Abstract
We developed a nested reverse transcription-PCR (nRT-PCR) for the detection of noroviruses in stools, using random primers for RT, the JV12/JV13 primer pair in the first round of nPCR, and a set of nine inner primers for the second, comprising the reverse sequences of primers SR46, SR48, SR50, and SR52, and five novel oligonucleotide sequences (113-1, 113-2, 115-1, 115-2, and 115-3). The specificity of the nRT-PCR was confirmed by testing 61 stools containing enteric viruses other than noroviruses. In comparative assays on either stools or RNA dilutions from two genogroup I and three genogroup II (GII) norovirus-positive samples, nRT-PCR was always at least as sensitive as RT-PCR and Southern hybridization. With some of the samples tested, the increase in sensitivity was 10-fold or higher. For GII viruses, the detectable range of nRT-PCR was estimated to be 8.4 × 104 to 2 RNA viral particles. When used on 85 stools from pediatric patients with acute gastroenteritis negative for viruses by electron microscopy and cell culture, the nRT-PCR detected norovirus in 19 samples (22.3%), while it failed to detect one reference RT-PCR-positive sample containing a Desert Shield strain. Sixteen of the 19 nRT-PCR-positive samples gave concordant results with reference RT-PCR and Southern hybridization, and all with sequence analysis. Partial sequencing of the polymerase region revealed that from January to April 2000 all GII strains except two (Rotterdam- and Leeds-like viruses) formed a tight cluster related to Hawaii virus. The nRT-PCR described could prove suitable for large epidemiological studies and for specialized clinical laboratories performing routine molecular testing.
Human caliciviruses have been considered one of the most common causes for both nonbacterial acute gastroenteritis outbreaks and sporadic cases in adults and children worldwide (7, 9, 29, 34, 38). The family Caliciviridae can be divided into four genera: “Norwalk-like virus,” recently renamed Norovirus, with the prototype strain Norwalk virus (NV); Sapovirus, formerly known as “Sapporo-like virus,” more often responsible for sporadic cases of gastroenteritis in humans; and the genera Lagovirus and Vesivirus, which naturally infect animals only (15, 25).
Norovirus infection generally has acute onset characterized by nausea, vomiting, abdominal pains, diarrhea, headache, and also fever and myalgia. Vomiting is more common among children, whereas diarrhea is more likely to predominate among adults. Symptoms generally last 1 to 3 days, and dehydration remains the most common complication of the disease, especially in young children and elderly patients (8).
The frequent asymptomatic infections, the stability of the virus in the environment, and the lack of permanent immunity in the infected subject favor the widespread outbreaks of gastroenteritis associated with person-to-person transmission through spattering and aerosol, as well as with ingestion of contaminated water and food, such as oysters (shellfish) and vegetables (3, 4, 6, 11, 21, 31). Epidemic outbreaks often occur in enclosed settings such as nursing homes, day care centers, schools, cruise ships, and hospitals (10, 16, 22, 26).
The primary reason for the underestimation of the disease burden of norovirus infection has been the difficulty in developing and applying sensitive, easy-to-perform diagnostic assays. The virus cannot be cultivated from clinical samples, and no animal models are available to study norovirus. Until recently, the primary diagnostic methods were electron microscopy (EM), which is a relatively insensitive technique and requires expertise in recognizing the small viral particles (12), and immunoelectron microscopy, which needs antisera or monoclonal antibodies available only in reference laboratories (2).
Only in the past decade has the knowledge of the nucleotide sequence and the genomic organization of several strains of noroviruses led to the development of nucleic acid amplification methods through reverse transcription-PCR (RT-PCR) (1, 14, 20, 24).
Norovirus has a single-stranded, positive-sense RNA of 7.6 kb coding for three open reading frames (ORF1 to -3). Based on the sequence of the RNA polymerase region (ORF1), the comparison of the complete genome sequences of different strains of human noroviruses has shown that they can be divided into at least two genogroups: genogroup I (GI), with Norwalk virus as the prototype strain, and GII, with Snow Mountain virus and Hawaii virus as the prototype strains. Both genogroups can be further divided into several genetic clusters or antigenic types (5, 15).
The high genetic polymorphism of noroviruses is responsible for the difficulties encountered in the choice of primer pairs suited to the development of a molecular diagnostic method with good sensitivity and specificity. RT-PCR, using generic primers and Southern hybridization or sequence analysis of the PCR products, is currently the most sensitive and commonly applied method for the detection of noroviruses (1, 13, 19). However, this method is time-consuming and costly for epidemiological and diagnostic purposes when large numbers of specimens are examined, especially during an epidemic.
In this report, we describe a nested RT-PCR (nRT-PCR) assay for reliable detection of the genetically diverse noroviruses in stool specimens. For this purpose, in addition to published primers, we have also designed a new broadly reactive primer set on the basis of nucleotide sequences within the RNA polymerase region of a variety of norovirus strains.
MATERIALS AND METHODS
Stool samples.
A panel of 85 stool samples was collected from 83 children (median age, 2 years 7 months, ranging from 51 days to 12 years 11 months, for the 71 patients whose ages were available) admitted with acute gastroenteritis to the Development Age Department of the Major Hospital of Parma. These samples were selected out of 185 stool samples (belonging to 177 patients) submitted to the Unit of Virology of the Department of Pathology and Laboratory Medicine for diagnostic purposes in the period January to April 2000, covering the norovirus epidemic period of the year in temperate countries (26, 28). The samples were selected because they had proved negative for viral agents by EM and conventional cell culture. A second panel of 61 specimens positive for viruses other than noroviruses served as negative controls in the specificity testing. This panel included rotavirus (19 samples); astrovirus (5 samples); poliovirus types 1 (1 sample), 2 (3 samples), and 3 (1 sample); coxsackievirus types B2, B4, and B5 (1 sample each); echovirus types 1 (1 sample), 24 (2 samples), 25 (1 sample), and 30 (5 samples); adenovirus (19 samples); and cytomegalovirus (1 sample). Norovirus stock aliquots, prepared from five fecal samples containing different norovirus genotypes (Malta and Sindlesham genotypes for GI and Bristol, Leeds, and GIIb genotypes for GII) as determined by sequence analysis, were used for sensitivity testing. All specimens were prepared in 10% suspension in phosphate-buffered saline (PBS; pH 7.2), clarified by low-speed centrifugation (3,100 × g), and stored at −20°C until further processing.
EM.
EM was performed essentially as previously described using standard techniques (40). Briefly, for routine EM investigation the suspension was concentrated with hygroscopic polyacrylamide absorbent gel (0.1 g in 550 μl of clarified stool suspension for 15 to 20 min; Sigma); a drop (4 to 4.5 μl) of the suspension was incubated for 25 s on a 400-mesh Formvar carbon-coated grid, then stained with 2% phosphotungstic acid (pH 6.5) for 25 s. To increase the sensitivity, RT-PCR-positive stools with sufficient volume (2 to 2.5 ml) were ultracentrifuged for 1 h at 25,000 rpm by using a Beckman SW41 rotor at 4°C (Beckman Coulter). The supernatant was discarded, and the pellet was resuspended in 50 μl of PBS (40- to 50-fold concentration factor), then stained as described above. For quantification of norovirus particles by EM and latex beads, fecal suspensions were used after 200- or 50-fold concentration, depending on the quality of the fecal sample.
Quantification of norovirus particles by EM and latex beads.
The concentration of norovirus virions of Bristol, Leeds, and GIIb genotypes used in sensitivity testing was determined by the “loop drop method” with some modifications (39). Briefly, 110-nm-diameter latex beads (Polysciences, Inc.) of known concentration (4.73 × 1013/ml; at final dilutions of 1:300 and 1:600 in the mixture for grids) were mixed (1:1:1) with ultracentrifuged clarified fecal suspension (200- or 50-fold concentration factor) and 0.5% bovine serum albumin. A drop of this mixture was placed onto a Formvar-coated grid and stained as previously described. The concentration of norovirus virions (v) in the original fecal suspension was determined by comparison with the number of latex beads (LB) counted (LBC) per grid and the number of virions counted (VPC) per grid, using the equation adapted from that described by Mittereder et al. (27), v = (VPC/LBC) × (4.73 × 1013 LB ml−1/LBd) × 3/μ, where LBd is the dilution factor of latex beads in the mixture for the grid and μ is the concentration factor of the fecal sample after ultracentrifugation. In each grid, the counting of virions and latex beads continued until 1,000 latex beads had been counted.
Reference positive materials.
RNA of NV (GI.1; kindly supplied by the Istituto Zooprofilattico Sperimentale of Lombardia and Emilia-Romagna, Brescia, Italy) and fecal suspensions containing Malta (GI.4), Sindlesham (GI.6), Winchester (GI.7), Hawaii (GII.1), Melksham (GII.2), Bristol (GII.4), Leeds (GII.7), and GIIb (GII. not assigned) viruses, characterized by sequencing and available from our viral strain collection, were used as reference positive materials.
RNA extraction.
Viral RNA was rapidly extracted from 250 μl of stool suspension and resuspended in 20 μl of nuclease-free water (equivalent to 200 μl of stool suspension; 10-fold concentration), using the EXTRAgen kit (Amplimedical, Bioline Division group) according to the manufacturer's instructions. The method is based on the lysis and denaturation activity of guanidinium hydrochloride.
Nested RT-PCR.
RT was carried out in a final volume of 25 μl by using random primer RT-Kit random primers (Amplimedical), starting from 10 μl of extracted RNA. The RT was performed at 37°C for 1 h 30 min, followed by inactivation at 96°C for 5 min. For the nPCR, we chose primers specific to the polymerase gene of both GI and II noroviruses (Table 1). The first round was performed with primers JV12 and JV13 (36) and the second round with nine inner primers, four of which were the reverse sequences of SR46, SR48, SR50, and SR52, described by Ando et al. in 1995 (1). The other five primers were selected for their broad-range reactivity against norovirus based on the sequence alignment using FASTA (30) and CLUSTAL W (35), between the reference sequence of NV (M87661) and 280 sequences of both GI and GII norovirus strains present in the GenBank database. This new set of primers shares two primers with those described by Yuen et al. (41) with some modifications: a 2-nucleotide deletion in the 5′ end and an additional nucleotide in the 3′ end. All primers used in this study were synthesized by the phosphoramidite technique.
TABLE 1.
Sequences and locations of primers used in this study
| Primer | Polaritya | Sequence 5′-3′ | Location (nucleotides)b | Reference or source |
|---|---|---|---|---|
| JV12 | + | ATA CCA CTA TGA TGC AGA TTA | 4552-4572 | 36 |
| JV13 | − | TCA TCA TCA CCA TAG AAA GAG | 4878-4858 | 36 |
| 113-1 | + | GTG CAG CCA TGG AGG TAA TG | 4611-4630 | This study |
| 113-2 | + | CAG AAT CCT TCT CCA TCA TG | 4611-4630 | This study |
| 115-1c | + | GCA GCA CTT GAA ATC ATG GT | 4613-4632 | This study |
| 115-2c | + | GCA GCC CTA GAA ATC ATG GT | 4613-4632 | This study |
| 115-3 | + | GAG TCC TTA GCA ATC ATG TG | 4613-4632 | This study |
| revSR46d | − | CCA GTG GGC GAT GGA ATT CCA | 4786-4766 | 1 |
| revSR48d | − | CCA GTG ATT TAT GCT GTT CAC | 4786-4766 | 1 |
| revSR50d | − | CCA GTG GTT TAT ACT GTT CAC | 4786-4766 | 1 |
| revSR52d | − | CCA ATG GTT TAT ACT GTT CAC | 4786-4766 | 1 |
+, virus sense; −, virus antisense.
Corresponds to the equivalent location within the Norwalk virus genomic sequence (accession no. M87661).
Corresponds to some modifications of NV4611 primer described by Yuen et al. in 2001 (41).
Reverse primer obtained from the original primer sequence of SR46, SR48, SR50, or SR52.
Briefly, for the nPCR 10 μl of cDNA was added to 40 μl of the first-round mixture, which contained 50 mM KCl, 10 mM Tris HCl (pH 8.4), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.1% Triton X-100, 0.5 μM primers JV12 and JV13, and 1 U Taq polymerase (DyNAzymeII; Finnzymes). The first-round PCR cycling was performed in a Progene thermal cycler (Techne) as follows: 94°C for 2 min; 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. Then 1 μl of the first-round product was added to 49 μl of reaction mixture containing the nested primers instead of the outer primers. After an analogous series of 30 cycles with an annealing temperature of 55°C, amplification products were electrophoresed at 120 V for 35 min in 4% agarose gel and stained with ethidium bromide. Bands were visualized on a UV transilluminator, and gel images were digitally recorded with GelAnalyzer (Clonit). The selected inner primers yielded a product of 174 or 176 bp.
Internal RNA standard control.
To verify the suitability of stool samples for amplification (presence of cDNA and absence of inhibitors), 10 μl of plasmid RNA from phage MS2 (CPE-RNA; 2,000 copies/μl; Amplimedical) was added to each sample during extraction, reverse transcribed by random RT, and coamplified with specific primers (RECK; Amplimedical) at a final concentration of 0.1 μM together with the viral target by norovirus nRT-PCR. The amplified product specific to the internal RNA standard control had an expected size of 610 bp.
RT-PCR and Southern hybridization.
The RT-PCR and subsequent hybridization by Southern blotting previously described by Vinjé et al. (36, 38) were used as the method of reference. Amplification products of the expected size of 327 bp obtained with the primer pair JV12/JV13 were confirmed by Southern hybridization using a pool of 5′-biotin-labeled probes.
Sequencing.
Agarose gel electrophoresis bands of the amplification products were purified with QIAquick gel extraction (QIAGEN), and both strands were sequenced using dye terminator chemistry on an automated sequencer (ABI Prism 373 DNA sequencer; Applied Biosystems). Database searches were conducted in GenBank using the BLAST program, and/or in the Food-borne Viruses in Europe database (EU contract QLK1-CT-1999-00594).
RESULTS
Specificity of nRT-PCR.
The specificity of the primers used in the newly developed nRT-PCR was assessed by demonstrating the absence of amplification from 61 stool specimens containing enteric viruses different from noroviruses. Conversely, an nRT-PCR product of 174 or 176 bp was obtained by using norovirus reference RNAs. All samples devoid of norovirus-specific PCR bands were able to produce the PCR band from the internal control, indicating the absence of reaction inhibitors.
Analytical sensitivity of nRT-PCR.
To assess the analytical sensitivity of nRT-PCR, RNAs from an aliquot of each norovirus stock preparation (see Materials and Methods) were 10-fold serially diluted and assayed by nRT-PCR and RT-PCR and Southern hybridization. The nRT-PCR method was as sensitive as RT-PCR and Southern hybridization in detecting GI norovirus genotypes (Table 2). In the case of GII strains, nRT-PCR showed a 10-fold-higher sensitivity for the detection of Leeds and GIIb genotypes and a 100-fold-higher detection limit for Bristol genotype. Less-consistent positive dilutions (showing only one positive out of three tests) were omitted from comparisons. These findings indicate that the detection range of nRT-PCR was 2.1 × 105 to 1.45 × 103 particles/ml versus 3.9 × 106 to 1.45 × 104 particles/ml by RT-PCR and Southern hybridization.
TABLE 2.
Comparison between nRT-PCR and conventional RT-PCR and Southern hybridization for detection of norovirus RNAs serially diluted after extraction
| Genotype (cluster classification) | Virus stock concn (particles/ml)a | Dilution of test RNA | nRT-PCR |
RT-PCR/SHg for: |
|||
|---|---|---|---|---|---|---|---|
| norovirus |
ICb (%) | norovirus |
|||||
| No. of positive samples | % | No. of positive samples | % | ||||
| Malta (GI.4) | Lc | 100 | 3 | 100 | 100 | 3 | 100 |
| 10−1 | 0 | 0 | 66 | 0 | 0 | ||
| Sindlesham (GI.6) | L | 10−2 | 3 | 100 | 0 | 3 | 100 |
| 10−3 | 0 | 0 | 0 | 0 | 0 | ||
| Bristol (GII.4) | 3.9 × 108 | 10−2 | 3 | 100 | 100 | 3 | 100 |
| 10−3 | 2 | 66 | 0 | 0 | 0 | ||
| 10−4 | 2 | 66 | 0 | 0 | 0 | ||
| 10−5 | 0 | 0 | 0 | 0 | 0 | ||
| Leeds (GII.7) | 2.1 × 108 | 10−2 | 3 | 100 | 33 | 3 | 100 |
| 10−3 | 3 | 100 | 0 | 1 | 33 | ||
| 10−4 | 0 | 0 | 0 | 0 | 0 | ||
| GIIb (GII.NAd) | 1.45 × 108 | 10−4 | 3 | 100 | 33 | 3 | 100 |
| 10−5 | 3 | 100 | 0 | 0 | 0 | ||
| 10−6 | 1 | 33 | 0 | 0 | 0 | ||
| 10−7 | 0 | 0 | 0 | 0 | 0 | ||
| Negative controle | 0 | 100 | 0 | 0 | 100 | NDf | ND |
| 10−1 | 0 | 0 | 66 | ND | ND | ||
| 10−2 | 0 | 0 | 66 | ND | ND | ||
| 10−3 | 0 | 0 | 33 | ND | ND | ||
| 10−4 | 0 | 0 | 0 | ND | ND | ||
Viral concentrations in each norovirus genotype stock were estimated by EM and latex beads. See Materials and Methods.
IC, internal RNA standard control.
L, lower than electron microscopy sensitivity (absence of visible calicivirus-like particles on norovirus genotype stock).
GII.NA, GII classification not assigned.
Norovirus-negative fecal specimen subjected to RNA purification.
ND, not done.
RT-PCR/SH, RT-PCR and Southern hybridization.
Detection limit for nRT-PCR on stools.
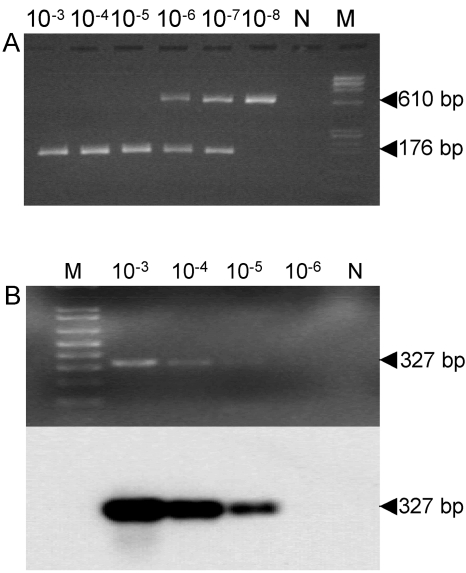
In order to evaluate the detection limit of nRT-PCR on stool specimens, the RNAs from three separate sets of serial 10-fold dilutions of each norovirus stock preparation were tested in comparison with reference RT-PCR and Southern hybridization. The overall results for each dilution are listed in Table 3, and a representative norovirus detection by nRT-PCR compared to RT-PCR and Southern hybridization in a 10-fold dilution series is shown in Fig. 1.
TABLE 3.
Comparison between nRT-PCR and conventional RT-PCR and Southern hybridization for detection of norovirus genomic RNA in stoolsd
| Genotype (cluster classification) | Virus stock concn (particles/ml)a | Dilution of stools | nRT-PCR |
RT-PCR/SH |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Result for series: |
% Positive | Result for series: |
% Positive | |||||||
| a | b | c | a | b | c | |||||
| Malta (GI.4) | Lb | 100 | + | + | + | 100 | + | + | + | 100 |
| 10−1 | + | + | + | 100 | − | + | + | 66 | ||
| 10−2 | − | − | + | 33 | − | − | + | 33 | ||
| 10−3 | − | − | − | 0 | − | − | − | 0 | ||
| Sindlesham (GI.6) | L | 10−1 | + | + | + | 100 | + | + | + | 100 |
| 10−2 | + | + | + | 100 | + | − | − | 33 | ||
| 10−3 | − | − | + | 33 | − | − | − | 0 | ||
| 10−4 | − | − | − | 0 | − | − | − | 0 | ||
| Bristol (GII.4) | 3.9 × 108 | 10−3 | + | + | + | 100 | + | + | + | 100 |
| 10−4 | + | + | + | 100 | + | − | + | 66 | ||
| 10−5 | − | + | + | 66 | + | − | + | 66 | ||
| 10−6 | − | − | + | 33 | − | − | − | 0 | ||
| 10−7 | − | − | + | 33 | − | − | − | 0 | ||
| 10−8 | − | − | − | 0 | NTc | NT | NT | 0 | ||
| Leeds (GII.7) | 2.1 × 108 | 10−2 | + | + | + | 100 | + | + | + | 100 |
| 10−3 | − | − | + | 33 | − | − | − | 0 | ||
| 10−4 | − | − | + | 33 | − | − | − | 0 | ||
| 10−5 | − | − | − | 0 | NT | NT | NT | 0 | ||
| GIIb (GII.NA) | 1.45 × 108 | 10−4 | + | + | + | 100 | + | + | + | 100 |
| 10−5 | + | + | + | 100 | − | − | − | 0 | ||
| 10−6 | − | − | + | 33 | − | − | − | 0 | ||
| 10−7 | − | − | − | 0 | NT | NT | NT | 0 | ||
Viral concentrations in each norovirus genotype stock were estimated by EM and latex beads. See Materials and Methods.
L, lower than electron microscopy sensitivity (absence of visible calicivirus-like particles on norovirus genotype stock).
NT, not tested.
RT-PCR/SH and GII.NA are as defined for Table 2.
FIG. 1.
Representative norovirus detection by nRT-PCR compared to RT-PCR and Southern hybridization in a 10-fold dilution series from a positive stool sample containing a Bristol genotype at a concentration of 3.9 × 108 virus particles/ml and the internal RNA standard control. (A) Agarose gel electrophoresis of nRT-PCR products from 10−3 to 10−8 dilutions. (B) Results of ethidium bromide staining (top) and corresponding Southern hybridization (bottom) of conventional RT-PCR products from 10−3 to 10−6 dilutions. N, zero RNA; M, DNA molecular size markers.
In the case of GI genotypes, nRT-PCR provided detection limits which were similar to, or more sensitive (10-fold or more) than, the reference method.
With regard to GII genotypes, the detection limit of nRT-PCR ranged from 2.1 × 106 virus particles/ml (corresponding to 8.4 × 104 virus particles/reaction for Leeds strain) down to 39 virus particles/ml (corresponding to 1.56 virus particles/reaction for Bristol strain), resulting in sensitivity ratios varying from 1 (identical sensitivity) to at least 10 (10-fold more sensitive) in comparison with RT-PCR and Southern hybridization.
All samples yielding a negative result for the viral target produced a visible band corresponding to the internal standard control.
Evaluation of nRT-PCR on clinical samples.
To evaluate nRT-PCR on clinical samples in comparison with reference RT-PCR and Southern hybridization, RNA from 85 stool specimens was extracted and analyzed by both methods. Sixteen of these samples (18.8%) were found positive by nRT-PCR and the reference method, whereas 4 (4.7%) gave discordant results, among which 3 samples were positive only by nRT-PCR and 1 only by reference RT-PCR.
In order to confirm the specificity of the nRT-PCR positive results for the three samples which had appeared to be negative by the reference RT-PCR, it would have been reasonable to directly sequence the nested amplicons obtained. Due to the difficulties in sequencing the nRT-PCR final product generated by the nine-inner-oligonucleotide mixture and the absence of visible bands after the first-round amplification, we chose to reamplify the first-round product of nRT-PCR with primers JV12 and JV13 for 35 further cycles, under the same conditions. These amplification products could be sequenced on both strands, revealing nucleotide sequences related to those published for GII noroviruses. The amplified product obtained from the only nRT-PCR-negative and reference RT-PCR-positive sample contained a nucleotide sequence corresponding to GI norovirus.
Nucleotide sequencing.
DNA sequencing was performed on the 20 norovirus-positive samples. Sequence analysis using the database Food-borne Viruses in Europe revealed 17 strains belonging to the Hawaii virus cluster, 1 to the Rotterdam genotype, and 1 to the Leeds virus cluster, all GII noroviruses, as well as 1 strain belonging to the Desert Shield genotype of GI. Among the 17 sequences of Hawaii genotype, the comparison of a consensus 140-nucleotide stretch revealed variation in a maximum of 9 bases, and in 12 strains an identical sequence. The analysis of the longest sequence obtained showed identity with a human calicivirus (GenBank accession number AF472568) described in Hungary for 262 out of 263 bases. We have no evidence to indicate that laboratory contamination accounted for the close relationship between norovirus strains. All RTs and PCRs were strictly controlled with zero-RNA samples, confirming that no cross-contamination occurred.
Comparison of nRT-PCR and EM.
Four out of the 16 stool samples which had appeared to be positive either by nRT-PCR or by RT-PCR and Southern hybridization were examined by EM following ultracentrifugation procedure. Only two out of the four samples (50%) showed the presence of viral particles related to the family Caliciviridae, in agreement with the higher sensitivity of molecular methods.
DISCUSSION
We have established a sensitive and broadly reactive norovirus RNA detection system. The nRT-PCR assay described in this paper uses a new primer set for nested amplification and represents a further development in the application of molecular assays to clinical services. Although sensitivity and specificity of tests may not be critical when analyzing high numbers of samples in the course of epidemic outbreaks, the reliability of diagnostic assays is of great importance in pursuing proper management and therapeutic treatment in the case of severely affected patients, particularly in infancy.
Firstly, the specificity of the nRT-PCR assay was demonstrated by negative results when the assay was performed on 61 fecal specimens known to be positive for enteric viruses other than norovirus (rotavirus, adenovirus, astrovirus, enterovirus, and cytomegalovirus).
The detection limit for the nRT-PCR assay in comparison with RT-PCR and subsequent Southern hybridization (36, 38), which we assumed as the reference method, was determined in stools by testing RNAs from 10-fold serial dilutions of five fecal samples known to contain GI and GII noroviruses of different genotypes. At least for GII strains the results showed a great degree of variability between genotypes, ranging from 2.1 × 106 (Leeds strain) to 39 (Bristol strain) virus particles/ml by nRT-PCR versus 2.1 × 106 virus particles/ml (corresponding to 3.4 × 104 virus particles) to 3.9 × 103 virus particles/ml (corresponding to 62 virus particles) revealed by the reference method, the detection limit of which was estimated on Hawaii strain to be 3 to 30 virus particles (23). Overall for both genogroups, nRT-PCR appeared to be at least as sensitive as RT-PCR and Southern hybridization. In some cases, the sensitivity of nRT-PCR appeared to be 10-fold higher or more. For both tests, replica experiments showed a considerable degree of variability between runs, reflecting the inevitable variation in the virus amount between sample aliquots. When using fecal material, uniformity is difficult to achieve due to the possible presence of virus aggregates. In fact, tests conducted on serial dilutions of viral RNA from a same extraction yielded a better reproducibility by both methods, with consistently positive results at each dilution. Nevertheless, a wide range of detection limits was observed, suggesting a genotype-dependent correlation. In these experimental conditions, nRT-PCR proved as sensitive as RT-PCR and Southern hybridization for GI norovirus strains and reached a 1- or 2-log- higher positive dilution for GII norovirus strains, which supports the results obtained on stool dilutions.
The examination of 85 stool samples, negative by routine EM and cell culture assays, by nRT-PCR and by the norovirus reference assay yielded overall positive results for 20 samples. Of these, 19 (22.3%) were detected by nRT-PCR and 17 (20%) by the RT-PCR/Southern hybridization, with a positive result in both assays in 16 cases. These samples belonged to pediatric patients hospitalized with acute gastroenteritis in the major epidemic period of the year (winter to early spring) (26, 28).
A conclusive confirmation of positive reaction for norovirus was obtained for the 17 RT-PCR/Southern hybridization-positive samples by direct DNA sequencing, which showed nucleotide sequences closely related to norovirus sequences deposited in GenBank. The sequence analysis of amplicons obtained upon repeated DNA amplification from the three stool samples, positive by nRT-PCR and negative by reference RT-PCR, also confirmed the presence of DNA related to norovirus.
The sensitivity and specificity of RT-PCR assays depend largely on primer selection. Several factors affect the ability of a primer pair to detect a given norovirus strain, including primer sequence, the amount of virus present in the sample to be assayed, and the temperature used for primer annealing during the PCR amplification process.
All primers used in this study were selected in the region coding for the RNA polymerase, which has the highest conservation among norovirus strains of both genogroups. The generic primer pair JV12/JV13 and primers SR46, SR48, SR50, and SR52, which we adapted to reverse priming, were chosen among the most frequently used primers (2). The five new forward nested primers (113-1, 113-2, 115-1, 115-2, and 115-3) were designed for their broad-range reactivity against noroviruses in order to increase sensitivity in detecting different viral strains. It must be stressed that at present no single assay detects all variants of noroviruses (37). However, our nRT-PCR can detect strains representing GI and GII noroviruses related to at least 10 different genotypes, including the Rotterdam genotype revealed in this study in a clinical sample.
It is unclear why one strain belonging to the Desert Shield genotype could not be detected by our nRT-PCR, since the same minimal number of mismatches between the nested primers and Desert Shield sequence were also observed with the efficiently detected Leeds strains (data not shown). However, this event is probably due to a combination of multiple factors, such as a small viral amount (as also suggested by the lowest detection limit for Leeds genotype) and primer homology.
The failure to detect norovirus by reference RT-PCR and Southern hybridization in 3 out of the 19 cases which were positive by nRT-PCR may be due to the lower detection limit of the former assay.
To improve the chances for successful amplification using the single primer pair JV12/JV13, the reference RT-PCR was based on a primer annealing temperature as low as 37°C. However, this strategy has the disadvantage of an increased production of nonspecific amplicons. For this reason, it requires confirmation of PCR products by hybridization and/or sequencing procedures, which are cumbersome and time-consuming and also require additional equipment.
Based on the present results, our nRT-PCR, which uses two rounds of PCR amplification in stringent conditions, may guarantee both high sensitivity and specificity in the absence of additional confirmation tests. Moreover, nRT-PCR is an easy-to-perform, rapid assay with a turnaround time of approximately 8 h. The main argument against nested protocols is that they are subject to the possibility of carryover or amplicon contamination. Although real, this possibility can be considerably reduced by strictly adhering to sterile technique and performance of the various procedures in designated areas. We used four designated areas in four different laboratory rooms and operated a strict one-way system to reduce the possibility of amplicon contamination. Moreover, positive results were considered valid only when confirmed twice.
The combination of random RT with nPCR for norovirus detection makes the assay advantageous for molecular diagnostic laboratories where RT procedures are routinely used to obtain cDNA from different RNA viruses in clinical samples and/or nPCR is extensively performed. One-step real-time PCR has been developed in recent years for the detection of noroviruses in stools as a potential candidate for diagnostic use combining high sample throughput, reproducibility, and powerful carryover prevention. However, one recent study (17) comparing the detection rate of real-time PCR to that of nRT-PCR failed to detect 7% of norovirus nRT-PCR-positive samples, probably containing a virus load below its detection limit. Because noroviruses are usually shed in stools in low concentrations (1), highly sensitive methods in nPCR format should be preferred for their detection in specialized molecular clinical laboratories. Moreover, thanks to its high sensitivity, our nRT-PCR might prove useful for the detection of norovirus in environmental, water, or food samples, where the concentration of the virus is expected to be extremely low and possibly well under the capability of conventional methods.
To our knowledge, this is the first report on the use of a generic internal RNA standard control subjected to simultaneous amplification together with the norovirus target by specific primers in the nRT-PCR assay. Amplicons from the internal standard control can be distinguished from the target viral amplicon on the basis of the different sizes (610 bp versus 174 to 176 bp). In this approach, RT-PCR amplification is optimized only for the norovirus target and the reaction for the internal control follows the one for the specific target; i.e., optimization of the internal control amplification is not a necessary part of the assay development as long as the internal control amplicon is detected (18). In addition, in order to limit the competition of the target- and internal control-specific reactions for oligonucleotides and enzymes, the concentration of the internal control primers is kept to a suboptimal level (at least one-fifth of that of the target-specific primers). Moreover, the larger size of the internal control product compared to the target sequence drives the reaction kinetics towards the latter product. This strategy for detecting the presence of RT-PCR inhibitors in fecal samples is currently used in our molecular laboratory for other amplification protocols and can be successfully adapted when using random RT and single or nested PCR with annealing temperatures under 60°C, according to the producer's instructions.
The predominance of norovirus GII strains (95%) during the period of study is in agreement with previous European reports of a higher prevalence of GII compared to GI strains (32, 33). DNA sequencing revealed 1 strain (5%) related to Leeds virus, 1 strain (5%) related to the Rotterdam cluster, and 17 strains (85%) related to the prototype Hawaii virus, closely similar to one another. A final norovirus strain was found to belong to the Desert Shield cluster. Hawaii-like viruses also circulated as a predominant strain in Hungary in 1999 to 2000 (32). Interestingly, the longest sequence obtained from our samples was found to be virtually identical (262 of 263 bases) to norovirus Hawaii from Hungary, which further suggests the transnational circulation of norovirus strains. Even if our present data do not allow us to determine the overall prevalence of calicivirus in our community or to exclude a possible common source of exposure linking any of the cases, the observation of a predominant strain among the pediatric patients submitted to norovirus detection may suggest the endemicity of this strain in northern Italy during the year of investigation.
The greatly enhanced sensitivity of the nRT-PCR assay should facilitate the detection of norovirus in clinical samples and provide valuable information on the incidence and transmission of the virus in public settings.
Acknowledgments
This work was supported by the grant “Programma per la ricerca finalizzata 1999,” art. 12 d.lgs 502/92, IZSLER PRF92202, from the Italian Ministry of Health.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Fun, M. D. Sobsey, O. D. Simmons III, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well: international consequences. JAMA 278:563-568. [PubMed] [Google Scholar]
- 4.Berg, D. E., M. A. Kohn, T. A. Farley, and L. M. McFarland. 2000. Multi-state outbreaks of acute gastroenteritis traced to fecal-contaminated oysters harvested in Louisiana. J. Infect. Dis. 181(Suppl. 2):S381-S386. [DOI] [PubMed] [Google Scholar]
- 5.Berke, T., B. Golding, X. Jiang, D. W. Cubitt, M. Wolfaardt, A. W. Smith, and D. O. Matson. 1997. Phylogenetic analysis of the caliciviruses. J. Med. Virol. 52:419-424. [DOI] [PubMed] [Google Scholar]
- 6.Boccia, D., A. E. Tozzi, B. Cotter, C. Rizzo, T. Russo, G. Buttinelli, A. Caprioli, M. L. Marziano, and F. M. Ruggeri. 2002. Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg. Infect. Dis. 8:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresee, J. S., M.-A. Widdowson, S. S. Monroe, and R. I. Glass. 2002. Foodborne viral gastroenteritis: challenges and opportunities. Clin. Infect. Dis. 35:748-753. [DOI] [PubMed] [Google Scholar]
- 9.de Wit, M. A. S., M. P. G. Koopmans, L. M. Kortbeek, W. J. B. Wannet, J. Vinjé, F. van Leusden, A. I. M. Bartelds, and Y. T. H. P. van Duynhoven. 2001. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: incidence and etiology. Am. J. Epidemiol. 154:666-674. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 11.Girish, R., S. Broor, L. Dar, and D. Ghosh. 2002. Foodborne outbreak caused by a Norwalk-like virus in India. J. Med. Virol. 67:603-607. [DOI] [PubMed] [Google Scholar]
- 12.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., C. I. Gallimore, J. P. Norcott, D. Lewis, and D. W. G. Brown. 1995. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J. Med. Virol. 47:392-398. [DOI] [PubMed] [Google Scholar]
- 14.Green, J., J. P. Norcott, D. Lewis, C. Arnold, and D. W. G. Brown. 1993. Norwalk-like viruses: demonstration of genomic diversity by polymerase chain reaction. J. Clin. Microbiol. 31:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 16.Grohmann, G., R. I. Glass, J. Gold, M. James, P. Edwards, T. Borg, S. E. Stine, C. Goldsmith, and S. S. Monroe. 1991. Outbreak of human calicivirus gastroenteritis in a day-care center in Sidney, Australia. J. Clin. Microbiol. 29:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höhne, M., and E. Schreier. 2004. Detection and characterization of norovirus outbreaks in Germany: application of a one-tube RT-PCR using a fluorogenic real-time detection system. J. Med. Virol. 72:312-319. [DOI] [PubMed] [Google Scholar]
- 18.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., P. N. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83:145-154. [DOI] [PubMed] [Google Scholar]
- 21.Johansson, P. J. H., M. Torvén, A.-C. Hammarlund, U. Björne, K.-O. Hedlund, and L. Svensson. 2002. Food-borne outbreak of gastroenteritis associated with genogroup I calicivirus. J. Clin. Microbiol. 40:794-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, S., T. Morishita, T. Yamashita, K. Sakae, O. Nishio, T. Miyake, Y. Ishihara, and S. Isomura. 1991. A large outbreak of gastroenteritis associated with small round structured virus among schoolchildren and teachers in Japan. Epidemiol. Infect. 107:81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koopmans, M., J. Vinjé, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262-S269. [DOI] [PubMed] [Google Scholar]
- 24.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small-round structured (Norwalk-like) virus. Science 200:319-325. [DOI] [PubMed] [Google Scholar]
- 25.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 26.Meakins, S. M., G. K. Adak, B. A. Lopman, and S. J. O'Brien. 2003. General outbreaks of infectious intestinal disease (IID) in hospitals, England and Wales, 1992-2000. J. Hosp. Infect. 53:1-5. [DOI] [PubMed] [Google Scholar]
- 27.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mounts, A. W., T. Ando, M. Koopmans, J. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S284-S287. [DOI] [PubMed] [Google Scholar]
- 29.Parks, C. G., C. L. Moe, D. Rhodes, A. Lima, L. Barrett, F. Tseng, R. Baric, A. Talal, and R. Guerrant. 1999. Genomic diversity of “Norwalk like viruses” (NLVs): pediatric infections in a Brazilian shantytown. J. Med. Virol. 58:426-434. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pönkä, A., L. Maunula, C. H. von Bonsdorff, and O. Lyytikäinen. 1999. Outbreak of calicivirus gastroenteritis associated with eating frozen raspberries. Euro Surveill. 4:66-69. [DOI] [PubMed] [Google Scholar]
- 32.Reuter, G., T. Farkas, T. Berke, X. Jiang, D. O. Matson, and G. Szücs. 2002. Molecular epidemiology of human calicivirus gastroenteritis outbreaks in Hungary, 1998 to 2000. J. Med. Virol. 68:390-398. [DOI] [PubMed] [Google Scholar]
- 33.Schreirer, E., F. Doring, and U. Kunkel. 2000. Molecular epidemiology of outbreaks of gastroenteritis associated with small round structured viruses in Germany in 1997/98. Arch. Virol. 145:443-453. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, M. B., S. Parker, W. O. Grabow, and W. D. Cubitt. 1996. An epidemiological investigation of Norwalk virus infection in South Africa. Epidemiol. Infect. 116:203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinjé, J., and M. P. G. Koopmans. 1996. Molecular detection and epidemiology of small round structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 37.Vinjé, J., H. Vennema, L. Maunula, C.-H. von Bonsdorff, M. Hoehne, E. Schreier, A. Richards, J. Green, D. Brown, S. S. Beard, S. S. Monroe, E. de Bruin, L. Svensson, and M. P. G. Koopmans. 2003. International collaborative study to compare reverse transcriptase PCR assays for detection and genotyping of noroviruses. J. Clin. Microbiol. 41:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinjé, J., S. Altena, and M. P. Koopmans. 1997. The incidence and genetic variability of small-round structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 39.Watson, D. H., W. C. Russell, and P. Wildy. 1963. Electron microscopic particle counts on herpes virus using the phosphotungstate negative staining technique. Virology 19:250-260. [DOI] [PubMed] [Google Scholar]
- 40.Whitby, H. J., and F. G. Rodgers. 1980. Detection of virus particles by electron microscopy with polyacrylamide hydrogel. J. Clin. Pathol. 33:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen, L. K. W., M. G. Catton, B. J. Cox, P. J. Wright, and J. A. Marshall. 2001. Heminested multiplex reverse transcription-PCR for detection and differentiation of Norwalk-like virus genogroups 1 and 2 in fecal samples. J. Clin. Microbiol. 39:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]