). Purified DNA preparations from the two isolates were used for PCR and sequencing of genes such as glmM (ureC), babB, and oipA as described previously (10). The amplified products of the ureC fragment and oipA (HP0638) and babB genes were gel eluted and purified with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany). Antimicrobial susceptibility testing by Epsilometer test (Etest; AB Biodisk, Solna, Sweden) was performed for clarithromycin and metronidazole.
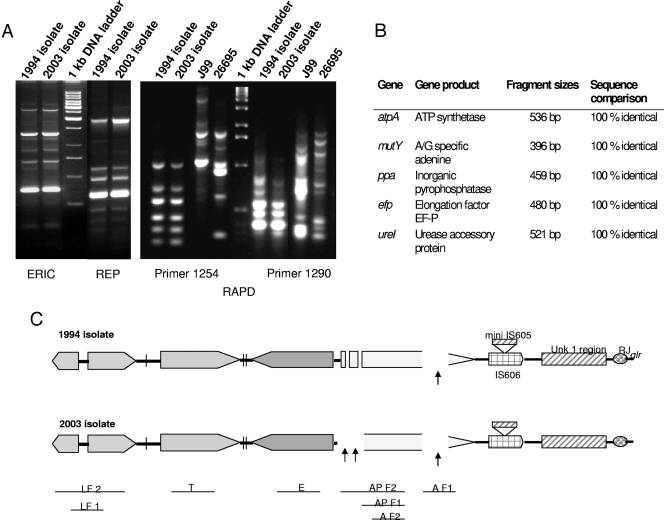
Genome-wide profiling studies, such as ERIC, REP, and RAPD-PCR, showed the same pattern for the 1994 isolate and the 2003 isolate, indicating that they were derived from the same parental strain (Fig. 1). The second isolate, obtained 9 years after the first isolation, did not present any mutation over the 2,400 bp of the five housekeeping genes sequenced. Both the isolates possessed the cagA gene and the s1 allele of the vacA gene. The allele of the vacA middle sequence was m1 for the more recent strain but that for the older one could not be determined. Indeed, a 1,072-bp fragment of the vacA gene ranging from nucleotide 1435 to 2506 (compared to J99) was absent in the 1994 isolate. This deletion creates a frameshift resulting in a truncated nonfunctional VacA protein. The 1,394-bp sequence surrounding the deletion was identical in both the isolates. Our molecular clock studies involving the extreme right junction of the cag PAI revealed nearly similar profiles except for some rearrangements at two places due to substitution polymorphisms (Fig. 1; Table 1). The cag PAIs of the two isolates were nearly identical except for rearrangements at the cagA promoter region that prevented amplification of this locus probably due to change at the primer binding sites in the 2003 isolate. This region, however, was reproducibly amplified for the 1994 isolate.
FIG. 1.
(A) ERIC, REP, and RAPD analyses of the two isolates; (B) multilocus sequence typing analysis revealing 100% identical sequences of the 5 housekeeping genes, indicating the undisputed descent of the original strain; (C) PCR-based analysis of insertion, deletion, and substitution polymorphisms within and at the extreme right junction of the cag PAI. Precise locations of rearrangements have been marked with arrows. LF 1, left end of the cag PCR product 1 (384 bp); LF 2, left end of the cag PCR product 2 (877 bp); T, cagT gene (301 bp); E, cagE gene (329 bp); AP F1, cagA promoter PCR product 1 (730 bp); AP F2, cagA promoter PCR product 2 (1,181 bp); A F1, cagA gene PCR product 1 (349 bp); A F2, cagA gene PCR product 2 (701 bp); Unk 1, unknown region 1 as described by Kersulyte et al. (17); RJ, right junction of cagA; glr, glutamate racemase.
TABLE 1.
Genotyping resultsa
| Parameter no. | Locus/method details | Genotyping status |
Microevolution | Conclusion | |
|---|---|---|---|---|---|
| 1994 isolate | 2003 isolate | ||||
| 1 | oipA gene presence | + | + | NC | |
| 2 | OipA frame status | In frame | In frame | NC | |
| 3 | No. of dinucleotide CT repeats in the oipA signal sequence | 09b | 09b | NC | |
| 4 | Plasticity region cluster | + | + | Yes | Incomplete cluster |
| 5 | JHP912 | + | + | No | |
| 6 | JHP927 | − | − | No | |
| 7 | JHP931 | − | − | No | |
| 8 | JHP926 | + | − | Yes | Deletion/rearrangement |
| 9 | JHP945 | − | − | No | |
| 10 | JHP944 | − | − | No | |
| 11 | JHP947 | − | − | No | |
| 12 | HP986 | − | − | No | |
| 13 | cag-PAI cluster | + | + | Yes | Nearly complete cluster |
| 14 | cagA promoter region 1 | + | − | Yes | Deletion/rearrangement |
| 15 | cagA promoter region 2 | + | − | Yes | Deletion/rearrangement |
| 16 | cagA gene region 1 | − | − | NC | |
| 17 | cagA gene region 2 | + | + | NC | |
| 18 | cagE gene | + | + | NC | |
| 19 | cagT gene | + | + | NC | |
| 20 | Left end of cag (LEC1) | + | + | NC | |
| 21 | Left end of cag (LEC2) | + | + | NC | |
| 22 | cagA-glr motif typing | + | + | Yes | Instability |
| 23 | cagF4584(1) plus CagR5280(3) | − | − | NC | |
| 24 | cagF4584(1) plus mIS605.R(8) | + | + | ||
| 25 | fcn unk 6 plus CagR5280(3) | − | − | NC | |
| 26 | fcn unk 6a plus CagR5280(3) | − | + | Yes | Possible acquisition |
| 27 | cagR5280(3) plus CagF4856(4) | + | + | NC | |
| 28 | cagR5280(3) plus IS606-1692(5) | − | + | Yes | Possible acquisition |
| 29 | cagF4584(1) plus Xins.R(7) | + | + | NC | |
| 30 | iceA gene | + | + | NC | |
| 31 | iceA1 | − | − | NC | |
| 32 | iceA2 | + | + | NC | |
| 33 | glmM gene | + | + | NC | |
| 34 | vacA gene | + | + | NC | |
| 35 | s1 allele | + | + | NC | |
| 36 | s2 allele | − | − | NC | |
| 37 | m1a allele | − | + | Yes | Possible acquisition |
| 38 | m1b allele | − | − | NC | |
| 39 | m1c allele | − | − | NC | |
| 40 | m2 allele | − | − | NC | |
| 41 | MLST analysis (all five genes) | + | + | NC | All were amplified |
| 42 | atpA gene sequence | Identical | Identical | NC | Conserved |
| 43 | mutY gene sequence | Identical | Identical | NC | Conserved |
| 44 | ppA gene sequence | Identical | Identical | NC | Conserved |
| 45 | efp gene sequence | Identical | Identical | NC | Conserved |
| 46 | urel gene sequence | Identical | Identical | NC | Conserved |
| 47 | RAPD method 1254 | Identical | Identical | NC | Stable molecular clock |
| 48 | RAPD method 1290 | Identical | Identical | NC | Stable molecular clock |
| 49 | ERIC typing | Identical | Identical | NC | Stable molecular clock |
| 50 | REP PCR typing | Identical | Identical | NC | Stable molecular clock |
NC, no change; +, region intact; −, region rearranged or deleted; MLST, multilocus sequence typing.
Values are numbers of dinucleotide CT repeats.
Both the isolates carried exactly the same genotypes with respect to the sequence typing of oipA, glmM, and iceA gene loci. Also, seven of the eight loci from the strain-specific plasticity region cluster reflected similar profiles for both the isolates. In addition, the two isolates exhibited the same antimicrobial susceptibilities for metronidazole (MIC, 32 mg/liter) and clarithromycin (MIC, 0.03 mg/liter). Globally, these data indicate that both the isolates in fact originated from the same strain (Fig. 1; Table 1).
Despite the fact that an acquisition of the strain from the same source cannot be ruled out, the most likely scenario emerging from the genotyping results presented here is that the strain might have persisted in the patient during the 9 years. These data show a great genomic stability for the five housekeeping genes despite an expectedly high number of DNA replications that occurred during the entire duration of colonization. Such a study is important to get insight into the evolution of H. pylori in vivo, as the analysis of two clinical isolates 9 years apart has rarely been performed. Here, several elements prove undoubtedly that it was the same bacterial strain studied over the years, but some differences however exist between the two serial isolates. One gene of the plasticity region cluster was absent in the 2003 isolate, and the promoter region of the cagA gene could not be amplified anymore in this isolate. But more surprisingly, new fragments of genes are present in the more recent isolate; it is, for example, the case for the middle part of vacA. This acquisition could have happened during coinfection with another strain, but if another strain was present in the same host, it is likely that more sequence diversity would have been found in the 2003 isolate (16). We can rather think that vacA was present in the parental strain and that in 1994 we obtained an isolate representing a subpopulation having lost this gene. In such a hypothesis, a larger sample of bacteria in 1994 might have shown a heterogeneous population for this gene. This phenomenon has been previously observed for a Mexican patient, for whom identical H. pylori strains with the same RAPD and AFLP pattern but with m1 or m2 alleles of vacA were simultaneously present (5). Typing techniques like RAPD cannot detect small variations occurring at the gene level (11). Israel et al. (14) compared several isolates of the reference strain J99 taken 6 years apart from the same patient, and despite the fact that RAPD profiles were identical, microarray analysis revealed acquisition and deletion of genes over the years. Some studies have focused on strain diversity through family exploration (16), but no concept of chronological evolution was reported.
In conclusion, we used multilocus genotyping and fingerprinting approaches to look into the molecular evolution of the two isolates obtained 9 years apart. From a practical point of view, our results hint that the organism H. pylori has a tremendous potential to survive and reemerge, despite eradication therapy. We suggest that performing diagnostic tests 6 months after H. pylori eradication therapy may be important to increase the accuracy of the posttreatment follow-up.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank under the following accession numbers: for strain 908 (the 1994 isolate), DQ076757; and for strain 2017 (the 2003 isolate), DQ076758.
Acknowledgments
We are thankful to Seyed E. Hasnain for his help and support.
REFERENCES
- 1.Ahmad, A., Y. Govil, and B. B. Frank. 2003. Gastric mucosa-associated lymphoid tissue lymphoma. Am. J. Gastroenterol. 98:975-986. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., K. A. Eaton, and D. E. Berg. 1995. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect. Immun. 63:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atherton, J. C., T. L. Cover, R. J. Twells, M. R. Morales, C. J. Hawkey, and M. J. Blaser. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aviles-Jimenez, F., D. P. Letley, G. Gonzalez-Valencia, N. Salama, J. Torres, and J. C. Atherton. 2004. Evolution of the Helicobacter pylori vacuolating cytotoxin in a human stomach. J. Bacteriol. 186:5182-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkholm, B., M. Sjölund, P. G. Falk, O. G. Berg, L. Engstrand, and D. I. Andersson. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. Morain, C. M. Habibullah, and Cyril J. Smyth. 2004. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J. Med. Microbiol. 53:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghose, C., G. I. Perez-Perez, M. G. D. Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser. 2002. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA 99:15107-15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustavsson, A., M. Unemo, B. Blomberg, and D. Danielsson. 2005. Genotypic and phenotypic stability of Helicobacter pylori markers in a nine-year follow-up study of patients with noneradicated infection. Dig. Dis. Sci. 50:375-380. [DOI] [PubMed] [Google Scholar]
- 12.Hussain, M. A., F. Kauser, A. A. Khan, S. Tiwari, C. M. Habibullah, and N. Ahmed. 2004. Implications of molecular genotyping of Helicobacter pylori isolates from different human populations by genomic fingerprinting of enterobacterial repetitive intergenic consensus regions for strain identification and geographic evolution. J. Clin. Microbiol. 42:2372-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikenoue, T., S. Maeda, K. Ogura, M. Akanuma, Y. Mitsuno, Y. Imai, H. Yoshida, Y. Shiratori, and M. Omata. 2001. Determination of Helicobacter pylori virulence by simple gene analysis of the cag pathogenicity island. Clin. Diagn. Lab. Immunol. 8:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Israel, D. A., N. Salama, U. Krishna, M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauser, F., A. A. Khan, M. A. Hussain, I. M. Carroll, N. Ahmad, S. Tiwari, Y. Shouche, B. Das, M. Alam, S. M. Ali, C. M. Habibullah, R. Sierra, F. Megraud, L. A. Sechi, and N. Ahmed. 2004. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J. Clin. Microbiol. 42:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 17.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuipers, E. J., D. A. Israel, J. G. Kusters, M. M. Gerrits, J. Weel, A. van der Ende, R. W. van der Hulst, H. P. Wirth, J. Hook-Nikanne, S. A. Thompson, and M. J. Blaser. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 16:1311-1315. [DOI] [PubMed] [Google Scholar]
- 20.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 68:6240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori BabA and BabB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman, M., A. K. Mukhopadhyay, S. Nahar, S. Datta, M. M. Ahmad, S. Sarker, I. M. Masud, L. Engstrand, M. J. Albert, G. B. Nair, and D. E. Berg. 2003. DNA-level characterization of Helicobacter pylori strains from patients with overt disease and with benign infections in Bangladesh. J. Clin. Microbiol. 41:2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tummuru, M. K. R., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61:1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]