Abstract
Recent epidemiological studies suggested that cpb2-positive Clostridium perfringens isolates are associated with gastrointestinal (GI) diseases in horses. These putative relationships, indicated by PCR genotyping, were tested in the present study by further genotyping and phenotyping of 23 cpb2-positive C. perfringens isolates from horses with GI disease (referred to hereafter as horse GI disease isolates). Our beta2-toxin (CPB2) Western blot analyses demonstrated that all of the tested isolates were unable to produce detectable levels of CPB2. However, Southern blot and nucleotide sequencing analyses identified intact cpb2 open reading frames in all of our surveyed horse GI disease isolates. Furthermore, reverse transcriptase PCR and Northern blot analyses showed that cpb2 genes in all of our surveyed horse GI disease isolates were transcriptionally active, i.e., an ∼1.2-kb cpb2-specific mRNA was identified in total RNA from our surveyed isolates. The levels of cpb2 mRNA in CWC245 (a high-CPB2-producing pig strain) and our surveyed horse GI disease isolates differed to such an extent (35-fold) that this difference could be considered as a major cause of the difference in levels of CPB2 production by CWC245 and horse GI disease isolates. This finding received further support from our observation that the complementing strain 106902(pMRS140), which produced significantly higher levels of mRNA than strain 106902, produced high levels of CPB2. Collectively, our results indicated that there is a positive correlation between cpb2 transcription levels and the amount of CPB2 produced by a C. perfringens cell and that decreased transcription and/or message instability may be involved, at least in part, in the low CPB2 production noted for horse GI disease isolates in comparison to that noted for pig GI disease isolate CWC245.
Clostridium perfringens is an important cause of histotoxic and gastrointestinal (GI) diseases in both humans and animals (20, 26). The virulence of this bacterium results largely from its ability to produce at least 15 different toxins (21). C. perfringens isolates are commonly classified (21) into one of five types, A to E, based upon their ability to produce the four major lethal toxins (i.e., alpha-, beta-, epsilon-, and iota-toxins). The major lethal toxins, however, are not the only biomedically important toxins; some C. perfringens isolates (mostly those belonging to type A) produce C. perfringens enterotoxin (20), and many isolates of both types A and C produce beta2-toxin (CPB2) (5, 9, 11, 29)
The 28-kDa CPB2 was first purified from C. perfringens type C strain CWC245 and shown to be cytotoxic for Chinese hamster ovary cells (15). Fisher et al. (9) recently demonstrated that CPB2 produced by a non-food-borne human GI disease isolate is cytotoxic for CaCo-2 cells. Although cpb2 was initially identified in C. perfringens type C isolates, a recent study (5) demonstrated the presence of cpb2 sequences in all types of C. perfringens. The deduced amino acid sequence of CPB2 is highly conserved in type A and C strains of C. perfringens isolated from diarrheic pigs (11, 29). However, the variability in CPB2 sequences in type A human isolates (9) coupled with CPB2 sequence differences in isolates from nonporcine animals (16) indicate that CPB2 is an unusual C. perfringens toxin.
Several epidemiological studies suggested that cpb2-positive C. perfringens isolates are highly associated with enteric diseases in domestic animals, particularly enteritis in piglets (10, 11, 18), typhlocolitis in horses (2, 13), diarrhea in dogs (27), ulcerative enteritis in an African elephant (1), and enterotoxemia in calves (19). These putative associations between cpb2-positive isolates and GI diseases in animals remained tentative because this conclusion was drawn on the basis of PCR genotyping, in which only the presence of cpb2 sequences and not the expression of cpb2 was demonstrated. Although a recent study (29), by demonstrating that all of the surveyed cpb2-positive isolates from pigs with GI disease (referred to hereafter as pig GI disease isolates) can, in fact, produce CPB2, confirmed the association of cpb2-positive isolates with GI diseases in piglets, no such evidence with a large number of clinical isolates has been provided in favor of an association of cpb2-positive isolates with GI diseases in nonporcine animals. Even though a recent study (5) demonstrated the production of CPB2 in 100% (n = 2), 50% (n = 10), and 40% (n = 5) of the surveyed horse, bovine, and avian GI disease isolates, respectively, the correlation between PCR genotype and CPB2 production in horse GI disease isolates still remained quite tentative due to the minute surveyed population.
The putative associations thus noted between cpb2-positive isolates and CPB2-associated horse GI diseases clearly require verification by analyzing larger numbers of horse GI disease isolates. In response to this issue, our present study has performed the first in-depth molecular genetic and phenotypic analysis of a large number of European cpb2-positive C. perfringens type A isolates obtained from horses with GI diseases. Results from the present study indicate that there is a positive correlation between cpb2 transcription levels and the amount of CPB2 produced by a C. perfringens cell and that decreased transcription and/or message instability may be involved, at least in part, in the low cpb2 expression levels observed in horse isolates compared to the level in pig isolate CWC245.
MATERIALS AND METHODS
Bacterial strains.
The C. perfringens isolates used in this study are listed and described in Table 1. The cpb2-positive C. perfringens horse GI disease isolates used in this study were identified earlier by multiplex PCR analysis (10, 13) as cpe-negative type A isolates. In this study, by using the reverse CAMP test (12), we reconfirmed that all cpb2-positive type A isolates surveyed in this study produced (data not shown) the arrow-shaped zone of synergistic hemolysis indicative of alpha-toxin expression.
TABLE 1.
Summary of genotypic and phenotypic characterization results obtained in this study for cpb2-positive C. perfringens isolates from horses with GI diseases
| Strain | Beta2-toxin-specific Western blotting resultsa | cpb2 RFLP pattern (kb) with HpaI | cpb2 location determined by PFGEb | Northern blotting resultsc | Reference |
|---|---|---|---|---|---|
| cpb2-positive CWC245 | + | ∼5 | Plasmid | + | 11 |
| cpb2-negative 106527 | − | − | 11 | ||
| Diarrheic-horse isolates | |||||
| 106888 | − | >20 | Plasmid | + | 13 |
| 106889 | − | >20 | Plasmid | + | 13 |
| 106902 | − | >20 | Plasmid | + | 13 |
| 106903 | − | >20 | Plasmid | + | 13 |
| 100 | − | >20 | ND | + | 13 |
| 101 | − | >20 | ND | + | 13 |
| 170 | − | >20 | ND | + | 13 |
| 1991 | − | >20 | ND | + | 13 |
| 1992 | − | >20 | ND | + | 13 |
| 1993 | − | >20 | ND | ND | 13 |
| 1994 | − | >20 | ND | + | 13 |
| 1995 | − | >20 | ND | + | 13 |
| 1996 | − | >20 | ND | ND | 13 |
| AHT327 | − | >20 | ND | + | 10 |
| AHT2600 | − | ∼8 | ND | + | 10 |
| AHT2779 | − | ∼14 | ND | + | 10 |
| AHT2911 | − | >20 | ND | + | 10 |
| AHT3553 | − | >20 | ND | + | 10 |
| AHT3992 | − | >20 | Plasmid | + | 10 |
| AHT4578 | − | >20 | Plasmid | + | 10 |
| AHT5195 | − | >20 | ND | ND | 10 |
| AHT5773 | − | >20 | Plasmid | + | 10 |
| AHT45771 | − | >20 | ND | ND | 10 |
+, presence of 28-kDa CPB2 immunoreactive band; −, absence of 28-kDa CPB2 immunoreactive band.
ND, not determined.
+, presence of ∼1.2-kb cpb2 transcript; −, absence of ∼1.2-kb cpb2 transcript.
Growth conditions.
A starter culture (6 ml) of each C. perfringens isolate was prepared by overnight growth at 37°C in fluid thioglycolate broth (Difco) as described previously (8). For DNA and RNA isolation or culture supernatant protein preparation, an aliquot (0.2 ml) of each fluid thioglycolate broth culture was inoculated into 10 ml of TGY broth (3% Trypticase, 2% glucose, 1% yeast extract, 0.1% cysteine [8]), which was then incubated at 37°C for 3 to 24 h without shaking. (Note that, because of moderate aerotolerance, C. perfringens can grow in liquid medium without anaerobic incubation). For selection of C. perfringens transformants carrying pMRS140, C. perfringens cultures were plated onto a brain heart infusion agar plate containing erythromycin (50 μg/ml) and incubated at 37°C for 18 h in an anaerobic jar with BBL GasPak (Becton Dickinson and Company).
Protein preparations and Western blot analysis.
Culture supernatant fluid was separated by centrifugation at 500 × g from 20-ml TGY cultures of C. perfringens isolates grown for 18 h at 37°C. This culture supernatant fluid was used to isolate supernatant proteins by ammonium sulfate precipitation (29) or to concentrate to 20-fold at 4°C using Amicon Centricon Ultra centrifugal filter devices (Millipore). Total cell protein was prepared from the bacterial pellet of a 20-ml TGY culture which was used to prepare culture supernatant proteins as previously described (29). Prepared proteins were electrophoresed on a 0.1% sodium dodecyl sulfate-12% polyacrylamide gel and transferred onto a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad), and CPB2 Western blot analysis was performed as previously described (29).
Restriction fragment length polymorphism (RFLP) and Southern blot analyses.
A 318-bp digoxigenin (DIG)-labeled cpb2-specific DNA probe was prepared by using a two-step PCR amplification method as previously described (29). Isolated C. perfringens DNA samples, prepared as previously described (8, 23), were digested with EcoRV or HpaI (New England Biolabs), separated by electrophoresis on 1% agarose gels, and Southern transferred. The blots were hybridized with the DIG-labeled cpb2 probe, which was then detected using a DIG chemiluminescence detection system utilizing CSPD [disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5-chloro)tricyclo[3,3.1.13.7]decane}-4-yl)phenyl phosphate] ready-to-use substrate (Roche) as described earlier (23).
PFGE and Southern blot analyses.
Cells from overnight TGY cultures were collected by centrifugation and used to prepare agarose plugs containing genomic C. perfringens DNA as previously described (4, 6, 7, 24, 25). The undigested total unsheared genomic DNA in 100 μl of an agarose plug was electrophoresed at 200 V with a Bio-Rad CHEF-DRII apparatus, with a pulse ramped from 50 to 90 s over 20 h (24, 25). These pulsed-field gel electrophoresis (PFGE) gels were then subjected to cpb2 Southern blot analysis using the same procedure described above.
Cloning and sequencing of the cpb2-containing fragment from various cpb2-positive isolates.
A 1,373-bp DNA fragment carrying cpb2 open reading frame (ORF) and 262-bp upstream and 312-bp downstream sequences from each of three horse GI disease isolates (106902, 106903, and AHT3553) was PCR amplified using primers 5′-GCTCTAGAGGATATCTTAAATTTAGCACAG-3′ and 5′-CCGGAATTCTTTTTTAAGCTCAATTTTTACTGG-3′ as previously described (29). These PCR products were then cloned into the pCR-XL-TOPO vector using the TOPO XL cloning kit (Invitrogen). Both strands of the cpb2-containing DNA insert, from two clones for each isolate, were then sequenced using M13 forward and reverse primers.
RT-PCR analysis.
Total RNA was isolated from TGY cultures of C. perfringens strains as previously described (8, 14). The primer set 5′-AAACTGAATTTTTAAATGGTGC-3′ and 5′-TCCACATCCAATGATCTACAA-3′, which amplified a 318-bp cpb2 internal fragment, was used to detect cpb2-specific mRNA in total RNA preparations by reverse transcriptase (RT)-PCR analysis with the commercially available Access RT-PCR kit (Promega). After RT-PCR, the presence of an amplified product was analyzed by subjecting an aliquot of each sample to agarose gel electrophoresis followed by ethidium bromide staining and visualization under UV illumination. The presence of an amplified product of the expected size was indicative of the presence of specific mRNA transcripts in total RNA preparations.
Northern blot analysis.
A 560-bp internal cpb2 DNA fragment was amplified from CWC245 by PCR using primer set 5′-GGAGCAATAAGTCCAATGAA-3′ and 5′-TCCACATCCAATGATCTACAA-3′ and labeled with alkaline phosphatase using the Gene Images AlkPhos direct labeling and detection system (Amersham Biosciences). Total RNA isolated from C. perfringens strains grown in TGY broth at 37°C for different time periods as previously described (8, 14) was separated by electrophoresis on 1% agarose gels and Northern transferred. The blots were hybridized with the alkaline phosphatase-labeled cpb2 probe, and hybridized probe was then detected by CDPstar chemiluminescence (Amersham Biosciences). The relative levels of cpb2 mRNA in horse GI disease isolates were determined from a calibration curve, which was made using various amounts (0.1 to 5 μg) of RNA prepared from high-CPB2-producing pig strain CWC245. The densitometric analysis was performed on a Macintosh computer using the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
Construction of complementing strains 106902(pMRS140) and 106903(pMRS140).
A 1,373-bp KpnI-XhoI fragment was extracted from pMW1 and cloned into KpnI/SalI sites of pJIR751 (3). Plasmid pMW1, which is a derivative of the pCR-XL-TOPO vector carrying the 106902 cpb2 ORF and its upstream and downstream sequences, was constructed for nucleotide sequencing analyses as described above. The plasmid pMRS140 was introduced into C. perfringens 106902 and 106903 electrocompetent cells by electroporation as previously described (8, 23).
RESULTS
CPB2 production by cpb2-positive horse GI disease isolates.
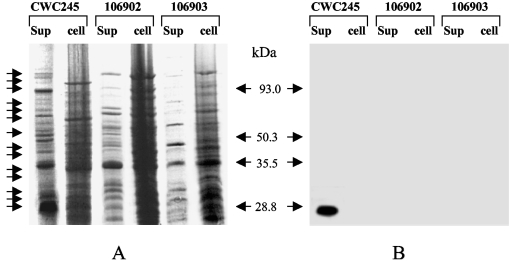
Our collection of European horse GI disease isolates has been previously identified (10, 13) as cpb2 positive by using a multiplex PCR assay. Since a recent study (5) reported that only 50% of nonporcine cpb2-positive C. perfringens isolates can produce CPB2, in this study we first evaluated whether our collection of European horse GI disease isolates positive for cpb2 by PCR (cpb2 PCR-positive isolates) could, in fact, produce CPB2. We first analyzed culture supernatant proteins from cpb2-positive C. perfringens. When CPB2 Western blotting was performed with supernatant proteins from a control CPB2-producing C. perfringens pig isolate, CWC245 (11), an ∼28-kDa immunoreactive band was obtained (Fig. 1), consistent with previous reports (11, 29). However, no CPB2 production could be detected in the culture supernatant of a cpb2-negative isolate, 106527 (data not shown).
FIG. 1.
Western blot analysis of CPB2 production by selected horse GI disease isolates. (A) Culture supernatant (sup) or total cell (cell) proteins, prepared from each of the specified C. perfringens isolates, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Migration of the supernatant proteins is shown by arrows. (B) Western blot of the gel shown in panel A. The blot was probed with CPB2 antibodies and developed by chemiluminescent detection to identify immunoreactive species. Results shown for control isolates include those for CWC245 (a cpb2-positive type C strain). Results shown for representative horse GI disease isolates include those for isolates 106902 and 106903. Molecular mass markers are shown in the middle.
When similar CPB2 Western blot analyses were performed on culture supernatant proteins prepared from our unknown 23 horse GI disease isolates, no CPB2-specific immunoreactive band was detected (Table 1; representative results are shown in Fig. 1). To investigate whether this lack of CPB2 production in our surveyed cpb2-positive horse GI disease isolates was due to impaired secretion of CPB2, we compared the production of CPB2 in culture supernatant with that in total cell protein preparations (Fig. 1) using CPB2 Western blotting analysis. Again, no CPB2-specific immunoreactive band was observed in the assays with either supernatant or total cell proteins of horse GI disease isolates (Fig. 1). These results suggested that all of our surveyed horse GI disease isolates were incapable of producing detectable levels of CPB2.
RFLP genotyping of cpb2-positive C. perfringens isolates.
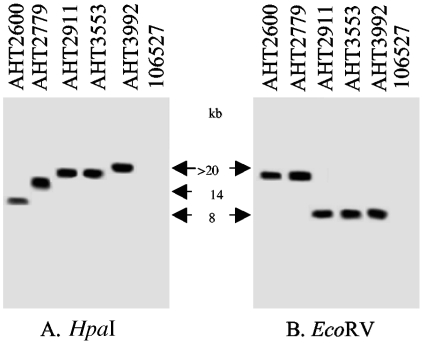
To evaluate whether CPB2 Western blotting-negative isolates were truly cpb2 positive, our collection of horse GI disease isolates was subjected to RFLP analysis. We first performed RFLP-Southern blot analysis on representative cpb2-positive (CWC245) and cpb2-negative (106527) C. perfringens isolates using HpaI or EcoRV restriction enzymes which have no cleavage site within the cpb2 ORF. As expected, a DIG-labeled cpb2 probe hybridized to an ∼5-kb HpaI or ∼7.5-kb EcoRV DNA fragment of CWC245, respectively (reference 29 and data not shown). However, no hybridizing band was observed with DNA from cpb2-negative isolate 106527 (Fig. 2).
FIG. 2.
RFLP-Southern blot analysis of HpaI- or EcoRV-digested DNA from horse GI disease isolates. Total DNA isolated from each of the specified C. perfringens strains was digested with HpaI (A) or EcoRV (B) and then Southern transferred. The Southern blots were probed with a 318-bp DIG-labeled cpb2-specific probe. Results shown for control isolates include those for 106527 (a cpb2-negative type C strain). Representative cpb2-positive horse GI disease isolates include AHT2600, AHT2779, AHT2911, AHT3553, and AHT3992. Migration of the hybridizing band derived from each strain is indicated between the two blots.
When the same HpaI RFLP-Southern blot assay was applied to our collection of horse GI disease isolates, the cpb2 probe hybridized to >20-kb HpaI DNA fragments from 21 out of 23 surveyed isolates (Table 1; representative results are shown in Fig. 2). Two isolates, AHT2600 and AHT2779, produced hybridizing bands of ∼8 kb and ∼14 kb, respectively (Table 1 and Fig. 2). In order to confirm that these differing HpaI RFLP results were not simply due to the loss of an HpaI restriction site, further RFLP analyses were performed on EcoRV-digested DNA (Fig. 2). These EcoRV RFLP results confirmed our HpaI RFLP results, demonstrating that the EcoRV RFLP patterns of these two isolates are different from that of the other surveyed horse GI disease isolates (Fig. 2). These results suggest that all of our surveyed horse GI disease isolates carry the cpb2 sequences.
PFGE evidence supporting the plasmid localization of cpb2 in isolates from horses with GI diseases.
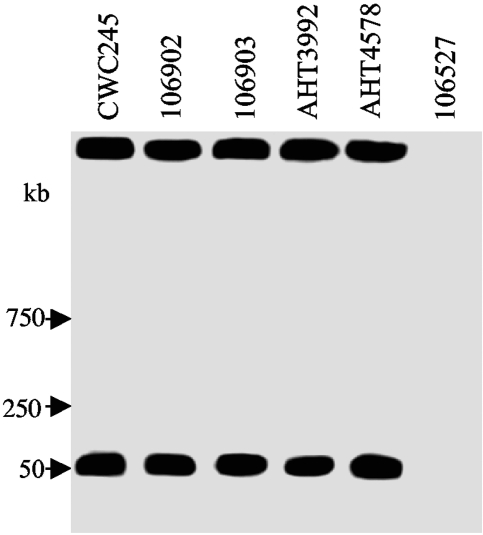
To determine whether cpb2 is located on the chromosome or plasmid in horse GI disease isolates, representative cpb2-positive C. perfringens isolates were subjected to PFGE-Southern blot analysis, which has been successfully used (4, 6, 7, 24, 29) to formally establish the chromosomal or plasmid localization of cpe and cpb2. As expected, the cpb2-containing DNA from control strain CWC245 entered the pulsed-field gel in the absence of restriction enzyme digestion and produced a hybridizing band of ∼50 kb (Fig. 3). These PFGE results confirmed previous observations that cpb2 is located on the plasmid of cpb2-positive isolate CWC245 (11, 29).
FIG. 3.
PFGE evidence supporting the plasmid localization of cpb2 in horse GI disease isolates. PFGE and Southern hybridizing analysis of undigested DNA, prepared in agarose plugs, from each of the specified C. perfringens isolates. Blots were probed with a 318-bp cpb2-specific probe. Results shown for control isolates include those for CWC245 (a cpb2-positive type C isolate carrying the cpb2 gene on a large plasmid) and 106527 (a cpb2-negative type C isolate). Representative results shown for cpb2 horse GI disease isolates include those for 106902, 106903, AHT3992, and AHT4578. The pulsed-field gel was calibrated with λ DNA markers, whose migration is shown at the left of the blot.
When similar PFGE genotyping analyses were performed on DNA of representative cpb2-positive horse GI disease isolates, their cpb2-containing plasmid DNA entered the pulsed-field gels and produced hybridizing bands that comigrated with the cpb2-containing plasmid DNA from CWC245 (Table 1; representative results are shown in Fig. 3). These results indicated that the cpb2-positive C. perfringens horse GI disease isolates carry cpb2 on a plasmid.
Comparison of the cpb2 ORFs present in different cpb2-positive horse GI disease isolates.
To examine whether horse GI disease isolates carry intact cpb2 ORFs, both strands of a 1,373-bp PCR-amplified DNA insert carrying the cpb2 ORFs from three different horse GI disease isolates were nucleotide sequenced. These analyses (data not shown) revealed that no frameshift mutations or premature termination codons were found in the cpb2 ORF sequences of the horse GI disease isolates which we surveyed. The cpb2 ORF sequences present in horse GI disease isolates were identical to one another. Furthermore, the cpb2 ORF sequences present in three of these isolates match the previously determined (11) cpb2 ORF sequence from CWC245 (a type C strain isolated from a diseased pig in France) except for one nucleotide change. The replacement of G with A at position 408 (from ATG) in cpb2 ORFs of surveyed horse GI disease isolates creates a replacement of valine with isoleucine. These results suggest that our surveyed three horse GI disease isolates carry a consensus, but not atypical, cpb2 gene (16). Comparison of the cpb2 upstream and downstream sequences present in all three isolates revealed that both upstream and downstream sequences were identical in all three horse GI disease isolates and matched the previously determined cpb2 upstream and downstream sequences of CWC245 (11).
RT-PCR analysis of cpb2-positive horse GI disease isolates.
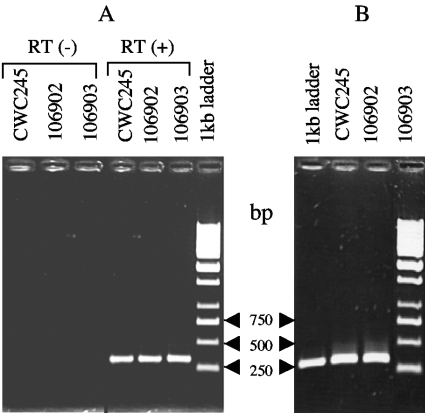
Having obtained evidence for the presence of intact cpb2 ORFs in all of our surveyed CPB2 Western blotting-negative horse GI disease isolates, we performed RT-PCR analyses to evaluate whether the cpb2 ORFs in these isolates were transcriptionally active. When control experiments were performed to confirm the specificity and reliability of the RT-PCR assay, no detectable RT-PCR-amplified product was observed with RNA of a cpb2-positive isolate, CWC245, in the absence of RT (Fig. 4A), indicating that no contaminating DNA was present in the RNA preparation. However, as expected, a 318-bp amplified product was detected in RNA of CWC245 in the presence of RT (Fig. 4A). The size of the RT-PCR-amplified product exactly matched the size of the product obtained in the control PCR using CWC245 chromosomal DNA as a template and the same primers (Fig. 4B). These results indicated that the cpb2 mRNA is present in total RNA of CWC245.
FIG. 4.
RT-PCR analysis of C. perfringens strains. (A) Total RNA prepared from each specified strain was subjected to RT-PCR analysis using cpb2-specific internal primers as described in Materials and Methods. RT (+) and RT (−) indicate the presence and absence, respectively, of RT in the PCR. The RT-PCR-amplified products were analyzed by agarose (1%) gel electrophoresis and photographed under UV light. Molecular sizes of the DNA markers are given on the right. (B) Total DNA isolated from each specified strain was subjected to PCR analysis using the same cpb2-specific internal primers as those used for RT-PCR. The PCR-amplified products were analyzed by agarose (1%) gel electrophoresis and photographed under UV light. Molecular sizes of the DNA markers are given on the left.
When the same RT-PCR analysis was applied to RNA from our four representative horse GI disease isolates (106902, 106903, AHT327, and AHT3992), the 318-bp RT-dependent amplified product was detected in total RNA of all four surveyed isolates (Fig. 4A and data not shown). The size of each RT-PCR-amplified product exactly matched the size of the PCR product obtained with DNA of each surveyed isolate (Fig. 4B and data not shown). These results suggest that the horse GI disease isolates are able to produce cpb2-specific transcripts and that the negative CPB2 production in these isolates may not be due to the transcriptional inactivation of cpb2.
Comparative Northern blot analysis of cpb2-positive horse GI disease isolates.
After we demonstrated the presence of cpb2 transcripts in RNA of our surveyed isolates, it became of interest to compare the level of cpb2 mRNA synthesis in CPB2 Western blotting-negative horse GI disease isolates with that in the CPB2-producing pig isolate CWC245 by Northern blot analysis. As shown in Fig. 5, our cpb2-specific probe hybridized to an ∼1.2-kb mRNA species present in RNA samples extracted from CWC245. The size of the CWC245 cpb2 transcript matched the size of the cpb2 transcript (∼1.4 kb) from strain 13 (22) and was in close agreement with the cpb2 coding sequence from CWC245 (10). However, as expected, this probe showed no hybridization to RNA samples extracted from a cpb2-negative isolate, 106527 (Fig. 5), even though ethidium bromide staining (data not shown) confirmed that equivalent amounts of RNA samples from both CWC245 and 106527 had been electrophoresed. These results indicated that our Northern blot results were reliable and specific.
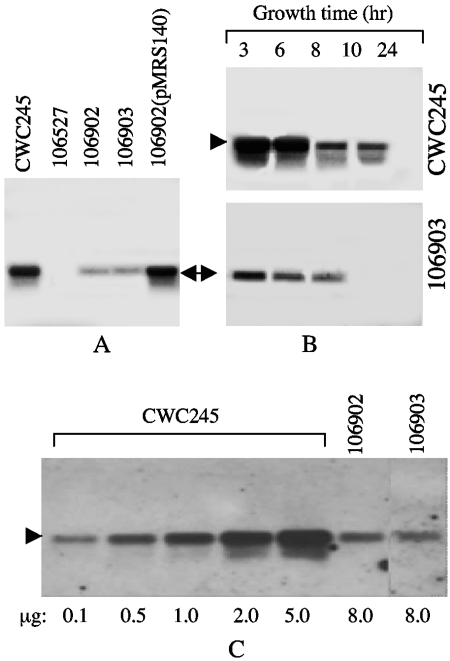
FIG. 5.
Northern blot analysis of cpb2 mRNAs from C. perfringens. (A) Expression of cpb2 mRNAs from horse GI disease isolates. RNA was extracted from specified C. perfringens cultures grown at 37°C for 3 h, and 8 μg of RNA from each was subjected to Northern blot analysis using a 560-bp alkaline phosphatase-labeled cpb2-specific probe. Results shown for control isolates include those for CWC245 (a known cpb2-positive type C strain) and 106527 (a cpb2-negative type C strain). Representative results shown for cpb2-positive horse GI disease isolates include those for 106902 and 106903. The cpb2 mRNA (1.2 kb) is indicated by an arrow. (B) Time course expression of cpb2 mRNAs. RNA was extracted from CWC245 and 106903 cultures grown at 37°C for specified time periods, and 8 μg of RNA from each was subjected to Northern blot analysis using a cpb2-specific probe. The cpb2 mRNA (1.2 kb) is indicated by arrowheads. (C) Comparative expression of cpb2 mRNAs from representative horse GI disease isolates and that from CWC245. Various amounts of RNA (0.1 to 8 μg), extracted from CWC245 (0.1 to 5 μg), 106902 (8 μg), and 106903 (8 μg) cultures grown at 37°C for 3 h, were subjected to Northern blot analysis using a 560-bp alkaline phosphatase-labeled cpb2-specific probe. The relative levels of cpb2 mRNA in horse GI disease isolates were determined as described in Materials and Methods. The cpb2 mRNA (1.2 kb) is indicated by an arrowhead. The various amounts of RNA loaded onto the gel are indicated at the bottom of the blot.
When these Northern blot analyses were applied to RNA prepared from C. perfringens horse GI disease isolates, a single 1.2-kb band, which comigrated with the cpb2 probe-reactive band of CWC245, was detected in RNA from all our surveyed isolates (Fig. 5 and Table 1). However, the expression of cpb2 in horse GI disease isolates was significantly low compared to that in pig isolate CWC245 (Fig. 5A). Over three experimental repetitions, the 1.2-kb cpb2 probe-reactive species was reproducibly less abundant in RNA samples extracted from our surveyed horse GI disease isolates than in RNA samples extracted from CWC245, possibly because these strains produced too little cpb2 message, as would be consistent with Western blotting results indicating no detectable CPB2 production by horse GI disease isolates. Time course experiments demonstrated that the maximal cpb2 transcription in 3-h growth cultures of both pig isolate CWC245 and horse isolate 106903 and the transcription level gradually decreased over the time period. Although expression of cpb2 was continued up to 10 h in the growth culture of CWC245, no cpb2 transcript could be detected in the 106903 culture after 8 h (Fig. 5B). When the relative cpb2 mRNA levels were determined by semiquantitative Northern blot analysis, an average difference of 35-fold in cpb2 transcription levels was observed between a pig strain (CWC245) producing high levels of CPB2 and five surveyed horse GI disease strains (170, AHT327, AHT3992, 106902, and 106903) producing undetectable levels of CPB2 (Fig. 5C and data not shown). These results suggest that decreased transcription and/or message instability may be involved, at least in part, in the low CPB2 production levels noted for the horse GI disease isolates compared with that for pig GI disease isolate CWC245.
Overexpression of cpb2 in horse GI disease isolates 106902 and 106903 by a recombinant plasmid carrying the cpb2 gene from 106902.
To confirm that undetectable CPB2 production in horse GI disease isolates was due to low expression of cpb2 mRNA, we introduced plasmid pMRS140, which is a derivative of multicopy shuttle vector pJIR751 and carries the 106902 cpb2 ORF with its own promoter, into 106902 and 106903 cells. After erythromycin-resistant colonies were selected, PCR analysis using vector-specific M13 forward and reverse primers confirmed the presence of pMRS140 in 106902 and 106903 transformants, which were then named 106902(pMRS140) and 106903(pMRS140). When total RNA extracted from TGY cultures of either 106902(pMRS140) or 106903(pMRS140) was subjected to Northern blot analyses, a single 1.2-kb cpb2 probe-specific band was detected (Fig. 5A and data not shown). The 1.2-kb cpb2 probe-reactive species was reproducibly more abundant in RNA samples extracted from 106902(pMRS140) and 106903(pMRS140) than in RNA samples extracted from host strains 106902 and 106903, respectively (Fig. 5A and data not shown). There were no differences in the abundance of the 1.2-kb species in RNA samples extracted from 106902(pMRS140), 106903(pMRS140), and CWC245 (Fig. 5A and data not shown). These results suggest that the cpb2 gene present on the multicopy plasmid pMRS140 could overexpress cpb2 mRNA in 106902 and 106903.
CPB2 Western blot analyses (Fig. 6) demonstrated that the plasmid pMRS140 could overproduce CPB2 in 106902 and 106903. pMRS140 transformants were able to produce CPB2 in a secretion-dependent manner reminiscent of CPB2 production by CWC245; that is, neither pMRS140 transformant produced detectable CPB2-specific immunoreactivity in total cell proteins (data not shown), but the transformants did produce CPB2-specific immunoreactivity in culture supernatant proteins (Fig. 6). CPB2 Western blots also demonstrated, as expected, that CPB2 production (Fig. 6 and data not shown) was absent from both total cell and culture supernatant proteins when the shuttle vector pJIR751 alone was introduced into 106902 or 106903, confirming that the observed overproduction of CPB2 in 106902(pMRS140) and 106903(pMRS140) was specifically caused by transcomplementation of the wild-type cpb2 gene present in pMRS140. Collectively, these results indicated that the cpb2 gene present in horse GI disease isolate 106902 was functional and that there was a positive correlation between cpb2 message levels and the amount of CPB2 produced by C. perfringens.
FIG. 6.
Overproduction of CPB2 in horse GI disease isolates with a recombinant plasmid carrying cpb2 from 106902. Culture supernatant proteins, prepared from each of the specified C. perfringens isolates, were subjected to Western blot analysis. The blot was probed with CPB2 antibodies and developed by chemiluminescent detection to identify immunoreactive species. Molecular mass markers are shown on the left.
DISCUSSION
Our present study reports an array of evidence supporting the hypothesis that all of our surveyed horse GI disease isolates carry a functional cpb2 gene and that the lack of CPB2 production is not due to the inactivation or loss of the cpb2 gene. (i) RFLP- and PFGE-Southern blot analyses reconfirmed the presence of cpb2 sequences on a large plasmid in all of our surveyed horse GI disease isolates. (ii) Nucleotide sequencing identified intact cpb2 ORFs in our surveyed CPB2 Western blotting-negative isolates. The cpb2 ORF sequences present in three surveyed horse GI disease isolates are identical to one another and to previously determined (11, 29) cpb2 ORF sequences. (iii) Finally, complementation studies demonstrated that a recombinant plasmid carrying 106902 cpb2 increased CPB2 production in 106902 cells to a detectable level (Fig. 6), indicating that the 106902 cpb2 gene is fully functional. Collectively, these results suggested that the differences in CPB2 production levels observed between CWC245 and our surveyed horse GI disease isolates may be due to differences in cpb2 transcription levels.
The most significant finding of this study is the presentation of the first evidence that cpb2 is transcriptionally active in all of our surveyed CPB2 Western blotting-negative isolates and the observation of an average of 35-fold-lower mRNA levels in horse GI disease isolates than in pig GI disease isolate CWC245. This lower level of mRNA synthesis might be a major cause of the undetectability of CPB2 production in horse GI disease isolates. This idea received further support from our observation that the complementing strain 106902(pMRS140), which produced significantly higher levels of mRNA than its parent strain, 106902 (Fig. 5A), produced CPB2 at a level similar to CWC245 (Fig. 6). These findings provide a genetic basis, at least in part, for the lack of a detectable level of CPB2 production in vitro in C. perfringens horse GI disease isolates, which may also be applicable to other nonporcine isolates characterized previously (5), where only 50% of cpb2-positive nonporcine isolates produced CPB2.
Our complementation studies also suggest that there is a positive correlation between cpb2 transcription levels and the amount of CPB2 produced by a C. perfringens cell and that decreased transcription and/or message instability may be involved, at least in part, in the lower CPB2 production noted for the horse GI disease isolates than for pig isolate CWC245. We did not examine stabilities of cpb2 mRNAs and therefore cannot rule out the possibility that the cpb2 mRNAs of horse GI disease isolates are less stable than that of CWC245. However, no degraded cpb2 transcript was detected on the Northern blot of our surveyed isolates (Fig. 5), suggesting that the difference in CPB2 production levels in vitro between CWC245 and horse GI disease isolates may have arisen due to different rates of cpb2 transcription. The differential level of alpha-toxin gene (plc) transcription in C. perfringens was previously reported; the levels of plc transcripts differ up to 40-fold from one C. perfringens type to another, and the greater production of alpha-toxin by the type A strains is due mainly to an increased level of plc transcripts (17, 28). Further transcriptional analyses of a large number of cpb2-positive type C isolates should address whether the difference in the levels of cpb2 transcription in CWC245 (type C pig strain) and horse GI disease isolates (type A) observed in our present study is typically due to isolates' type (type A versus type C) or origin (pig versus horse).
What could be responsible for the different rates of cpb2 transcription between CWC245 and horse GI disease isolates? An inverted repeat sequence identified in the downstream region of the CWC245 cpb2 sequence (11) is also present in our surveyed horse GI disease isolates. The cpb2 upstream promoter region is identical in both pig and horse isolates. The 1,373-bp sequenced fragment does not contain any features which would account for different cpb2 expression levels between pig strain CWC245 and surveyed horse GI disease isolates. Therefore, it can be envisioned that the relevant difference(s) might be present outside of this fragment and be involved in regulating cpb2 expression.
The association of cpb2-positive C. perfringens type A isolates with GI diseases in horses raises an important, but still unanswered, question. Is this low-level expression of cpb2 enough to cause any GI symptoms of CPB2-associated GI diseases, or do these symptoms also result from expression of other C. perfringens toxin(s)? Our present study does not rule out the possibility that these cpb2-positive type A horse GI disease isolates acquire high-level expression of cpb2 in vivo, which could be enough to cause symptoms of CPB2-associated GI diseases in horses. All of the type A horse GI disease isolates surveyed in this study were classified as cpe-negative type A isolates, indicating that the production of C. perfringens enterotoxin or beta-, iota-, or epsilon-toxin is not required for a CPB2-producing type A isolate to cause GI diseases in horses. Since the present study confirmed that cpb2-positive type A horse GI disease isolates can produce both CPB2 and alpha-toxin, it is possible that alpha-toxin also can be a major contributor to the pathogenesis of diseases caused by these isolates. Further CPB2 and alpha-toxin knockout studies should address the relative contributions of CPB2 and alpha-toxin in cpb2-positive C. perfringens type A-associated GI diseases in animals.
Acknowledgments
This research was supported by a grant from the N. L. Tartar Foundation of Oregon State University, by a grant from the Medical Research Foundation of Oregon Health Science University, by a grant from the Agricultural Research Foundation of Oregon State University, and by USDA grant 2002-02281 from the Ensuring Food Safety Research Program (all to M.R.S.).
We gratefully acknowledge the provision of isolates by Neil Chanter at the Animal Health Trust, United Kingdom. We thank Nahid Mahfuz for technical assistance and I-Hsiu Huang for his editorial comments.
REFERENCES
- 1.Bacciarni, L. N., O. Pagan, J. Frey, and A. Grone. 2001. Clostridium perfringens beta2-toxin in an African elephant (Loxodonta africana) with ulcerative enteritis. Vet. Rec. 149:618-620. [DOI] [PubMed] [Google Scholar]
- 2.Bacciarni, L. N., P. Boerlin, R. Straub, J. Frey, and A. Grone. 2003. Immunohistochemical localization of Clostridium perfringens β2-toxin in the gastrointestinal tract of horses. Vet. Pathol. 40:376-381. [DOI] [PubMed] [Google Scholar]
- 3.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors carrying single antibiotic resistance determinants. Plasmid 229:233-235. [DOI] [PubMed] [Google Scholar]
- 4.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bueschel, D. M., B. H. Jost, S. J. Billington, H. T. Trinh, and J. G. Songer. 2003. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet. Microbiol. 94:121-129. [DOI] [PubMed] [Google Scholar]
- 6.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 8.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, D. J., K. Miyamoto, B. Harrison, S. Akimoto, M. R. Sarker, and B. A. McClane. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56:747-762. [DOI] [PubMed] [Google Scholar]
- 10.Garmory, H. S., N. Chanter, N. P. French, D. Bueschel, J. G. Songer, and R. W. Titbal. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibert, M., C. Jolivet-Reynaud, and M. R. Popoff. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203:65-73. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, M. V., and L. P. Elliott. 1980. New presumptive identification test for Clostridium perfringens: reverse CAMP test. J. Clin. Microbiol. 12:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herholz, C., R. Miserez, J. Nicolet, J. Frey, M. R. Popoff, M. Gibert, H. Gerber, and R. Straub. 1999. Prevalence of β2-toxigenic Clostridium perfringens in horses with intestinal disorders. J. Clin. Microbiol. 37:358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, I., M. Waters, R. R. Grau, and M. R. Sarker. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 234:234-240. [DOI] [PubMed] [Google Scholar]
- 15.Jolivet-Reynaud, C., M. R. Popoff, M. A. Vinit, P. Ravisse, H. Moreau, and J. E. Alouf. 1986. Enteropathogenicity of Clostridium perfringens beta toxin and other clostridial toxins. Zentbl. Bakteriol. S15:145-151. [Google Scholar]
- 16.Jost, B. H., S. J. Billington, H. T. Trinh, D. M. Bueschel, and J. G. Songer. 2005. Atypical cpb2 genes, encoding beta2-toxin in Clostridium perfringens isolates of nonporcine origin. Infect. Immun. 73:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, S., O. Matsushita, J. Minami, S. Mizobushi, and A. Okabe. 1993. Comparison of the alpha-toxin genes of Clostridium perfringens type A and C strains: evidence for extragenic regulation of transcription. Infect. Immun. 61:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaasen, H. L. B. M., M. J. C. H. Molkenboer, J. Bakker, R. Miserez, H. Hani, J. Frey, M. R. Popoff, and J. F. Van den Bosch. 1999. Detection of the β2 toxin gene of Clostridium perfringens in diarrhoeic piglets in The Netherlands and Switzerland. FEMS Immunol. Med. Microbiol. 24:325-332. [DOI] [PubMed] [Google Scholar]
- 19.Manteca, C., G. Daube, T. Jauniaux, A. Linden, V. Pirson, J. Detilleux, A. Ginter, P. Coppe, A. Kaeckenbeeck, and J. G. Mainil. 2002. A role for the Clostridium perfringens beta2-toxin in bovine enterotoxaemia? Vet. Microbiol. 86:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClane, B. A. 2001. Clostridium perfringens, p. 351-372. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 21.Mcdonel, J. L. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p. 477-517. In F. Dorner and H. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom.
- 22.Ohtani, K., H. I. Kawsar, K. Okumura, H. Hayashi, and T. Shimizu. 2003. The Vir/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence gene in Clostridium perfringens strain 13. FEMS Microbiol. Lett. 222:137-141. [DOI] [PubMed] [Google Scholar]
- 23.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 24.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Songer, J. G. 1996. Clostridial enteric disease of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiede, S., R. Goethe, and G. Amtsberg. 2001. Prevalence of beta2-toxin gene in Clostridium perfringens type A from diarrheic dogs. Vet. Rec. 149:273-274. [DOI] [PubMed] [Google Scholar]
- 28.Tsutsui, K., J. Minami, O. Matsushita, S. Katayama, Y. Taniguchi, S. Nakamura, M. Nishioka, and A. Okabe. 1995. Phylogenetic analysis of phospholipase C genes from Clostridium perfringens type A to E and Clostridium novyi. J. Bacteriol. 177:7164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waters, M., A. Savoie, H. S. Garmory, D. Bueschel, M. R. Popoff, J. G. Songer, R. W. Titball, B. A. McClane, and M. R. Sarker. 2003. Genotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J. Clin. Microbiol. 41:3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]