Abstract
Five repetitive-element PCR (rep-PCR) techniques [primer sets ERIC1R-ERIC2 and REP1R-REP2I and primers ERIC2, BOXA1R, and (GTG)5] were evaluated for the discrimination of Salmonella enterica isolates at the serotype level. On the basis of number, even distribution over the whole fingerprint, and clarity of bands in the fingerprints, the enterobacterial repetitive intergenic consensus (ERIC) primer set and the (GTG)5 primer were chosen for use in the following experiments. For these two primer sets, reproducibility was tested on different lysates of five selected serotypes of Salmonella in the same PCR by using three different PCR runs. Reproducibility was poor between different PCR runs but high within the same PCR run. Furthermore, 80 different serotypes and five isolates which were not typeable by serotyping were fingerprinted. All strains were typeable by the ERIC primer set and the (GTG)5 primer and generated unique fingerprints, except for some strains with incomplete antigenic codes. Finally, 55 genetically different strains belonging to 10 serotypes were fingerprinted to examine the genetic diversity of the rep-PCR within serotypes. This experiment showed that one serotype did not always correlate to only one ERIC or (GTG)5 fingerprint but that the fingerprint heterogeneity within a serotype was limited. In epidemiological studies, ERIC- and/or (GTG)5-PCR can be used to limit the number of strains that have to be serotyped. The reproducibility of isolates in one PCR run, the discriminatory power, and the genetic diversity (stability) of the fingerprint were similar for the Eric primer set and the (GTG)5 primer, so both are equally able to discriminate Salmonella serotypes.
Salmonella enterica is one of the major causes of human gastroenteritis worldwide. In Belgium, 12,894 human Salmonella isolates were received in 2003 by the National Reference Centre for Salmonella and Shigella (12). The most common serotypes isolated were Salmonella serotype Enteritidis, Salmonella serotype Typhimurium, Salmonella serotype Virchow, Salmonella serotype Derby, and Salmonella serotype Brandenburg. The internationally used method for characterizing Salmonella is serotyping of the isolates according to the Kauffmann-White scheme (13). Each Salmonella serotype is characterized by the combined expression of particular lipopolysaccharides, or O antigens, and flagellar proteins, or H antigens. Currently, more than 2,500 serotypes are recognized (14). In most countries, serotyping is restricted to national reference laboratories, to which clinical and food microbiology laboratories send Salmonella isolates. Serotyping, especially of less abundant types, is expensive and time-consuming. In addition, according to the National Reference Centre for Salmonella and Shigella (12) and the Belgian Reference Laboratory for Salmonella (1), respectively, 0.12% of the human isolates and 4% of the isolates of animal origin were not typeable in 2003 (by autoagglutination).
PCR-based molecular techniques are easy to perform and rapid. Repetitive-element PCR (rep-PCR) uses primers complementary to naturally occurring, highly conserved, repetitive DNA sequences. These noncoding sequences are present in multiple copies in the genomes of most gram-negative and several gram-positive bacteria (10). Examples of these repetitive elements are the repetitive extragenic palindromic (REP) sequences, the enterobacterial repetitive intergenic consensus (ERIC) sequences, the BOX sequences, and the polytrinucleotide (GTG)5 sequence (18).
According to Van Lith and Aarts (17), it is possible to use the primer set ERIC1R-ERIC2 to discriminate Salmonella serotypes. Burr et al. (3) and Milleman et al. (11) tested the same primer set and concluded that the fingerprints obtained were not correlated with serotypes. Two studies (8, 9) showed that elevated annealing temperatures combined with the use of a commercial PCR mix improve the reproducibility and the resolving power of rep-PCR with the ERIC2 and BOXA1R primers.
The studies performed yielded conflicting results and evaluated only the ERIC primer set, the ERIC2 primer, and/or the BOX primer on a limited number of serotypes and/or on a limited number of strains per serotype. The purpose of this study was to evaluate five different rep-PCR techniques for the discrimination of Salmonella isolates, including the (GTG)5 primer (5, 18). Selected rep-PCR techniques were further evaluated for their powers of discriminating between as many as 80 different serotypes as well as for genetic diversity within several serotypes.
MATERIALS AND METHODS
Salmonella isolates.
The Salmonella isolates were isolated from the following sources: human feces, poultry, meat, eggs, pigeons, cattle, swine, deer, reptiles, water, farm environments, and farm equipment. Almost all isolates were isolated in Belgium from the year 1998 until 2003 (Table 1). They were serotyped at the Belgian Salmonella reference laboratories according to the Kauffman-White scheme (13).
TABLE 1.
Salmonella isolates used in the experiments
| Serotype | Isolate no. | Source | Serotype | Isolate | Source | |
|---|---|---|---|---|---|---|
| Aba | VVG03/0027 | Pork | ||||
| Aberdeen | MB2555 | Human | ||||
| Adelaidea | MB526 | Not known | ||||
| Agona | KS002 | Broiler's feces | ||||
| Agona | MB1230 | Poultry meat | ||||
| Agona | MB2506 | Industrial surface | ||||
| Agona | 5.22RV | Broiler's carcass | ||||
| Altona | MB2549 | Human | ||||
| Anatum | KS187 | Overshoes in poultry house | ||||
| Apapa | MB2560 | Not known | ||||
| Bareilly | MB1253 | Not known | ||||
| Blockley | 3.1D | Broiler's feces | ||||
| Blockley | 10.4E | Broiler's feces | ||||
| Blockley | KS109 | Feed in broiler house | ||||
| Blockley | KS163 | Overshoes in poultry house | ||||
| Blockley | VVG02/703 | Undetermined food | ||||
| Bovismorbificans | VVG02/1518 | Pig's carcass | ||||
| Braenderup | KS113 | Overshoes in poultry house | ||||
| Brandenburg | KS181 | Cecal dropping of broiler | ||||
| Brandenburg | MB1720 | Pig slaughterhouse | ||||
| Brandenburg | MB1722 | Pig slaughterhouse | ||||
| Brandenburg | MB1724 | Pig slaughterhouse | ||||
| Brandenburg | VVG02/0928 | Pork | ||||
| Bredeney | VVG02/0784 | Chicken fillet | ||||
| Cerro | MB2368 | Environment of food factory | ||||
| Chester | MB2543 | Not known | ||||
| Coeln | MB1080 | Not known | ||||
| Concord | VVG03/0546 | Human | ||||
| Derby | MB1531 | Pig | ||||
| Derby | MB1736 | Pig's carcass | ||||
| Derby | MB1737 | Pig slaughterhouse | ||||
| Derby | MB1739 | Pig | ||||
| Derby | MB1745 | Pig slaughterhouse | ||||
| Derby | VVG02/1145 | Pork | ||||
| Dublin | VVG01Z/1018 | Cattle carcass | ||||
| Enteritidis | KS104 | Egg | ||||
| Enteritidis | KS157 | Overshoes in poultry house | ||||
| Enteritidis | KS585 | Paper tray liners (transport) | ||||
| Enteritidis | MB1208 | Human feces | ||||
| Enteritidis | MB1221 | Tiramisu | ||||
| Enteritidis | MB1409 | Egg | ||||
| Enteritidisc | MB1419 | Egg | ||||
| Enteritidisc | MB1420 | Egg | ||||
| Enteritidisb | MB1432 | Egg white powder | ||||
| Enteritidisd | MB1450 | Human case | ||||
| Enteritidis | MB1535 | Deer | ||||
| Enteritidis | MB1677 | Human | ||||
| Enteritidis | MB2350 | Chicken | ||||
| Gallinarum | MB2499 | Reptile | ||||
| Give | VVG01/0873 | Pig's carcass | ||||
| Goldcoast | VVG01/0646 | Pork | ||||
| Hadar | KS077 | Cecal dropping of broiler | ||||
| Hadar | KS106 | Feed tray in poultry house | ||||
| Hadar | MB1134 | Poultry breeder animals | ||||
| Hadar | MB1148 | Pig | ||||
| Hadar | MB1149 | Poultry | ||||
| Hadar | VVG02/0777 | Poultry meat | ||||
| Havana | VS219353a | Pig | ||||
| Idikan | MB1235 | Not known | ||||
| Indiana | 6.4 | Broiler's feces | ||||
| Indiana | 5.35 | Broiler's carcass | ||||
| Indiana | VVG02/0038 | Poultry meat | ||||
| Infantis | KS001 | Equipment in broiler house | ||||
| Infantis | MB1146 | Feed | ||||
| Infantis | MB1729 | Pig slaughterhouse | ||||
| Infantis | MB1730 | Pig slaughterhouse | ||||
| Infantis | MB1735 | Overshoes in the pig sty | ||||
| Infantis | VVG02/0398 | Beef | ||||
| Isangi | MB1092 | Not known | ||||
| Kedougoub | MB1433 | Egg white powder | ||||
| Kentucky | KS192 | Overshoes in poultry house | ||||
| Kiambu | VVG00/1507 | Poultry meat | ||||
| Kingstone | VVGN077 | Mesenteric lymph node of pig | ||||
| Kintambo | MB2507 | Surface isolation | ||||
| Kottbus | MB2546 | Human | ||||
| Larochelle | MB2544 | Human | ||||
| Litchfield | MB2548 | Not known | ||||
| Livingstone | VVG01/0925 | Pork | ||||
| London | VVG02/0819 | Pork | ||||
| Manhattan | MB1260 | Not known | ||||
| Mbandaka | KS61 | Environment of poultry house | ||||
| Meleagridis | VVG00/1923 | Pig | ||||
| Minnesota | MB2558 | Human | ||||
| Montevideo | VVG13.20K | Broiler's feces | ||||
| Muenchen | MB2547 | Not known | ||||
| Muenster | VVGN049 | Mesenteric lymph node of pig | ||||
| Newport | MB1246 | Not known | ||||
| Ohio | VVG01/0822 | Pig's carcass | ||||
| Oranienburg | VVGN179 | Mesenteric lymph node of pig | ||||
| Panama | VVG02/0923 | Pork | ||||
| Paratyphi A | MB2541 | Human | ||||
| Paratyphi B | VVG02/0726 | Beef | ||||
| Plymouth | MB2553 | Human | ||||
| Poona | VS821475c | Mesenteric lymph node of pig | ||||
| Pullorum | MB2349 | Poultry feathers during plucking | ||||
| Putten | VVGP109M | Processing of broilers | ||||
| Rissen | VVGK015 | Poultry | ||||
| Rubislaw | MB2556 | Not known | ||||
| Sandiego | VVG02/0235 | Undetermined food | ||||
| Senftenberg | MB1559 | Feed from broiler house | ||||
| Stanleyville | MB1312 | Egg | ||||
| Stourbridge | MB2550 | Human | ||||
| Sundsvall | VVGN485 | Mesenteric lymph node of pig | ||||
| Swartzengrund | VS112916c | Pig's feed trough | ||||
| Telelkebir | MB2557 | Not known | ||||
| Tennessee | MB1198 | Poultry feed | ||||
| Thompson | VVG01/0010 | Poultry meat | ||||
| Typhimurium O5− | MB1217 | Human | ||||
| Typhimurium O5− | MB1780 | Pigeon | ||||
| Typhimurium O5− | MB1786 | Pigeon | ||||
| Typhimurium O5− | VVG01/0922 | Pig | ||||
| Typhimurium O5+ | MB1241 | Not known | ||||
| Typhimurium O5+ | MB2177 | Pig carcass | ||||
| Typhimurium O5+ | MB2199 | Overshoe on pig farm | ||||
| Typhimurium O5+ | MB2249 | Human | ||||
| Typhimurium O5+ | MB2274 | Human | ||||
| Typhimurium O5+ | MB2299 | Human | ||||
| Urbana | VVGN198 | Mesenteric lymph node of pig | ||||
| Virchow | KS087 | Equipment in broiler house | ||||
| Virchow | MB2339 | Broilers | ||||
| Virchow | MB2341 | Chicken fillet | ||||
| Virchow | MB2342 | Chicken fillet | ||||
| Virchow | MB2396 | Not known | ||||
| Virchow | VVG02/1504 | Poultry meat | ||||
| Waycross | MB2559 | Human | ||||
| Wien | VVG02/0527 | Undetermined food | ||||
| 4:i:− | VVGN385 | Mesenteric lymph node of pig | ||||
| 47:z4z23:− | VS219188a | Pig | ||||
| 6,7:−:5 | 5.60RV | Broiler's carcass | ||||
| 6,7:r:− | MB2529 | Industrial surface | ||||
| 6,8:−:1,2 | VVG00/0669 | Pig's carcass | ||||
| 9:−:− | MB2551 | Not known | ||||
| IV48:g,z51:− | MB2561 | Not known | ||||
| Not typeable | VVG02/0042 | Pork | ||||
| Not typeable | KS128 | Overshoes in poultry house | ||||
| Not typeable | MB2562 | Not known | ||||
| Not typeable | MB2563 | Not known | ||||
| Not typeable | MB2564 | Not known |
Originates from Slovakia.
Originates from The Netherlands.
Originates from Austria.
Originates from the United Kingdom.
DNA isolation.
The bacteria were grown overnight on tryptone soy agar plates (CM0131; Oxoid, Basingstoke, United Kingdom) at 37°C. The cells were harvested and resuspended in 300 μl 0.05 M NaOH-0.125% (wt/vol) sodium dodecyl sulfate and heated at 90°C for 17 min. The lysates were stored at −20°C until use, which was approximately 3 weeks later. The lysates were used only once. Before use in the PCR, the lysates were centrifuged for 2 min at 10,000 rpm.
Only when elevated annealing temperatures were tested as described by Johnson and Clabots (8) were DNA extracts used instead of lysates. DNA was extracted using a commercial genomic DNA purification kit (AquaPure genomic DNA isolation kit 732-6340; Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer's instructions.
Selection of primers for rep-PCR.
Salmonella strains belonging to 22 serotypes were typed with the primer sets ERIC1R-ERIC2 and REP1R-REP2I and primers ERIC2, BOXA1R, and (GTG)5. The best primer or primer set and the best corresponding annealing temperatures were selected on the basis of the number, distribution, and clarity of bands in the obtained fingerprints.
Reproducibility of rep-PCR.
In the second experiment, three different lysates were made on three separate days, starting from different bacterial cultures of five selected serotypes (Fig. 1). Reproducibility was evaluated on the different lysates by using three different PCR runs on the same thermal cycler.
FIG. 1.
Cluster of the composite data set and ERIC and (GTG)5 fingerprints of three different lysates made on three different days (t1, t2, and t3) and run in three different PCR runs in the same thermal cycler. The similarities between the fingerprints were calculated using the Pearson correlation (optimization, 1%; position tolerance, 1%), and the fingerprints were grouped according to their similaritiesby use of the UPGMA algorithm. The vertical grey line shows the delineation level of 92.5%. The last column shows the strain numbers. The grey bar at the top of the figure shows the part (79.3% to 84.3%) of the ERIC fingerprints that is not taken into account to calculate the cluster, as explained in the text.
rep-PCR on isolates belonging to different serotypes.
Eighty serotypes and five isolates which were not typeable by serotyping were characterized to test the typeability and the discriminatory power of the selected primer sets.
rep-PCR on isolates belonging to the same serotypes.
Strains belonging to the same serotype, but of different origins and genetically different, were fingerprinted to examine the genetic diversity (stability) of the rep-PCR within serotypes with the selected primer or primer set. The aim of this experiment was to investigate whether genetically different strains of the same serotype resulted in the same fingerprint. The genetic diversity of the strains had been tested by pulsed-field gel electrophoresis using XbaI as the restriction enzyme (2). Fifty five strains of 10 different serotypes were fingerprinted: 13 serotype Enteritidis, 9 serotype Typhimurium, 5 serotype Hadar, 5 serotype Derby, 5 serotype Virchow, 5 serotype Infantis, 4 serotype Blockley, 4 serotype Brandenburg, 3 serotype Agona, and 2 serotype Indiana strains.
PCR.
PCR amplifications were performed in a Perkin-Elmer 9700. The sequences of the primers used and the amplification protocols were those described by Versalovic et al. (18). The reaction mixtures for primers ERIC1R, ERIC2, REP1R, REP2I, and BOXA1R were as described by Rademaker and de Bruijn (15), but Tween 20 (0.5%) and gelatin (0.01%) (6) were added when lysates were used. The reaction mixture for the (GTG)5 primer contained the following: 10 mM Tris HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 60 pmol (GTG)5 (Eurogentec, Seraing, Belgium), 0.5% (vol/vol) Tween 20, 0.01% (wt/vol) gelatin, 1 U Taq DNA polymerase (YellowStar Taq; Eurogentec, Seraing, Belgium), and 1 μl crude cell lysate with a final volume of 25 μl.
Elevated annealing temperatures were also tested in the first experiment. An annealing temperature of 70°C for ERIC2 and BOXA1R and an amplification protocol of 35 cycles without an initial touch-down were used (8), but with the reaction mixture of Rademaker and de Bruijn (15) instead of Ready to Go PCR beads (8). Elevated annealing temperatures were also tested using modified amplification protocols of Versalovic et al. (18). Modifications consisted of annealing temperatures of 57°C for ERIC2 and 65°C for BOXA1R.
The PCR products were size separated in a 1.5% agarose gel in 1× Tris-borate-EDTA at 120 V for 4 h. The gels were stained with ethidium bromide and digitally captured under UV light. The gel images were visually compared and analyzed with GelCompar, version 3.0 (Applied Maths, Kortrijk, Belgium) using a mixture of a 100-bp (Invitrogen, Paisley, United Kingdom) and a 500-bp (Bio-Rad Laboratories, Hercules, Calif.) ladder as the normalization reference (15). The similarities between the fingerprints were calculated using the Pearson correlation (with an optimization of 1% and a position tolerance of 1%), and the fingerprints were grouped according to their similarities by use of the UPGMA (unweighted-pair group method using arithmetic averages) algorithm.
Statistical analysis of dendrograms.
To assess the variability introduced by the preparation of lysates and the PCR run in the clustering of the fingerprints, several indices were derived from the similarity matrix obtained from experiment 2 (reproducibility of rep-PCR) by the method of Johnson and Clabots (8). For all strains, a similarity index (SI) was calculated as the mean of all pairwise correlations between different replicates of a strain at a certain level of a factor (lysate or PCR run). A lower SI can be interpreted as more variability attributable to the factor. The differentiation index (DI) was defined as the maximum of all pairwise correlations between different strains, at a certain level of a factor in one strain and all possible combinations in the other; this was repeated for each strain. A higher DI can be interpreted as more variability attributable to the factor. Finally, the difference between the similarity index and the differentiation index (called “net discriminating power” by Johnson and Clabots [8]) can be considered to be a measure for the discriminatory power of the strain. A higher value, calculated as 100 − (SI − DI), which can be defined as the net variability index, means more variability attributable to the factor on the clustering.
RESULTS
Selection of primers and annealing temperature.
With the ERIC1R-ERIC2 primer set, profiles consisted of 13 to 22 bands, evenly distributed over the entire fingerprint. With the REP1R-REP2I primer set, 5 to 10 bands were obtained for each fingerprint, but most of the bands were weak (data not shown). When the (GTG)5 primer was used, 11 to 16 bands were visible for each fingerprint, most of them located between 1,000 and 2,500 bp. With the BOXA1R primer, the fingerprints consisted of more than 25 bands, which made visual comparison between fingerprints very difficult (data not shown). Elevated annealing temperatures of 70°C did not generate any bands for the ERIC2 primer and generated five bands, all lower than 500 bp, for the BOXA1R primer (data not shown). Annealing temperatures of 57°C for ERIC2 and 65°C for BOXA1R resulted in fingerprints with 10 to 16 bands and 10 to 14 bands, respectively (data not shown).
According to Versalovic et al. (18), the optimal number of bands for rep-PCR is 8 to 15. The upper limit of bands, however, depends on the resolution of the electrophoretic system; the higher the resolution, the more bands can be reliably separated and visualized. With a 1.5% agarose gel separation on 20-cm-long gels, as used in this study, the upper limit was judged to be around 20 distinct fragments. On the basis of the number and clarity of bands, their even distribution over the whole fingerprint, and discriminatory power, the ERIC primer set and the (GTG)5 primer were chosen for use in subsequent experiments. The above data indicate that this primer and this primer set have the greatest potential to discriminate Salmonella strains belonging to different serotypes.
Reproducibility.
All fingerprints obtained with the ERIC primer set had a band at 250 bp (Fig. 1). This high-intensity band was excluded from the calculation of the Pearson correlation in this and subsequent experiments. The Pearson correlation takes the whole profile into account, so exclusion of dense bands often allows more meaningful clustering of groups (4, 7).
Each of the five serotypes clustered together with a minimum similarity coefficient of 74% with the ERIC primer set (data not shown), 83% with the (GTG)5 primer (data not shown), and 80% for the composite data set [ERIC plus (GTG)5] (Fig. 1). The low similarity coefficients were the result of the three different PCR runs. In Table 2 a quantitative assessment is made of the variability attributable to the PCR-run and to the preparation of the lysate in the clustering of the combined ERIC- and (GTG)5-PCR fingerprints. These results clearly indicate that the variability attributable to the PCR run (with the lysate factor kept constant) is systematically higher; this can be deduced from the lower SI values and the higher net variability index [100 − (SI − DI)]. It is therefore advisable to compare results within the same PCR run. Within one PCR run, the minimum similarity coefficient between the three different lysates (i.e., lysates made on different days) was 95% for the ERIC primer set, except for 4 out of a total of 45 lysates (data not shown). The minimum similarity coefficient within one PCR run for the three different lysates was 94% for the (GTG)5 primer, with the exception of six lysates (data not shown). These exceptions were due to overall weaker patterns, locally weaker bands, or normalization errors. For the composite data set, the minimum similarity coefficient for the three different lysates within one PCR run was 92.5%, except for two lysates: KS077, made on the first day and processed in the first PCR run, and MB1720, made on the third day and processed in the third PCR run (Fig. 1). Based on these results, the delineation levels for serotype discrimination in subsequent experiments were set at 95% for the ERIC primer set, 94% for the (GTG)5 primer, and 92.5% for the composite data set.
TABLE 2.
Assessment of variability attributable to PCR and lysates in the clustering of rep-PCR fingerprints
| Variable factor | Constant factor | SI (%)a | DI (%)a | 100 − (SI − DI) (%)a |
|---|---|---|---|---|
| PCR run | Lysate t1b | 85.92 | 61.62 | 75.70 |
| Lysate t2b | 87.66 | 63.14 | 75.48 | |
| Lysate t3b | 86.10 | 62.62 | 76.52 | |
| Lysate | PCR run 1c | 94.34 | 57.11 | 62.77 |
| PCR run 2c | 97.12 | 61.51 | 64.39 | |
| PCR run 3c | 95.76 | 63.48 | 67.72 |
Indices were calculated based on the similarity matrix of the dendrogram of replicate combined ERIC- and (GTG)5-PCR fingerprints of five strains as shown in Fig. 1. The means over the five strains of the different indices are shown.
Measures variability attributable to three different PCR runs on the same lysate.
Measures variability attributable to three different lysates in the same PCR run.
Typeability and discriminatory power.
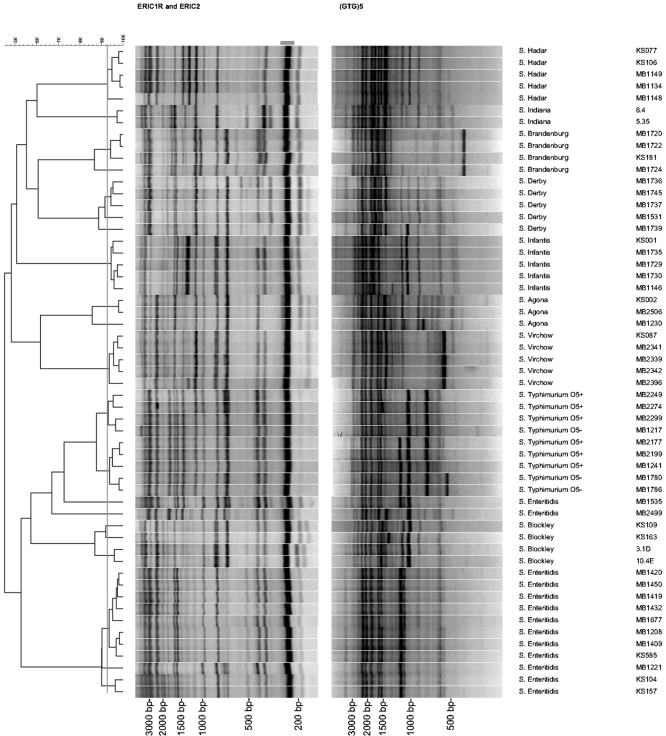
Figure 2 shows the fingerprints of the 80 different serotypes and the five isolates which were not typeable by serotyping, obtained with the ERIC primer set and the (GTG)5 primer, and the clustering of the composite data set. Six of the 85 fingerprints obtained with the ERIC primer set displayed no band at 250 bp. This band was excluded from the calculation of the Pearson correlation, as already mentioned. At a delineation level of 95% for the ERIC primer set, all serotypes generated unique fingerprints except for three pairs of two strains. Two strains (MB2563 and MB2564) which were not typeable by serotyping had a similarity coefficient of 98.8%. Serotype Typhimurium (MB2249) and serotype Typhimurium var. Copenhagen (VVG01/0922) had a similarity coefficient of 96.3%. Serotype Infantis (VVG02/0398) and strain MB2529, with antigenic formula 6,7:r:−, had a similarity of 95.6% (data not shown).
FIG. 2.
Cluster of the composite data set and ERIC and (GTG)5 fingerprints of 80 different serotypes and five isolates that were not typeable by serotyping. The similarities between the fingerprints were calculated using the Pearson correlation (optimization, 1%; position tolerance, 1%), and the fingerprints were grouped according to their similarities by use of the UPGMA algorithm. The vertical grey line shows the delineation level of 92.5%. The second column shows the antigenic formulas of the serotypes given in the first column. The last column shows the strain numbers. The grey bar at the top of the figure shows the part (79.3% to 84.3%) of the ERIC fingerprints that is not taken into account to calculate the cluster, as explained in the text.
At a delineation level of 94% for the (GTG)5 primer, all but nine strains had unique fingerprints. Strains MB2563 and MB2564, which were not typeable by serotyping, had a similarity coefficient of 99.2%. Serotype Enteritidis (MB1409), strain MB2562, which was not typeable by serotyping, and strain MB2551, with antigenic formula 9:−:−, had a similarity coefficient of 95%. Two other strains, one serotyped as serotype Paratyphi B (VVG02/0726) and strain KS128, which was not typeable by serotyping, had a similarity coeffient of 96.6%. Serotype Urbana (VVGN198) and serotype Sundsvall (VVGN485) had a similarity coefficient of 94.8%, although visually there was a difference of two bands (data not shown).
In the composite data set (Fig. 2), two pairs of strains were not discriminated from each other at the delineation level of 92.5%. The two nontypeable strains MB2563 and MB2564 had a similarity coefficient of 98.9%, and serotype Paratyphi B (VVG02/0726) and isolate KS128, which was not typeable by serotyping, had a similarity coefficient of 92.9%.
Genetic diversity (stability).
Figure 3 shows the fingerprints of the 55 isolates belonging to 10 serotypes obtained with the ERIC primer set and the (GTG)5 primer, as well as the clustering of the composite data set. With the ERIC primer set, 24 different clusters and/or separate strains were distinguished with 95% as the delineation level (data not shown). All strains belonging to serotype Agona or Indiana clustered together within the serotype. Serotype Blockley was divided into two clusters and/or separate strains, and serotype Hadar was divided into three clusters and/or separate strains, although the profiles within one serotype were visually the same for both clusters. Serotypes Derby, Brandenburg, and Virchow each had one strain (MB1736, MB1724, and MB2396, respectively) with an ERIC fingerprint that differed in one band from the other strains of the respective cluster. Serotype Infantis strains were divided into three clusters. The difference consisted of bands of more or less intensity at 400 bp. The nine strains of serotype Typhimurium clustered into two groups. The difference consisted of a double or single band at 700 bp. The serotype Enteritidis strains were divided into six clusters. Strain MB2499, strain MB1535, and strain MB1221 had each a different fingerprint from the other 10 strains. The other 10 strains were grouped together in three clusters, and the differences consisted in bands of different intensity at 2,000 bp.
FIG. 3.
Cluster of the composite data set and ERIC and (GTG)5 fingerprints of 55 genetically different strains belonging to 10 serotypes. The similarities between the fingerprints were calculated using the Pearson correlation (optimization, 1%; position tolerance, 1%), and the fingerprints were grouped according to their similarities by use of the UPGMA algorithm. The vertical grey line shows the delineation level of 92.5%. The last column shows the strain numbers. The grey bar at the top of the figure shows the part (79.3% to 84.3%) of the ERIC fingerprints that is not taken into account to calculate the cluster, as explained in the text.
With the (GTG)5 primer, 23 clusters and/or separate strains were distinguished when a delineation level of 94% was applied (data not shown). All strains belonging to serotypes Hadar, Virchow, and Indiana clustered together within the serotype. Each of the serotypes Infantis, Agona, Blockley, and Brandenburg had one strain (MB1146, MB1230, KS163, and KS181, respectively) with a (GTG)5 fingerprint profile that differed in one to three bands from the other strains of the cluster. Serotype Derby was divided into three clusters: two strains (MB1531 and MB1739) each had a fingerprint that differed in one high-intensity band from the other three strains. The nine serotype Typhimurium strains were divided into four clusters. The strains differed in the presence or absence of a band at 1,200 or 600 bp. Strain MB2274 had a band of higher intensity than the other strains at 1,600 bp. Eleven of the 13 strains of serotype Enteritidis were clustered together with a similarity coefficient of 94.6%, whereas the other two strains, MB1535 and MB2499, each had a different fingerprint.
Figure 3 shows the cluster of the composite data set of both primers and the fingerprints. With the delineation level of 92.5%, 21 different clusters and/or separate strains were distinguished. The strains belonging to serotypes Hadar, Indiana, Infantis, and Virchow clustered together within the serotype. The strains of serotypes Brandenburg, Typhimurium, and Agona were divided into two clusters. Serotypes Blockley and Derby were split up into three clusters, whereas serotype Enteritidis was divided into five clusters.
DISCUSSION
To our knowledge this is the first study that has compared five different rep-PCR primers with regard to the potential of discriminating Salmonella serotypes. It is also the first study of the ability of primer (GTG)5 to discriminate Salmonella serotypes. The reproducibility experiment indicated that the PCR run is more important for reproducibility than the lysates made at different times. This means that it is recommended to draw conclusions only from isolates that were processed in the same PCR run. An alternative is to include in the new PCR run some isolates (e.g., one from each cluster) that were processed in a previous PCR run.
All isolates generated fingerprints, including the five isolates which were not typeable by serotyping. All but a few serotypes had unique fingerprints. Strains MB2563 and MB2564, which were not typeable by serotyping, had identical fingerprints with both primers, meaning that they are probably genetically identical strains. Serotype Paratyphi B and one strain that was not typeable by serotyping also had identical fingerprints with both primers, although the similarity coefficient was below the delineation level for the ERIC primer set. Serotype Enteritidis, a strain that was not typeable by serotyping, and a strain with antigenic formula 9:−:− had a similarity coefficient of 95% with the (GTG)5 primer. This clearly shows that rep-PCR can reveal additional information when serotyping is not possible. Furthermore, it is possible that serotype Enteritidis and the strain with antigenic formula 9:−:− belong to the same serotype but that some somatic and flagellar factors of the latter were not expressed during serotyping. The same can be concluded for serotype Infantis and strain MB2529, with antigenic formula 6,7:r:−, which had identical fingerprints with the ERIC primer set. Serotype Typhimurium (O5+) and serotype Typhimurium var. Copenhagen (O5−) produced the same fingerprint with the ERIC primer set. With the (GTG)5 primer, a difference in one high-intensity band at 1,200 bp was observed among the serotype Typhimurium strains tested, which, however, did not correlate with the two varieties in this serotype.
The genetic diversity (stability) experiment further showed that not all isolates with the same serotype had the same fingerprint. However, the isolates with the same serotype still clustered together at a similarity coefficient of 85% or higher, except for two serotype Enteritidis strains. Strain MB2499 was isolated from a reptile, and strain MB1535 was isolated from a deer. Both were also atypical by other characterization methods such as randomly amplified polymorphic DNA and virulence typing (unpublished results). As mentioned by Torpdahl and Ahrens (16), serotype Enteritidis is a polyphyletic serotype. Although strains in this serotype are not genetically related, they share some characteristics such as the somatic and flagellar factors.
The discriminatory powers of the ERIC primer set and the (GTG)5 primer are similar, with 24 clusters (and/or separate strains) obtained by the former and 23 clusters (and/or separate strains) obtained by the latter for a collection of 55 strains belonging to 10 serotypes. Nevertheless, this experiment revealed that the ERIC primer set and the (GTG)5 primer are complementary, since they did not discriminate the same strains within certain serotypes. This experiment clearly shows that one serotype does not always correspond to only one ERIC or (GTG)5 fingerprint, but the fingerprint heterogeneity within a serotype seems to be limited to the absence or presence of mostly one and sometimes two bands for a primer or primer set or to differences in intensities of some bands. In a few restricted cases (e.g., MB1221 in ERIC-PCR of experiment 4 [Fig. 3]), an apparent fingerprint heterogeneity seemed to be due to normalization artifacts. Nevertheless, this experiment indicates that direct serotype identification by rep-PCR may be erroneous if only one reference fingerprint is included.
Other studies have also evaluated the Salmonella discriminating ability of rep-PCR. Most studies (3, 11, 17) tested only the ERIC primer set, with conflicting results. According to Van Lith and Aarts (17), it is possible to use the ERIC1R-ERIC2 primer set to discriminate Salmonella serotypes. Their study was performed on 65 Salmonella isolates of 49 serotypes. They concluded that all serotypes produced unique fingerprints and that the isolates within one serotype had identical patterns. According to Burr et al. (3), who tested the same primer set on 89 Salmonella isolates of 22 serotypes, the fingerprints obtained did not correlate with serotypes. Milleman et al. (11) also tested the ERIC primer set on 56 serotype Typhimurium and 14 serotype Enteritidis strains. They concluded that ERIC-PCR cannot be used to discriminate Salmonella serotypes, since all serotype Enteritidis isolates and some serotype Typhimurium isolates shared the same fingerprint. According to two other studies (13, 14), elevated annealing temperatures improve the reproducibility and resolving power of rep-PCR with the ERIC2 and BOXA1R primers. In the first study only 12 strains of 12 serotypes and in the second study 70 isolates of 15 serotypes were evaluated. We obtained more bands with the ERIC primer set than were obtained in the studies mentioned above. This is probably the reason why some studies revealed that no serotype-dependent fingerprints were obtained while other studies showed the opposite. The PCR conditions are probably critical factors, which also helps explain why in our study no serotype-dependent fingerprints were obtained at elevated annealing temperatures as described by Johnson and Clabots (8).
It can be concluded that in certain epidemiological studies, ERIC-PCR and/or (GTG)5 can be used to limit the number of strains that have to be serotyped, although it is useful only with isolates analyzed in one PCR run. Only one isolate of each cluster has to be sent to the national reference laboratories for serotyping. The reproducibility of isolates in one PCR run, the discriminatory power, and the genetic diversity (stability) of the fingerprint are very similar for the Eric primer set and the (GTG)5 primer, so both primers are equally able to discriminate Salmonella serotypes. These techniques also produce fingerprints for nontypeable strains, which can be molecularly serotyped on the basis of the relationship to known serotypes. Moreover, they are also able to reveal serotyping errors.
Acknowledgments
We thank Sandra Vangeenberghe, Annelies Wachtelaer, Elly Engels, and Soetkin De Wannemacker for excellent technical assistance.
REFERENCES
- 1.Belgian Reference Laboratory for Salmonella.2004. . Salmonella report of 2004. [Online.] http://www.var.fgov.be/pdf/Salmonella_Rapport_data_2004.pdf.
- 2.Botteldoorn, N., L. Herman, N. Rijpens, and M. Heyndrickx. 2004. Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses.Appl. Environ. Microbiol. 70:5305-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burr, M. D., K. L. Josephson, and I. L. Pepper. 1998. An evaluation of ERIC PCR and AP PCR fingerprinting for discriminating Salmonella serotypes.Lett. Appl. Microbiol. 27:24-30. [DOI] [PubMed] [Google Scholar]
- 4.Costas, M. 1992. Classification, identification and typing of bacteria by the analysis of their one-dimensional polyacrylamide gel electrophoretic protein patterns. Adv. Electrophor. 5:351-408. [DOI] [PubMed] [Google Scholar]
- 5.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 6.Heyndrickx, M., D. Vandekerchove, L. Herman, I. Rollier, K. Grijspeerdt, and L. De Zutter. 2002. Routes for Salmonella contamination of poultry meat: epidemiological study from hatchery to slaughterhouse. Epidemiol. Infect. 129:253-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyndrickx, M., K. Vandemeulebroecke, P. Scheldeman, K. Kersters, P. De Vos, N. A. Logan, A. M. Aziz, N. Ali, and R. C. W. Berkeley. 1996. A polyphasic reassessment of the genus Paenibacillus, reclassification of Bacillus lautus (Nakamura 1984) as Paenibacillus lautus comb. nov. and of Bacillus peoriae (Montefusco et al. 1993) as Paenibacillus peoriae comb. nov., and emended descriptions of P. lautus and of P. peoriae.Int. J. Syst. Bacteriol. 46:988-1003. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, J. R., and C. Clabots. 2000. Improved repetitive-element PCR fingerprinting of Salmonella enterica with the use of extremely elevated annealing temperatures. Clin. Diagn. Lab. Immunol. 7:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. R., C. Clabots, M. Azar, D. J. Boxrud, J. M. Besser, and J. R. Thurn.2001. . Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J. Clin. Microbiol. 39:3452-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes.J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milleman, Y., M.-C. Lesage-Descauses, J.-P. Lafont, and E. Chaslus-Dancla. 1996. Comparison of random amplified polymorphic DNA analysis and enterobacterial repetitive intergenic consensus-PCR for epidemiologic studies of Salmonella. FEMS Immunol. Med. Microbiol. 14:129-134. [DOI] [PubMed] [Google Scholar]
- 12.NRSS—National Reference Centre for Salmonella and Shigella. 2003. Annual report on Salmonella and Shigella in Belgium in 2003, Institute of Public Health. [Online.] http://www.iph.fgov.be/bacterio/iframes/rapports/SalmShig_2003_NL.pdf.
- 13.Popoff, M. Y., and L. Le Minor. 1997. Antigenic formulas of the Salmonella serotypes, 7th revision. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 14.Popoff, M. Y., J. Bockemühl, and L. L. Gheesling.2004. . Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155:568-570. [DOI] [PubMed] [Google Scholar]
- 15.Rademaker, J. L. W., and F. J. de Bruijn.1997. . Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis, p. 151-171. In G. Caetano-Anollés and P. M. Gresshoff (ed.), DNA markers: protocols, applications and overviews. J. Wiley & Sons, Inc., New York, N.Y.
- 16.Torpdahl, M., and P. Ahrens. 2004. Population structure of Salmonella investigated by amplified fragment length polymorphism. J. Appl. Microbiol. 97:566-573. [DOI] [PubMed] [Google Scholar]
- 17.Van Lith, L. A. J. T., and H. J. M. Aarts. 1994. Polymerase chain reaction identification of Salmonella serotypes. Lett. Appl. Microbiol. 19:273-276. [DOI] [PubMed] [Google Scholar]
- 18.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]