Abstract
The new species Candida nivariensis, isolated from the clinical samples of three patients in Spain over a 3-year period, is presented here. This species can be easily differentiated from Candida glabrata, the closest genetic species, by different colony color on CHROMagar and by its ability to ferment trehalose. The analyses of the internal transcribed spacer region and the D1-D2 region of the 26S rRNA gene sequences support a new species designation.
Candida species have become an important cause of nosocomial infection, primarily affecting immunocompromised patients (19). These infections show a high mortality rate, and Candida species can now be considered the fourth-most-common cause of hospital-acquired blood-borne infection (6, 15, 19, 20). Although Candida albicans remains the most common species, there has been an increase in the number of infections caused by non-Candida albicans species, such as Candida glabrata, Candida parasilopsis, Candida tropicalis, and Candida krusei, among others, which are emerging as opportunistic pathogens (2, 5, 15, 23). These species are generally more resistant to antifungal agents than C. albicans. For instance, C. glabrata is known to rapidly acquire increased resistance to fluconazole and C. krusei is considered to be inherently resistant to fluconazole (15, 16, 20, 23). Therefore, the rapid and correct identification of yeasts causing mycoses is fundamental in ensuring specific, early, and effective antifungal therapy.
We have recovered three yeast isolates from different clinical specimens of three different patients which were very similar to one another and which were identified as C. glabrata by commercial miniaturized methods. Further biochemical and molecular studies demonstrated that such identification was erroneous and that the three isolates belong to an as-yet-undescribed species.
In work presented here, this taxon is fully described, illustrated, and proposed as a new species of Candida.
MATERIALS AND METHODS
Isolates and patients.
The three isolates were recovered at the Hospital Universitario Nuestra Señora de Candelaria, Santa Cruz de Tenerife, Canary Islands, Spain, each of them from a different patient. These isolates were sent to the Microbiology Service of this hospital for identification, and they were identified as C. glabrata with the API ID 32C yeast identification system and ID-YST cards, VITEK 2 System (bioMérieux). The three patients presented the following clinical pictures.
Patient 1 was a 50-year-old woman presenting with anorexia, asthenia, fever, nocturnal swelling, cough with purulent expectoration, and moderate dyspnea (admitted 27 August 1999). Chest radiographic examination showed pulmonary nodes and bronchiechtasis. A diagnosis of multiple pulmonary abscesses was made. Microbiological cultures from bronchoalveolar lavage fluid showed the presence of the first of the three atypical isolates (HC5937-63) as a pure culture. There are no data about treatment of the patient and outcome of the illness.
Patient 2 was a 69-year-old woman admitted to the emergency ward with biliary pancreatitis and functional renal failure (admitted 26 November 2001). Her clinical history consisted of obesity, appendectomy, hypercholesterolemia, seizures, hypertension, hypothyroidism, and maniac depressive psychosis. After a laparotomy, pancreatic necrosis with pancreatic abscess was observed. The patient died within 5 days (on 1 December 2001) due to a multiorgan failure 1 day after treatment with Ambisome was commenced. A blood culture performed on the first day of hospitalization showed the presence of the second of the three atypical yeast isolates (HC4292-20). Moreover, an Enterococcus faecalis isolate was recovered from a femoral catheter.
Patient 3 was a 44-year-old woman complaining of lumbar pain and mild dyspnea after myomectomy (admitted 8 April 2002). When urine samples were analyzed, routine tests showed the presence of the third atypical isolate in pure culture (HC7609-30). No antifungal treatment was initiated. Follow-up is unknown.
The following isolates were used for comparative studies: C. albicans HC4029-91 and C. glabrata ATCC 90030 and HC8959-30.
Morphological and physiological studies.
All the isolates were inoculated onto Sabouraud chloramphenicol agar (Bio-Rad, Marnes-La-Coquette, France) at pH 5 to 6 and were incubated at 30°C, 37°C, and 45°C for up to 7 days. Surface growth on broth cultures was determined with yeast peptone dextrose (YPD) broth incubated at 37°C for 72 h. The strains were subcultured on CHROMagar Candida medium (Becton Dickinson, Heidelberg, Germany), incubated at 37°C, and examined after 24 to 72 h for colony color and morphology. Germ tube tests were performed by inoculating 2.0 ml of fresh, pooled, normal human serum with a fresh colony and incubation at 37°C for 2 h. To induce chlamydospores and pseudohyphal production, yeasts were incubated on potato carrot bile medium (Bio-Rad) and diluted milk medium (170 ml of natural milk, 1 liter of distilled water, and 0.25 g of chloramphenicol) for 24 to 48 h at 30°C (3, 4, 11, 13, 17). The induction of a possible ascosporic state was performed by using acetate ascospore agar (potassium acetate, 5.00 g; yeast extract, 1.25 g; dextrose, 0.50 g; agar, 15.00 g; distilled water, 500 ml), Gorodkowa medium (dextrose, 1.25 g; NaCl, 2.60 g; beef extract, 5.00 g; agar, 5.00 g; distilled water, 500 ml), and V-8 medium for ascospores (V-8 vegetable juice, 500 ml; dry yeast, 10 g; agar, 10 g; distilled water, 500 ml) and incubating yeasts for up to 1 month at 25°C (10). The ability of the isolates to assimilate carbohydrate source compounds was determined according to the manufacturer's instructions. Fermentation tests were performed with a standardized yeast nitrogen base (Difco, Detroit, Mich.).
The assimilation profiles of each isolate were examined on three independent occasions using different batches of ID 32C and ID-YST kits.
PCR.
Nuclear DNA extraction was performed as previously described (8). Since initially our isolates were tentatively identified as C. glabrata, these isolates and typical strains of C. glabrata were tested by PCR using previously described primers and conditions (12). The primers used were CGL1 (5′-TTATCACACGACTCGACACT-3′) and CGL2 (5′-CCCACATACTGATATGGCCTACAA-3′).
rRNA gene sequencing.
The internal transcribed spacer (ITS) region and D1-D2 region of the 26S rRNA gene were automatically sequenced from the PCR products with previously described primers (ITS5 and ITS4 and NL1 and NL4, respectively) (14, 22). All sequencing reactions were performed twice from independent PCR amplifications. A BLAST search for each sequence was performed to identify best matches.
Phylogenetic analysis.
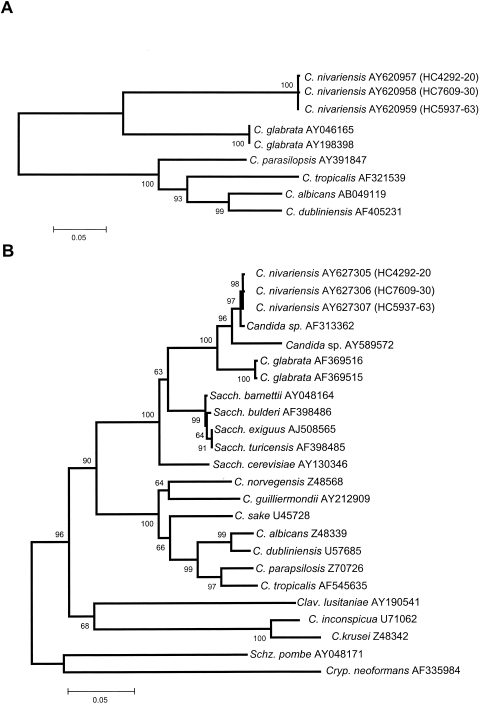
For phylogenetic analysis, the GenBank sequences indicated in Fig. 3A and B were used. The sequences were aligned using Clustal X software (1, 21), followed by manual adjustments with a text editor. Phylogenetic analyses using the neighbor-joining method (18) and based on Kimura's two-parameter corrected nucleotide distances were performed with MEGA2.1 software (7), including both transitions and transversions, and with pairwise deletion used to handle gaps and missing data. Confidence values for individual branches were determined by bootstrap analyses (1,000 replicates).
FIG. 3.
Neighbor-joining tree based on Kimura two-parameter corrected nucleotide distances among ITS region (A) and D1-D2 26S rRNA genes (B) of the Candida nivariensis isolates and other related species. Branch lengths are proportional to distance. Bootstrap replication frequencies (1,000 replications) are indicated on the nodes. C., Candida; K., Kluyveromyces; Sacch., Saccharomyces; Clav., Clavispora; Schz., Schizosaccharomyces; Cryp., Cryptococcus.
RESULTS AND DISCUSSION
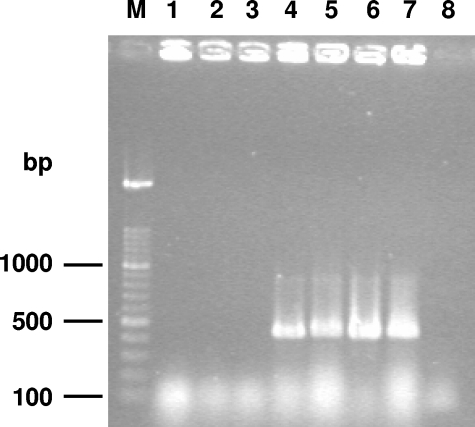
PCR using C. glabrata-specific primers yielded a 423-bp amplicon for all the reference C. glabrata strains. However, when this PCR identification was applied to the three supposed C. glabrata isolates recovered from the three patients, no amplicon was detected (Fig. 1). To ensure that this result was not due to inhibition of PCR, the same DNA preparations were used for PCR amplification of ITS or D1-D2 regions, and the three isolates yielded an amplicon, confirming that these isolates do not yield any fragment with the C. glabrata-specific primers.
FIG. 1.
Agarose gel electrophoresis of the PCR products obtained with the primers specific for C. glabrata. Lanes 1 to 3, atypical isolates (HC5937-63, HC7609-30, and HC4292-20, respectively); lanes 4 to 7, C. glabrata (HC3924-30, HC3279-29, HC1103-80, and ATCC 90030, respectively); lane 8, negative control (PCR mix without DNA template); lane M, DNA molecular size marker (100-bp ladder).
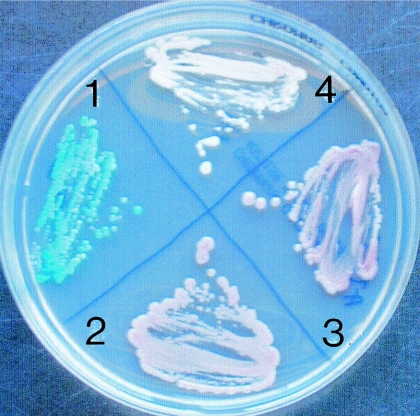
The three atypical Candida isolates shared a unique identical cluster of phenotypic properties (Table 1). These isolates were not able to form germ tubes, chlamydospores, pseudohyphae, or ascospores, even after a long incubation period. The three atypical Candida isolates and C. glabrata ATCC 90030 grew well on Sabouraud dextrose agar (SDA) and YPD broth at 30 and 37°C, but not at 45°C. However, in contrast to C. glabrata, our three isolates did not show turbidity on YPD broth. On CHROMagar, the atypical C. glabrata isolates yielded white colonies, while those of C. glabrata reference strains were typically pink (Fig. 2).
TABLE 1.
Physiological and morphological key features of the three atypical isolates, C. glabrata, and C. albicansa
| Feature | Atypical isolate |
C. glabrata |
C. albicans | |||
|---|---|---|---|---|---|---|
| HC4292-20 | HC7609-30 | HC5937-63 | HC8959-30 | ATCC 90030 | HC4029-91 | |
| Growth | ||||||
| Surface growth (37°C) | − | − | − | − | − | − |
| SDA cycloheximide (25°C) | − | − | − | − | − | + |
| SDA (37°C) | + | + | + | + | + | + |
| YPD (30°C) | + | + | + | + | + | + |
| YPD (45°C) | − | − | − | − | − | + |
| Turbidity YPD (37°C) | − | − | − | + | + | + |
| Assimilation | ||||||
| Dextrose | + | + | + | + | + | + |
| Maltose | − | − | − | − | − | + |
| Sucrose | − | − | − | − | − | + |
| Lactose | − | − | − | − | − | − |
| Galactose | − | − | − | − | − | + |
| Melibiose | − | − | − | − | − | − |
| Cellobiose | − | − | − | − | − | − |
| Inositol | − | − | − | − | − | − |
| Xylose | − | − | − | − | − | + |
| Raffinose | − | − | − | − | − | − |
| Trehalose | + | + | + | + | + | − |
| Dulcitol | − | − | − | − | − | − |
| KNO3 | − | − | − | − | − | − |
| 2-Keto-gluconate | − | − | − | − | − | + |
| Glicerol | + | + | + | + | + | − |
| Fermentation | ||||||
| Dextrose | + | + | + | + | + | + |
| Maltose | − | − | − | − | − | + |
| Sucrose | − | − | − | − | − | − |
| Lactose | − | − | − | − | − | − |
| Galactose | − | − | − | − | − | + |
| Trehalose | + | + | + | − | − | ± |
| Cellobiose | − | − | − | − | − | − |
| Other characteristics | ||||||
| Germ tubes | − | − | − | − | − | + |
| Urease (25°C) | − | − | − | − | − | − |
| Ascospores | − | − | − | − | − | − |
| Color of CHROMagar | White | White | White | Pink/violet | Pink/violet | Blue |
| Chlamydospores | − | − | − | − | − | + |
| Esculin hydrolysis | − | − | − | − | − | − |
| Pseudohyphae | − | − | − | − | − | + |
Abbreviations: +, positive; −, negative; ±, weak utilisation.
FIG. 2.
Colonies on CHROMagar Candida medium. 1, C. albicans 5039/95 blue colonies; 2, C. glabrata ATCC 90030; 3, C. glabrata 3924/30 pink colonies; 4, C. nivariensis HC4292-20 white colonies.
The three atypical isolates exhibited very similar carbohydrate source assimilation profiles with the API ID 32C and ID-YST cards of the VITEK 2 System, i.e., assimilation of trehalose, glucose, and glycerol, but they were unable to utilize the other carbon source and nitrogen source substrates tested. In contrast with C. glabrata, the atypical isolates were able to ferment trehalose. The atypical isolates were susceptible to cycloheximide and were not able to hydrolyze esculin. The API ID 32C code obtained was 0001-0101-01, which corresponded to C. glabrata but with an 85% ID which is a doubtful identification. By contrast, for the VITEK 2 the identification was 95 to 97%, which corresponded to a good identification.
Sequences of the two ribosomal regions (D1-D2 and ITS) were identical for the three Spanish atypical isolates. Their GenBank accession numbers are AY627305 and AY620957 for isolate HC4292-20, AY627306 and AY620958 for isolate HC7609-30, and AY627307 and AY620959 for isolate HC5937-63. A search of the GenBank database for the D1-D2 sequences revealed a high level of similarity (99% nucleotide identity, 546 of 548 bp) with the sequence of a Candida sp. AF313362 belonging to a strain isolated by Lachance et al. (9) on Hibiscus sp. flowers from the Northern Territories (Canada) and to a lesser degree with Candida sp. AY589572 (97% nucleotide identity) and C. glabrata AJ582736 (96% nucleotide identity). For the ITS region, the closest sequence was C. glabrata AY046165 (94% similarity).
In the ITS phylogenetic tree (Fig. 3A), two main clusters were distinguished. The first cluster included the three atypical Candida isolates, forming a subcluster with 100% bootstrap support, together with two strains of C. glabrata (100%). However, the group comprising the three atypical isolates was considerably distant from the rest. The second cluster included relevant clinical species of Candida, i.e., C. parasilopsis, C. tropicalis, C. albicans, and Candida dubliniensis, which were included for comparative purposes. In the phylogenetic tree obtained from analysis of the D1-D2 26S rRNA gene fragment sequences (Fig. 3B), the group formed by the three atypical Spanish isolates and the strain from Canada constituted a terminal branch with 97% bootstrap support of the subcluster, also containing the sequence Candida sp. AY589572 obtained from the GenBank and corresponding to a clinical isolate from Portugal. The two reference strains of C. glabrata included in the study were placed far from these sequences.
In this study, we have tried to demonstrate, using phenotypic and molecular methods, that three atypical clinical isolates recovered in Spain belong to an unknown species of Candida. Surprisingly, we noticed that another probable isolate of this species had been previously isolated in Canada (9). This isolate differed only by 2 bp for the D1-D2 fragment (a G and an A insertion in positions 10 and 18, respectively) from the Spanish isolates. Unfortunately, this isolate is not available for comparative studies. Our molecular data demonstrated that the closest species is C. glabrata, a species with which the atypical Spanish isolates had been previously confused. The main phenotypic differences between the Spanish atypical isolates and this species are the absence of turbidity on YPD broth and the ability to ferment trehalose, showed by the atypical isolates but not by C. glabrata. An additional feature that differentiates such isolates from C. glabrata is the colony color showed on the CHROMagar Candida medium (Fig. 2), which was white for the former and pink for C. glabrata.
The fact that three isolates of a single strain were recovered at the same hospital in a 3-year period from different patients and different clinical specimens is very interesting. The fact that a closely related isolate was found in flowers in Canada suggests that the hospital garden or potted plants could be the habitat of the Spanish strain and the source of infection or colonization.
We propose the three atypical isolates represent a new species, Candida nivariensis.
Description of Candida nivariensis sp. nov.
Candida nivariensis Alcoba-Flórez, Méndez-Álvarez, Cano, Guarro, Pérez-Roth & Arévalo, sp. nov.
Etym.: nivaria (Tenerife) hoc nomen acceperit a perpetua nive.
In YPD, post horas 72 ad 25°C, cellulae ellipsoideae, ovoideae vel subsphaericae, 2.5-6 × 2-5.5 μm, singulae et binae aut in racemis brevibus; multipolares gemmas proferentes. Coloniae albicerae, rotundatae, convexae, blandae et planae in agaro dextroso Sabouraud nascuntur et marginem mycelialem non habent. Germina tubulata non formantur. Chlamydosporas et pseudohyphas non fert. Ascosporae non fiunt. Assimilat trehalosum, glucosum, et glycerolum. Glucosum et trehalosum fermentatur. Cycloheximido praesente non crescit neque aesculinum hydrolysit. Typus CBS 9983T ex sanguis, feminae, Tenerife, Hispania, 2001, J. Alcoba-Florez isolatus est (originaliter ut HC4292-20). In collectione zymotica Centraalbureau voor Schimmelcultures Utrecht Batavorum deposita est.
On YPD broth, at 25°C for 72 h, the cells are ellipsoidal or ovoid to subspherical, 2.5 to 6 μm by 2 to 5.5 μm, single or in pairs or in small groups with multipolar budding. On SDA, the colonies are round, convex, pasty, smooth, and white to cream colored, with no mycelial border. Germ tubes are not formed. Chlamydospores and pseudohyphae are not produced. Ascospores are not detected. Assimilation of trehalose, glucose, and glycerol is positive. Susceptible to cycloheximide. Unable to hydrolyze esculin.
CBS 9983T (CECT 11998T [Spanish Type Culture Collection]; originally strain HC4292-20) was isolated in Tenerife, Spain, in 2001 by J. Alcoba-Flórez from a blood culture of a Spanish woman presenting with biliary pancreatitis. The other isolates, HC5937-63 and HC7609-30, have been deposited in the Centraalbureau voor Schimmelcultures (CBS), under accession numbers CBS 9984 and CBS 9985, respectively.
Acknowledgments
This work was supported by the following grants: PI62/02 from the Fundación Canaria de Investigación y Salud (FUNCIS) and FIS 99/3060 from the Fondo de Investigaciones Sanitarias.
We thank F. Plaza (University of La Laguna) for the Latin diagnosis and L. Divasson and L. K. León for editorial review.
REFERENCES
- 1.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 2.Borst, A., B. Theelen, E. Reinders, T. Boekhout, A. C. Fluit, and P. H. M. Savelkoul. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 41:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casal, M., and M. J. Linares. 1981. Comparations of six media for production of chlamidospore by Candida albicans. Mycopathologia 76:125-128. [DOI] [PubMed] [Google Scholar]
- 4.Feo, M., and A. De Pacheco. 1976. Candida albicans: milk in the production of chlamydospores. Rev. Latinoam. Microbiol. 18:23-24. [PubMed] [Google Scholar]
- 5.Graf, B., T. Adam, E. Zill, and U. B. Göbel. 2000. Evaluation of the VITEK 2 system for rapid identification of yeast and yeast-like organisms. J. Clin. Microbiol. 38:1782-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajjeh, R. A., A. N. Sofair, L. H. Harrison, G. M. Lyon, B. A. Arthington-Skaggs, S. A. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. A. Ciblak, L. E. Benjamin, L. T. Sanza, S. Huie, S. F. Yeo, M. E. Brandt, and D. W. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 8.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachance, M. A., W. T. Starmer, C. A. Rosa, J. M. Bowles, J. S. F. Barker, and D. H. Janzen. 2001. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1:1-8. [DOI] [PubMed] [Google Scholar]
- 10.Larone, D. H. 2002. Media, p. 327-328. In D. H. Larone (ed.), Medically important fungi: a guide to identification, 4th ed. American Society for Microbiology, Washington, D.C.
- 11.Letscher-Bru, V., M. H. Meyer, A. C. Galoisy, J. Waller, and E. Candolfi. 2002. Prospective evaluation of the new chromogenic medium Candida ID, in comparison with Candiselect, for isolation of molds and isolation and presumptive identification of yeast species. J. Clin. Microbiol. 40:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer, M. H., V. Letscher-Bru, B. Jaulhac, J. Waller, and E. Candolfi. 2004. Comparison of Mycosis IC/F and Plus Aerobic/F media for diagnosis of fungemia by the Bactec 9240 system. J. Clin. Microbiol. 42:773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell, K. 1993. Fusarium and its near relatives, p. 225-233. In D. R. Reynolds and J. W. Taylor (ed.), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CBA International, Wallingford, England.
- 15.Pemán, J., E. Cantón, M. Gobernado, and the Spanish ECMM Working Group on Candidemia. 2005. Epidemiology and antifungal susceptibility of Candida species isolated from blood: results of a 2-year multicentre sudy in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 24:23-30. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., and D. J. Diekema. 2002. Role of setinel surveillance of candidemia: trends in species distribution and antifungal susceptibility. J. Clin. Microbiol. 40:3551-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radosavljevic, M., H. Koeing, V. Letscher-Bru, J. Waller, F. Maloisel, B. Lioure, and R. Herbrecht. 1999. Candida catenulata fungemia in a cancer patient. J. Clin. Microbiol. 37:475-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Steffan, P., J. A. Vazquez, D. Boikov, Ch. Xu, J. D. Sobel, and R. A. Akins. 1997. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J. Clin. Microbiol. 35:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St-Germain, G., M. Laverdière, R. Pelletier, A.-M. Bourgault, M. Libman, C. Lemieus, and G. Noël. 2001. Prevalence and antifungal susceptibility of 442 Candida isolates from blood and other normally sterile sites: results of a 2-year (1996 to 1998) multicenter surveillance in Quebec, Canada. J. Clin. Microbiol. 39:949-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to the methods and applications. Academic Press, New York, N.Y.
- 23.Yokoyama, K., S. K. Biswas, M. Miyaji, and K. Nishimura. 2000. Identification and phylogenetic relationship of the most common pathogenic Candida species inferred from mitochondrial cytochrome b gene sequences. J. Clin. Microbiol. 38:4503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]