High sequence diversity among TT virus (TTV) and TTV-like viruses (TTLVs) is an obstacle to PCR detection of the entire spectrum of existing genotypes. To determine which primers are needed to detect all know genotypes of these viruses, an alignment of commonly used primer sequences with sequences of all known viral genotypes was performed to predict theoretically amplifiable genotypes. The results predicted that all TTV genotypes (except genotype 21), all SEN virus (SENV) genotypes (A to H), and three known TTVL minivirus (TLMV) groups would be detectable by TTV primers NG054/NG147/133/132 (NG) and TT6/7/8/9 (TT), combined with two TLMV primer sets (TLMV-S and TLMV-L). To confirm these findings, we genetically characterized 420 clones of NG primer products from 32 thalassemia patients and 8 healthy individuals, as well as 50 PCR products amplified by TT and RD037/038/051/052 primers from 50 thalassemia patients and 96 TLMV-S primer product clones from 16 thalassemia patients. We detected TTV genotypes 1 to 3, 9 to 13, 14, 16, 18, 22, 23, and group 5 (JT33F/JT34F/CT39F/CT44F); SENV genotypes A, B, D, and H; and four TLMV groups (three known plus one newly identified). TTV genotypes 4 to 8, 15, 17, 19, and 20 including SENV (genotypes C and E to G) were not found, presumably due to their low prevalence in the test populations. Genotype 21 was also not detected, as it requires a dedicated primer set. Primer alignment and PCR product characterization consistently indicated that five primer sets, NG, TT, TLMV-S, TLMV-L, and a genotype 21-specific set of primers, are required to detect all known genotypes of TTV and TTLVs.
A small single-stranded DNA virus, named TT virus (TTV), was identified by Nishizawa et al. in Japan from patients with non-A-G transfusion-acquired hepatitis (22). Shortly after TTV was discovered, several TTV-like viruses (TTLVs) including TTV-like minivirus (TLMV) (33), SEN virus (SENV) (8), and PM virus (PMV) (10) were also reported. All TTLVs are recognized as divergent genotypes of TTV with the exception of TLMV (19). TLMV is a unique virus with a smaller genome that may constitute a genetic evolutionary link between chicken anemia virus and TTV (33). TTV and TTLVs are widely distributed geographically, with an extremely high prevalence of viremia in the general population, particularly in tropical countries (32) and parts of Asia (6). Mixed-genotype infections are therefore common, particularly in thalassemia patients, due both to the high frequency of viral transmission through repeated therapeutic blood transfusions and the persistent nature of the viruses (5, 9, 14, 15, 18, 23, 31). TTV and TTLVs demonstrate an extremely wide range of sequence divergence. At least four groups comprising 23 genotypes of TTV have been identified (24, 25, 34, 35, 37). In addition, four new genotypes have recently been identified and classified as TTV group 5 (30). All TLMV isolates are classified into three genetically distinct groups (3).
Currently, PCR is the only available means for detection of TTV and TTLVs. Commonly used primers are derived from either the coding region in open reading frame (ORF) 1 or the conserved 5′ untranslated region (UTR) (i.e., ORF and 5′UTR primers, respectively). ORF primers detect only a restricted number of genotypes (16, 28). For example, N22 primers are designed using the coding region sequences of genotypes 1 and 2, which can amplify genotypes 1 to 6 in group 1. Genotypes 7 and 8 in group 2 are amplifiable by N22 primers, but not as efficiently as the genotypes in TTV group 1 (7). However, TTV genotypes 9 to 16, 18, 19, and 20, in groups 3 and 4, and genotypes 17, 22, and 23, in group 2, cannot be amplified by N22 primers. 5′UTR primers are thought to have the potential to detect all known genotypes of TTV including the new genotypes of TTV group 5 (30). Three known groups of TLMV can be amplified by one set of TLMV-specific primers (36). In most studies only one or two sets of primers, either ORF primers or 5′UTR primers, are used for virus detection. As a consequence, the prevalence of these viruses has been underestimated in early studies (12, 20, 29). At the present time, no clinical manifestations have been unequivocally associated with these viruses; however, it is not clear whether all these viral agents or genotypes have been detected or discovered in test populations using current detection systems. In spite of incomplete detection of TTV and related viruses, previous data have indicated that transient and mild abnormalities in liver enzyme levels are associated with TTV and TTLV infection. Although several 5′UTR primer sets have been introduced in recent studies for detection of TTV (16, 24), these primers differ in their abilities to detect TTV and related viruses by PCR (29). Therefore, results have varied depending on the choice of primer sets employed for analysis. This has impeded a proper assessment of the viral prevalence and pathogenesis. In this study, we have sought to validate the commonly used PCR primer sets using the following three methods: (i) alignment of primer sequences with all known TTV and TTLV sequences available in GenBank to predict the amplifiable genotypes for each set of primers, (ii) genetic characterization of a large number of PCR product clones and isolates from transfusion-dependent thalassemia patients as well as healthy individuals to demonstrate the genotypes that can be detected by various primer sets, and (iii) comparative analysis of detection rates for each set of primers and a combination of multiple sets of primers to evaluate the ability of the primers to detect TTV and TTLVs in different populations. We demonstrated that five sets of primers were required for detection of all known groups and genotypes of TTV and TTLVs. In addition, we investigated the ability of specific primer sets to detect known genotypes and in the process have discovered a new group of TLMV amplified by NG054/147/133/132 primers. This study provides a means for molecular detection for all known groups and genotypes of TTV and TTLVs, thereby fulfilling a prerequisite for assessment of viral prevalence and pathogenesis.
MATERIALS AND METHODS
Study groups.
Human plasma samples were obtained from the United Arab Emirates; these included samples from 200 healthy individuals and 118 thalassemia patients who had been treated with regular blood transfusions (13 to 18 transfusions/year).
Isolation of viral DNA.
Viral DNA was isolated using the QIAamp blood kit (QIAGEN) in accordance with the procedure provided by the manufacturer.
Amplification and detection of viral DNA.
Purified viral DNA was amplified using five sets of nested and/or seminested primers derived from the conserved 5′UTR and coding regions of the TTV and TTLV genomes, including nested primers NG054/147 and NG133/132 (NG054/147/133/132) (27), RD037/038 and RD051/052 (RD037/038/051/052) (25), TT6/7 and TT8/9 (TT6/7/8/9) (11), and TLMV-S1/2 and TLMV-S3/4 (TLMV-S1/2/3/4) as well as seminested primers TLMV-LS2/LA2/LA3 (36) (Table 1). A genotype 21-specific primer set (16) was not used in this study. The procedure described by Okamoto et al. (29) was used for DNA amplification by PCR. Viral DNA was detected on a 1.5% agarose gel.
TABLE 1.
Positions and nucleotide sequences of oligonucleotide primers
| Primera | Polarity | Nucleotide position | Nucleotide sequence (5′-3′) |
|---|---|---|---|
| RD | |||
| RD037 | Sense | 2008-2027 | GCAGCAGCATATGGATATGT |
| RD038 | Antisense | 2258-2277 | TGACTGTGCTAAAGCCTCTA |
| RD051 | Sense | 2061-2080 | CATACACATGAATGCCAGGA |
| RD052 | Antisense | 2238-2257 | GTACTTCTTGCTGGTGAAAT |
| TT | |||
| TT6 | Sense | 1900-1919 | ACAGACAGAGGAGAAGGCAA |
| TT7 | Antisense | 2211-2228 | TACCATTTAGCTCTCATT |
| TT8 | Sense | 1918-1938 | AACATGTTATGGATAGACTGG |
| TT9 | Antisense | 2166-2185 | CTGGCATTTTACCATTTCCA |
| NG | |||
| NG054 | Sense | 3-22 | TTTGCTACGTCACTAACCAC |
| NG147 | Antisense | 211-233 | GCGAGTCCCCGAGCCCGAATTGCC |
| NG133 | Sense | 91-115 | GTAAGTGCACTTCCGAATGGCTGA |
| NG132 | Antisense | 204-223 | AGCCCGAATTGCCCCTTGAC |
| TLMV-S | |||
| TLMV-S1 | Sense | 30-48 | ATTTGAATTGCCGACCACA |
| TLMV-S2 | Antisense | 298-319 | CGCCAGACTGATCTAGCCCGAA |
| TLMV-S3 | Sense | 36-55 | ATTGCCGACACAAACTGAC |
| TLMV-S4 | Antisense | 284-305 | AGCCCGAATTGCCCCTAGTC |
| TLMV-L | |||
| LS2 | Sense | 33-51 | WGAATTGCCGACCACAAAC |
| LA2 | Antisense | 311-330 | AAGTTTCTTGCCCGKTCCGC |
| LA3 | Antisense | 280-299 | AGCCCGAATTGCCCCTAGAC |
A genotype 21-specific primer set (16) was not used in this study.
Direct DNA sequencing.
Direct DNA sequencing of PCR products was performed on 50 thalassemia patient samples using an automated DNA sequencer (Visible Genetics). PCR products (approximately 150 bases) amplified by RD037/038/051/052 from 20 of the patients were sequenced using 5′ Cy5.5-labeled sense primer RD051. The PCR products (approximately 150 bases) amplified from 30 of the patients using TT6/7/8/9 primers were sequenced using 5′ Cy5.5-labeled sense primer TT8.
Clone-based DNA sequencing.
Clone-based DNA sequencing was performed on 56 samples from 48 thalassemia patients and 8 healthy individuals using the TOPO TA vector system (Invitrogen Corp.). Purified PCR products were directly cloned into the TOPO TA vector (Invitrogen Corp.). Viral sequences were then amplified from positive colonies of Escherichia coli using M13 primers (Pharmacia Biotech). The reamplified PCR products were sequenced using the same procedure as for direct DNA sequencing. Ten to 12 TTV clones and 5 to 10 TLMV clones from each individual were randomly selected and sequenced.
Genotyping and computer analysis of nucleotide sequences.
DNA sequences derived from experimental samples were compared to an online database for the best possible match using the BLAST (Basic Local Alignment Search Tool) program found on the National Center for Biotechnology Information home page (http://www.ncbi.nlm.nih.gov/). All further sequence analyses and comparisons and construction of phylogenetic trees were performed using the DNA-STAR Lasergene '99 software package (DNAStar, Madison, WI).
RESULTS
Sequence alignment to predict the genotypes amplifiable by different sets of primers.
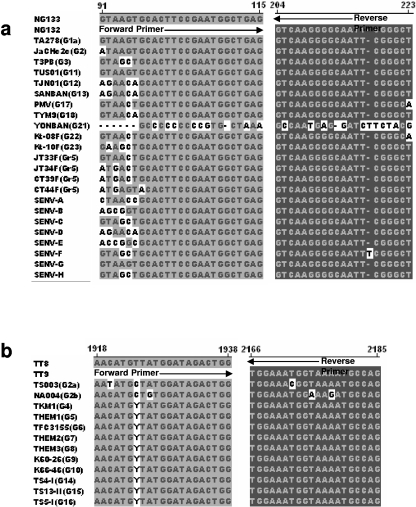
The partial or entire genome sequences are known for all published genotypes of TTV, eight genotypes of SENV, PMV, and isolates of the three known groups of TLMV (24, 30). Those genotypes detectable by various sets of primers should be predictable by multialignment of the primer sequences with the published reference sequences of TTV and TTLVs (13). Figure 1a shows that NG133/132 primer sequences are almost completely matched with the 5′UTR sequences of 14 known TTV genotypes (1, 2, 3, 11, 12, 13, 17 [PMV], 18, 22, 23, and four recently classified genotypes, JT33F, JT34F, CT39F, and CT44F) and all 8 genotypes of SENV (A to H). NG054/147 primer sequences matched these TTV genotype sequences as well as NG133/132 primer sequences (data not shown). We were unable to prepare multialignments for these primer sequences against the remaining TTV genotypes (4, 5, 6, 7, 8, 9, 10, 14, 15, and 16) because their 5′UTR sequences were not available in GenBank. Thus, at the present time, it is not known whether these primers match the 5′UTR sequences of those TTV genotypes. Fortunately, SENV-A, -B, -C, -D, -E, and -H are currently classified as variants of TTV genotypes and specifically correspond to genotypes 9, 10, 15, 12, 14, and 16, respectively (24). Therefore, the NG054/147/133/132 nested primer set should also be able to amplify these five additional TTV genotypes (9, 10, 14, 15, and 16). However, this nested primer set will not amplify genotype 21 (YONBAN) because there are only a few nucleotides at the middle of the primer sequences that are identical to the 5′UTR sequence of this genotype. Consequently, in total, at least 17 known TTV genotypes (1 to 3, 9 to 20, 22, and 23), 4 new TTV genotypes of group 5, and 8 known genotypes of SENV are predicted to be detectable by NG054/147/133/132 primers.
FIG. 1.
Sequence alignment of NG133/132 primers (a) and TT8/9 primers (b) with classified representatives of all known TTV genotypes and eight SENV genotypes (A to H) showing sequence identity between primers and viral genotypes. Representatives of known TTV genotypes and groups are indicated in parentheses as G and Gr, respectively.
TT8/9 primers and those that are similar to TT8/9 primers (e.g., NG059/NG063/NG061) (29) have been commonly used for recent studies (4, 21) because these primers are able to amplify more genotypes than other coding region-based primers for the detection of TTV. Figure 1b shows that TT8/9 primer sequences are almost 100% identical to the targeted sequences of TTV genotypes 2, 4, 5, 6, 7, 8, 9, 10, 14, 15, and 16. The one or two nucleotide mismatches at the middle of the primer sequences would be expected to have little impact on DNA amplification by PCR. The sequences from the other genotypes of TTV including the new genotypes of group 5 and all eight genotypes of SENV were found to have a low rate of nucleotide identity with TT8/9 primer sequences (data not shown) and therefore would not be amplified by TT8/9 primers. As with the NG054/147/133/132 primers, TT8/9 primers have only a few nucleotides that are identical to nucleotides on the primer binding sequences of TTV genotype 21 (data not shown). It is apparent that neither NG054/147/133/132 nor TT8/9 primers can detect TTV genotype 21. It was noted that five TTV genotypes (4, 5, 6, 7, and 8) of unknown detectability by NG054/147/133/132 primers are well matched by TT8/9 primers. It is expected that 22 of the 23 known TTV genotypes, 4 newly classified genotypes of group 5, and 8 genotypes of SENV can be amplified by a combination of the two sets of primers (NG054/147/133/132 and TT8/9). The TT6/7 primer sequences were found only to match the sequences of TTV genotypes 1, 2, and 3, but not other known TTV genotypes (data not shown). RD037/038/051/052 primers were well matched only with genotypes 1a and 1b and therefore would be restricted to TTV genotype 1.
The sequences of TLMV-S and TLMV-L primers are similar, and both were designed using the highly conserved repeat sequence within the 5′UTR. The two sets of TLMV primer sequences are well matched, with almost all the representative sequences (3) in the three known groups of TLMV (data not shown). There are single- or double-nucleotide mismatches found at the middle of the primer sequences in some isolates. A mismatch at the second nucleotide position of the 3′ end of the internal reverse primer TLMV-S4 occurred in some representatives of the known TLMV groups. This nucleotide mismatch may reduce the amplification efficiency for these viruses. However, Taq DNA polymerase should still be able to initiate DNA synthesis with only one mismatched nucleotide near the 3′ end of the primer (13). We predict that either the nested TLMV-S primer system or TLMV-L seminested primer system would be able to amplify all three known groups of TLMV. NG133/132 primers matched with some TLMV representatives in groups 1 and 3, but not in group 2, where several nucleotide mismatches occurred (data not shown). Thus, some strains of TLMV may be amplifiable by NG054/147/133/132 primers.
Genotypes of TTV and TTLVs amplified by different sets of primers.
To confirm the sequence alignment results, a total of 566 PCR product clones and PCR isolates were genetically characterized. Four hundred twenty clones of PCR products amplified by nested 5′UTR primers NG054/NG147/133/132 from 40 PCR-positive individuals including 32 thalassemia patients (equal representation of high and low alanine aminotransferase samples) and 8 randomly selected healthy individuals (controls) were subjected to clone-based sequencing. Fifty randomly selected PCR products amplified by ORF primers TT6/7/8/9 and RD037/038/051/052 from thalassemia patients were genotyped by direct DNA sequencing, and 96 TLMV-S primer product clones from 16 randomly selected thalassemia patients were genetically grouped for TLMV by clone-based sequencing.
Consistent with the primer alignments, the NG054/NG147/133/132 primers detected nine known TTV genotypes, 1, 2, 3, 11, 12, 13, 18, 22, and 23; four recently classified new genotypes (CT44F, CT39F, JT33F, and JT34F) of group 5; and four SENV genotypes (A, B, D, and H), as well as some strains of TLMV (Tables 2 and 3). A substantial proportion of the isolates could not be classified by nucleotide sequence alignment by BLAST. However, phylogenic analysis indicated that the unclassified isolates were related to the same genotypes as mentioned above (data not shown). Among the identified genotypes amplified by these primers, TTV types 1, 11, and SENV-H were predominant in both thalassemia patients and healthy individuals. TTV genotypes 17 (PMV) and SENV-C, -E, -F, and -G were not found among those isolates although they are amplifiable by this set of primers. This is presumably due to their low prevalence in the United Arab Emirates. There were no genotype 21 and/or closely related genetic variants found, in agreement with the sequence alignment (Fig. 1a). To further confirm the genotyping results determined by 5′UTR sequence alignment, sequence similarity or divergence between the representative TTV isolates from GenBank and our isolates was determined using DNAStar. The sequence identity for the representative TTV isolates and our classified isolates ranges from 90.0% to 100% (data not shown), which is consistent with nucleotide identity values within the 5′UTR commonly seen in known isolates of the same genotype, for example, 90.5% between TUS01 and TUPB for TTV genotype 11 and 92.9% between TJN01 and SENV-D for TTV genotype 12. The nucleotide identity values for unclassified strains amplified by NG054/147/133/132 primers and representative TTV isolates were 80.0% to 91.2% (data not shown), suggesting that some TTV isolates amplified by NG primers need to be classified as either genotypic variants or subtypes of TTV.
TABLE 2.
Genotypes of TTV and TTLV detected by NG 054/147/133/132 primers
| Identification no. | Matched strain(s)a | Genotype(s) identified |
|---|---|---|
| Thalassemia patients | ||
| 1 | CHN1 (1), JaCHCTC19 (1), JA20 (1), TUPB (11), TJN02, JT33F, JT34F, CT39F, CT44F, KAV, SENV-H | 1, 11, SENV-H |
| 2 | JA20 (1), CHN1 (1), JaCHC2c (2), TUPB (11) | 1, 2, 11 |
| 3 | JA20 (1), JaCHC2c (2), SENV-H, Th3j, Th3f2, Th3i2, CT39F | 1, 2, SENV-H |
| 4 | JaCHCTC19 (1), Th5d, Th5l | 1 |
| 5 | JA20 (1), TUPB (11), TUS01 (11), Kt-08F (22), SENV-A(P/1C1), JT34F, KC205/1-12G | 1, 11, 22 |
| SENV-A | ||
| 8 | TUS01 (11), Kt-10F (23), K183/1-12G, 624L, Th8h, Th8l | 11, 23 |
| 12 | TUPB (11), SENV-H | 11, SENV-H |
| 23 | TUPB (11) | 11 |
| 26 | JaCHCTC19 (1), JA1 (2), TUPB (11), TYM9 (18), K183/1-12G, Th6a | 1, 2, 11, 18 |
| 34 | TUPB (11), SENV-H, KC033/1-12G, TCHN-C2 | 11, SENV-H |
| 47 | TUPB (11), TLMV(CLC156), TLMV(A14), KC033/1-12G, TCHN-C2 | 11, TLMV |
| 54 | JaCHCTC19 (1), JA20 (1), KC033/1-12G, CT44F | 1 |
| 62 | JA20 (1), CHN1 (1), KC033/1-12G, Th84j | 1 |
| 70 | TUPB (11), TLMV(TGP96) | 11, TLMV |
| 87 | GH1(1), JA20 (1), TLMV(TGP96), KC183/1-12 G | 1, TLMV |
| 89 | JaCHCTC19 (1), JaCHC2c (2), TUS01(11), TJN01 (12), TLMV(CBD203), TT6393 | 1, 2, 11, 12 |
| TLMV | ||
| 90 | TUPB (11), TUS01 (11), TLMV(NLC023) | 11, TLMV |
| 91 | CHN2 (1), JaCHC2c (2), KC010/1-12G, TT6392, TLMV(CLC062) | 1, 2, TLMV |
| 92 | CHN1(1), KC183/1-12G, KC010/1-12G, TLMV(CLC156) | 1, TLMV |
| 93 | CHN1 (1), JA10 (3), KC010/1-12G, TT6393, KC033/1-12G | 1, 3 |
| 94 | TUS01(11), KC033/1-12G, KC183/1-12G, TLMV(CLC062), SENV-H | 11, SENV-H |
| TLMV | ||
| 95 | JA10 (3), TJN01 (12), KC033/1-12G, SENV-A(P/1C1), TLMV(CLC062) | 3, 12, SENV-A |
| TLMV | ||
| 96 | KC183/1-12G, SENV-B, TLMV(NLC026) | SENV-B, TLMV |
| 97 | CHN1 (1), TT6393, SENV-H(TCHN-A) | 1, SENV-H |
| 98 | JA20 (1), TUPB (11), TT6393 | 1, 11 |
| 99 | TCHN-C1, SENV-A(P/1C1), SENV-H(TCHN-A) | SENV-A, -H |
| 100 | TUS01 (11) | 11 |
| 101 | JaCHCTC19(1), JA1(2), JaCHC2c (2), KC183/1-12G, SC098, | 1, 2 |
| 102 | JaCHCTC19 (1), TT6393, KC033/1-12G | 1 |
| 103 | TUPB (11), JT34F, TTM5 | 11 |
| 116 | JA20 (1), JaCHCTC19 (1), CHN2 (1), TUS01 (11), TUPB (11), TJN01 (12), SANBAN (13), SENV-H(TCHN-A) | 1, 11 |
| 12, 13, SENV-H | ||
| 118 | CHN2 (1), TUS01 (11), TT6392, KC033/1-12G | 1, 11 |
| Healthy individuals (controls) | ||
| 76 | TUPB (11), SENV-H, KC010/1-12, KC033-12G | 11, SENV-H |
| 77 | CHN1(1), GH1 (1), KC183/1-12 | 1 |
| 78 | TUPB (11), SENV-H(TCHN-A), EH18B | 11, SENV-H |
| 79 | JA1 (2), JA10 (3), TUPB (11), SENV-D | 2, 3, 11, SENV-D |
| 80 | TUPB (11), SENV-H, CT39F, KCO33/1-12G, NH42a | 11, SENV-H |
| 81 | TUPB (11), TJN02, SENV-H, KC033/1-12, NH43b, NH43f | 11, SENV-H |
| 85 | TUPB (11), CT39F, TCHN-C1, NH45K | 11 |
| 86 | JaCHCTC19 (1), SENV-H, CT39F, CT44F, NH48d | 1, SENV-H |
Numbers in parentheses indicate the corresponding genotypes of the isolates if known. Recently classified genotypes of TTV, discovered by Peng et al. (30), in this study are indicated in boldface.
TABLE 3.
Prevalence of genotypes
| Genotypea | Prevalenceb (%) |
|---|---|
| Thalassemia patients | |
| 1 | 61.2 |
| 2 | 18.8 |
| 3 | 6.2 |
| 11 | 56.3 |
| 12 | 9.3 |
| 13 | 3.1 |
| 18 | 3.1 |
| 22 | 3.1 |
| 23 | 3.1 |
| JT33F | 3.1 |
| JT34F | 3.1 |
| CT39F | 6.2 |
| CT44F | 6.2 |
| SENV-A | 9.4 |
| SENV-B | 3.1 |
| SENV-H | 22.2 |
| TLMV | 31.3 |
| Healthy individuals (controls) | |
| 1 | 25.0 |
| 2 | 12.5 |
| 3 | 12.5 |
| 11 | 75.0 |
| CT39F | 25.0 |
| CT44F | 12.5 |
| SENV-D | 12.5 |
| SENV-H | 62.5 |
Recently classified genotypes of TTV, discovered by Peng et al. (30), in this study are indicated in boldface.
Prevalences of various genotypes of TTV and TLMVs are indicated by the frequencies of their appearance in the PCR product clones derived from 32 thalassemia patients and 8 healthy individuals.
Only TTV genotypes 1, 2, and 3 were found in the samples of 20 randomly selected TT6/7/8/9-amplified PCR products. We did not find other TTV genotypes, such as 4 to 8, that are expected to be detectable by this set of primers, perhaps due to the fact that the prevalence of those genotypes is low. The similarity between our classified isolates and the corresponding representative TTV isolates was 77.2% to 91.2% in the coding region (data not shown), which is similar to previous reports (77.8% to 100%) (29). It appears therefore that the TT6/7/8/9 primer set is group specific. Only genotype 1 (1a and 1b) was found in 30 RD037/038/051/052 primer-amplified PCR products, suggesting that this set of primers is genotype 1 specific. Direct DNA sequencing of PCR products amplified by the primers confirmed the sequence alignment results for the two sets of ORF primers.
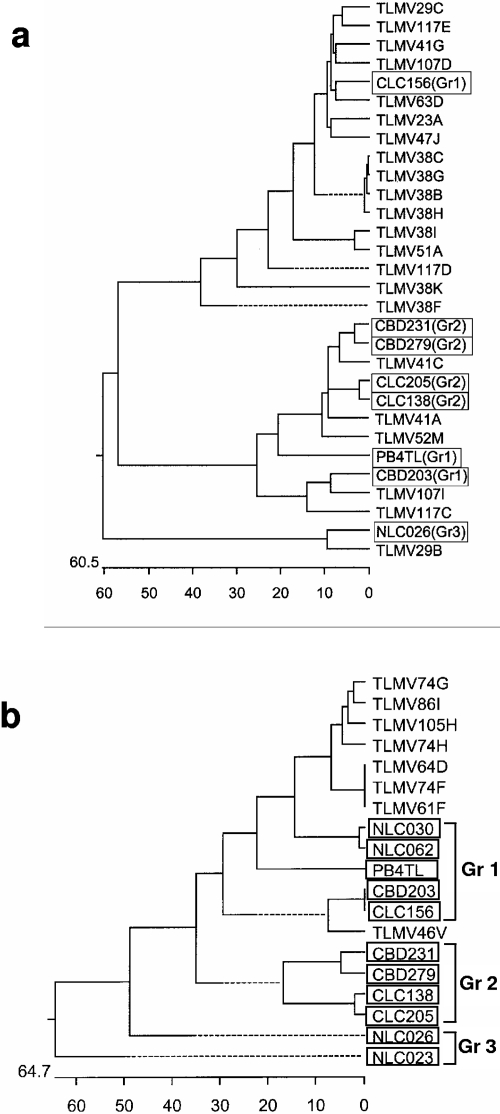
A total of 96 TLMV-S primer product clones derived from 16 thalassemia patients were sequenced, and 22 of them were randomly selected as representatives for phylogenetic analysis. Figure 2a shows that the isolates amplified by TLMV-S primers were genetically distinct from each other and thus distributed among the three known groups of TLMV (Fig. 2a). This has confirmed the ability of TLMV-S primers to detect the three known groups of TLMV. The isolates amplified by TLMV-L primers from this population were not sequenced; thus, classification was not determined for this study. However, we found that TTV 5′UTR primers NG054/147/133/132 were able to amplify some TLMV strains from both thalassemia patients and healthy individuals (Tables 2 and 3). Phylogenetic analysis indicates that these TLMV isolates were genetically closer to group 1 (identity with group 1 ranged from 57.5% to 65.9%), but they appear to be an independent new group (group 4), clustering on a branch of the phylogenetic tree (new isolates are labeled with prefix TLMV in Fig. 2b).
FIG. 2.
Phylogenetic trees constructed using published sequences of known TLMV isolates in boxes that are distributed among groups (Gr1 to Gr3) and sequences of cloned PCR products amplified by TLMV-S primers (a) and by NG057/147/133/132 primers (b) from thalassemia patients. Our TLMV isolates are indicated with prefix TLMV, and the GenBank TLMV isolates are shown in boxes with the group number labeled in brackets.
Detection of TTV and TTLVs using different sets of primers.
To determine the ability of each set of the primers to detect TTV and TTLVs, the five primer sets described above were tested on a panel of 118 thalassemia patients and 200 healthy individuals. Using the three nested TTV primer sets RD051/052/037/038, TT6/7/8/9, and NG054/147/133/132, TTV was positive in 45.8%, 76.3%, and 90.6% of thalassemia patients, respectively (Table 4). Using a combination of RD051/052/037/038 and TT6/7/8/9 primers, the detection rate was slightly increased, to 78.8%, in comparison with the detection by TT6/7/8/9 primers alone (76.3%) due to the fact that three samples that were negative with TT6/7/8/9 primers were positive with RD051/052/037/038 primers. This suggests that some TTV genotype 1 variants may not be amplifiable by TT6/7/8/9 primers. The detection rate was not changed when using NG054/147/133/132 combined with RD051/052/037/038 and TT6/7/8/9 primers compared with use of NG054/147/133/132 primers alone. This would appear to confirm our hypothesis that NG primers are able to detect most genotypes. For TLMV, 82.2% of thalassemia patients were positive using TLMV-S1/TLMV-S2 primers. All of the thalassemia patients (100%) were positive for TTV and/or TLMV when RD051/052/037/038, TT6/7/8/9, NG054/147/133/132, and TLMV-S primers were used in combination. For healthy individuals, the same pattern of detection for these primers was observed. As expected, the prevalence of the viruses was lower. Only one-half of healthy individuals (56.2%) were positive even when using multiple sets of primers (NG054/147/133/132, TLMV-S, and TLMV-L). The detection rate of TLMV-S primers (30.5%, 32/105) was higher than that of TLMV-L primers (21.9%, 23/105) for detecting TLMV in healthy individuals. However, among the 105 samples tested, four that were negative with TLMV-S primers were positive with TLMV-L primers. This indicated that both sets of primers are needed for effective detection of TLMV.
TABLE 4.
Detection of TTV and TTLVs with various sets of primers in thalassemia patients and healthy individuals
| Primer set | % Detection in samples from: |
|
|---|---|---|
| Thalassemiacs | Healthy individuals | |
| RD | 45.8 (54/118) | NDa |
| TT | 76.3 (90/118) | ND |
| RD/TT | 78.8 (93/118) | ND |
| NG | 90.6 (107/118) | 35.5 (67/200) |
| RD/TT/NG | 90.6 (107/118) | ND |
| TLMV-L | ND | 21.9 (23/105) |
| TLMV-S | 82.2 (97/118) | 30.5 (32/105) |
| NG/TLMV-S | ND | 51.4 (54/105) |
| RD/TT/NG/TLMV-S | 100.0 (100/100) | ND |
| NG/TLMV-S/TLMV-L | ND | 56.2 (58/105) |
ND, not done.
DISCUSSION
Although the genomes of TTV and TTLVs are extremely heterogeneous, sequence conservation is maintained for the 5′UTR localized downstream from the TATA box. This region is even conserved between TTV and some TLMVs, as well as TTVs from different species (20). In particular, for the purposes of PCR detection, it is important that all TTVs and TTLVs from humans and various animals possess two common 15-nucleotide sequences in their 5′UTRs, CGAATGGCTGAGTT and AGGGGCAATTCGGGC, that are the core regions of the NG 133 and NG 147 primer sequence binding sites (26, 29). Using both predictive theoretical modeling and practical testing, we demonstrated that it is possible to detect all known groups and genotypes of TTV and TTLVs including eight genotypes of SENV and PMV, and three known groups plus a new group of TLMV can be detected by using four sets of PCR primers combined with a genotype 21-specific set of primers. We found that using clone-based DNA sequencing made it possible to identify and quantify low-frequency mixed genotypes of TTV and TTLVs that were undetectable when using direct DNA sequencing of PCR products. The results generated by direct DNA sequencing of PCR products from thalassemia patients showed only the sequence representative of the predominate genotype and often were unable to provide clear readable DNA sequences due to mixed genotypes that exist in those samples, especially when 5′UTR primers that can result in the amplification of multiple genotypes were used (data not shown).
The extremely high worldwide prevalence of chronic TTV and TTLV infections presents a challenge to determining the natural history and pathogenic potential of these viruses. Furthermore, it is not possible to identify a specific genotype(s) that may be more pathogenic than others using a routine PCR method with one or two sets of primers due to the high rate of mixed infections by multiple genotypes (1, 12). It is thus crucial to develop a molecular diagnostic system that is not only capable of detecting and quantifying the full spectrum of existing TTV and TTLV genotypes and groups but that can also identify and quantify specific groups and genotypes of the viruses existing in individuals. It is known that the titers of TTV DNA vary depending on the chosen primers where viral loads determined by PCR with 5′UTR primers are 10 to 100 times higher than those with N22 primers (29). In this study, we have developed a detection system with multiple sets of primers that should be more accurate and specific in testing viral load for all prevalent genotypes or specific genotypes and groups of TTV and TTLVs.
Although the life cycle of the viruses is poorly understood, self-limited and persistent infections have been observed in both humans and chimpanzees (2, 24). It has been estimated that 1010 to 1011 virions are produced and cleared per day in chronically infected individuals (17). The ability of patients to clear TTV infections is currently unknown. We found that 100% of thalassemia patients were infected with TTV and TLMV; however, we also found that approximately 10% (11/118) of these patients aged 4 to 20 years (average, 9.45 years) were PCR negative when using the TTV primers RD037/038/051/052, NG054/147/133/132, and TT6/7/8/9, which are capable of detecting many common TTV genotypes. This suggests that TTV infection can be cleared or become undetectable, because this group of patients have been repeatedly exposed to multiple genotypes of virus present in blood transfusions (13 to 18 times/year) administered over a period of approximately 10 years. However, further evidence from epidemiological and immunological studies is needed to confirm that such patients have been infected by specific TTVs and TTLVs during the period of transfusion treatment.
In the present study, we focused on the establishment of a molecular detection system as an appropriate tool for assessment of TTV and TTLV pathogenesis and prevalence at the genotypic level. Several other important aspects of clinical virology such as viral persistence and reactivation, quasispecies shifting, viral immune escape, and host immune response should be addressed at both the genotypic and quasispecies levels in future investigations.
Acknowledgments
Financial support was provided by Bayer-Canadian Blood Services-Hema-Quebec Partnership Fund.
Informed consent was obtained from blood donors and thalassemia patients.
REFERENCES
- 1.Al Moslih, M. I., R. O. Abuodeh, and Y. W. Hu. 2004. Detection and genotyping of TT virus in healthy and subjects with HBV or HCV in different populations in the United Arab Emirates. J. Med. Virol. 72:502-508. [DOI] [PubMed] [Google Scholar]
- 2.Bendinelli, M., M. Pistello, F. Maggi, C. Fornai, G. Freer, and M. L. Vatteroni. 2001. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin. Microbiol. Rev. 14:98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J. Cantaloube, P. de Micco, and X. de Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82:379-383. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. K., K. W. Chik, K. F. To, C. K. Li, M. Hui, M. M. Shing, P. M. Yuen, J. S. Tam, and A. F. Cheng. 2001. Clearance of TT virus after allogeneic bone marrow transplantation. J. Pediatr. Hematol. Oncol. 23:57-58. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B. P., M. G. Rumi, M. Colombo, Y. H. Lin, L. Ramaswamy, J. Luna, J. K. Liu, D. Prati, and P. M. Mannucci. 1999. TT virus is present in a high frequency of Italian hemophilic patients transfused with plasma-derived clotting factor concentrates. Blood 94:4333-4336. [PubMed] [Google Scholar]
- 6.Cossart, Y. 2000. TTV—a virus searching for a disease. J. Clin. Virol. 17:1-3. [DOI] [PubMed] [Google Scholar]
- 7.Devalle, S., and C. Niel. 2004. Distribution of TT virus genomic groups 1-5 in Brazilian blood donors, HBV carriers, and HIV-1-infected patients. J. Med. Virol. 72:166-173. [DOI] [PubMed] [Google Scholar]
- 8.Fiordalisi, G., M. Bonelli, P. Olivero, D. Primi, L. Vaglini, S. R. L. Diasorin, S. Mattioli, F. Bonelli, A. DalCorso, G. Mantero, L. Giovanni, and A. Sottini. May 2000. Identification of SENV genotypes. Italian patent WO1999EP08566(WO0028039).
- 9.Gallian, P., Y. Berland, M. Olmer, D. Raccah, P. de Micco, P. Biagini, S. Simon, D. Bouchouareb, C. Mourey, C. Roubicek, M. Touinssi, J. F. Cantaloube, B. Dussol, and X. de Lamballerie. 1999. TT virus infection in French hemodialysis patients: study of prevalence and risk factors. J. Clin. Microbiol. 37:2538-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallett, R. L., J. P. Clewley, F. Bobet, P. J. McKiernan, and C. G. Teo. 2000. Characterization of a highly divergent TT virus genome. J. Gen. Virol. 81:2273-2279. [DOI] [PubMed] [Google Scholar]
- 11.Hohne, M., T. Berg, A. R. Muller, and E. Schreier. 1998. Detection of sequences of TT virus, a novel DNA virus, in German patients. J. Gen. Virol. 79:2761-2764. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Y. W. 2003. TT virus (TTV) and TTV-like viruses (TTLVs) in blood donors and blood transfusion dependent patients with thalassemia. In Proceedings of Cambridge Health Institute’s 9th Annual Meeting for Blood Safety, Washington, D.C. Cambridge Healthtech Institute, Newton Upper Falls, Mass.
- 13.Hu, Y. W., E. Balaskas, G. Kessler, C. Issid, L. J. Scully, D. G. Murphy, A. Rinfret, A. Giulivi, V. Scalia, and P. Gill. 1998. Primer specific and mispair extension analysis (PSMEA) as a simple approach to fast genotyping. Nucleic Acids Res. 26:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanda, Y., S. Chiba, Y. Tanaka, M. Kami, T. Saito, T. Asai, K. Izutsu, K. Yuji, S. Ogawa, H. Honda, K. Mitani, Y. Yazaki, and H. Hirai. 1999. TT virus in frequently transfused patients. Am. J. Med. 106:116-117. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, M., K. Chayama, Y. Arase, A. Tsubota, Y. Suzuki, I. Koida, S. Saitoh, N. Murashima, K. Ikeda, H. Koike, M. Hashimoto, and H. Kumada. 1999. Prevalence of TT virus before and after blood transfusion in patients with chronic liver disease treated surgically for hepatocellular carcinoma. J. Gastroenterol. Hepatol. 14:358-363. [DOI] [PubMed] [Google Scholar]
- 16.Maggi, F., M. Pifferi, C. Fornai, E. Andreoli, E. Tempestini, M. Vatteroni, S. Presciuttini, S. Marchi, A. Pietrobelli, A. Boner, M. Pistello, and M. Bendinelli. 2003. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J. Virol. 77:2418-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggi, F., M. Pistello, M. Vatteroni, S. Presciuttini, S. Marchi, P. Isola, C. Fornai, S. Fagnani, E. Andreoli, G. Antonelli, and M. Bendinelli. 2001. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J. Virol. 75:11999-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto, A., A. E. Yeo, J. W. Shih, E. Tanaka, K. Kiyosawa, and H. J. Alter. 1999. Transfusion-associated TT virus infection and its relationship to liver disease. Hepatology 30:283-288. [DOI] [PubMed] [Google Scholar]
- 19.Mushahwar, I. K. 2000. Recently discovered blood-borne viruses: are they hepatitis viruses or mere endosymbionts? J. Med. Virol. 62:399-404. [DOI] [PubMed] [Google Scholar]
- 20.Niel, C., and E. Lampe. 2001. High detection rates of TTV-like mini virus sequences in sera from Brazilian blood donors. J. Med. Virol. 65:199-205. [PubMed] [Google Scholar]
- 21.Nishiguchi, S., M. Enomoto, S. Shiomi, M. Tanaka, K. Fukuda, A. Tamori, T. Tanaka, T. Takeda, S. Seki, Y. Yano, S. Otani, and T. Kuroki. 2000. TT virus infection in patients with chronic liver disease of unknown etiology. J. Med. Virol. 62:392-398. [DOI] [PubMed] [Google Scholar]
- 22.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 23.Oguchi, T., E. Tanaka, K. Orii, M. Kobayashi, K. Hora, and K. Kiyosawa. 1999. Transmission of and liver injury by TT virus in patients on maintenance hemodialysis. J. Gastroenterol. 34:234-240. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto, H., and M. Mayumi. 2001. TT virus: virological and genomic characteristics and disease association. J. Gastroenterol. 36:519-529. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto, H., T. Nishizawa, N. Kato, M. Ukita, H. Ikeda, H. Iizuka, Y. Miyakawa, and M. Mayumi. 1998. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol. Res. 10:1-16. [Google Scholar]
- 26.Okamoto, H., T. Nishizawa, A. Tawara, Y. Peng, M. Takahashi, J. Kishimoto, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Species-specific TT viruses in humans and nonhuman primates and their phylogenetic relatedness. Virology 277:368-378. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto, H., T. Nishizawa, M. Ukita, M. Takahashi, M. Fukuda, H. Iizuka, Y. Miyakawa, and M. Mayumi. 1999. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology 259:437-448. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto, H., M. Takahashi, N. Kato, S. Fukuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Sequestration of TT virus of restricted genotypes in peripheral blood mononuclear cells. J. Virol. 74:10236-10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto, H., M. Takahashi, T. Nishizawa, M. Ukita, M. Fukuda, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1999. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology 259:428-436. [DOI] [PubMed] [Google Scholar]
- 30.Peng, Y. H., T. Nishizawa, M. Takahashi, T. Ishikawa, A. Yoshikawa, and H. Okamoto. 2002. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch. Virol. 147:21-41. [DOI] [PubMed] [Google Scholar]
- 31.Prati, D., Y. H. Lin, M. C. De, J. K. Liu, E. Farma, L. Ramaswamy, A. Zanella, H. Lee, P. Rebulla, J. P. Allain, G. Sirchia, and B. Chen. 1999. A prospective study on TT virus infection in transfusion-dependent patients with beta-thalassemia. Blood 93:1502-1505. [PubMed] [Google Scholar]
- 32.Simmonds, P. 1998. Transfusion virology: progress and challenges. Blood Rev. 12:171-177. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi, K., Y. Iwasa, M. Hijikata, and S. Mishiro. 2000. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 145:979-993. [DOI] [PubMed] [Google Scholar]
- 34.Takayama, S., S. Yamazaki, S. Matsuo, and S. Sugii. 1999. Multiple infection of TT virus (TTV) with different genotypes in Japanese hemophiliacs. Biochem. Biophys. Res. Commun. 256:208-211. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, H., H. Okamoto, P. Luengrojanakul, T. Chainuvati, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1998. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J. Med. Virol. 56:234-238. [DOI] [PubMed] [Google Scholar]
- 36.Vasconcelos, H. C., M. Cataldo, and C. Niel. 2002. Mixed infections of adults and children with multiple TTV-like mini virus isolates. J. Med. Virol. 68:291-298. [DOI] [PubMed] [Google Scholar]
- 37.Worobey, M. 2000. Extensive homologous recombination among widely divergent TT viruses. J. Virol. 74:7666-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]