Abstract
Spoligotyping was undertaken with 38 Mycobacterium tuberculosis isolates from Greek sarcoidosis patients and 31 isolates from patients with tuberculosis. Fifty percent of the isolates from sarcoidosis patients and 16.13% of the isolates from patients with tuberculosis were represented by a unique pattern, whereas the remaining isolates belonged to seven shared types. Interestingly, half of the isolates from sarcoidosis patients did not resemble the spoligotypes of the isolates from patients with tuberculosis, most of which pertained to shared spoligotypes.
Sarcoidosis is a granulomatous disorder of unknown cause that affects multiple organs. The cells of the granuloma are organized spatially as immune granulomas, the characteristic of sarcoidosis, as a result of an immunological response to an antigenic trigger. It has been suggested that infective agents, including mycobacteria, propionibacteria, parasites such as Schistosoma, and fungi such as Coccidioides, are likely triggers (but not as infection) in a genetically predisposed individual and that this initial event leads to the sarcoidosis granulomatous response (2). Other agents such as beryllium, zirconium, and aluminum can also trigger the granulomatous response (2). Most studies of a possible causal organism have focused on mycobacteria (8), but a convincing association between sarcoidosis and mycobacterial infections has yet to be established. Recently, using PCR techniques based on IS6110, MPB64, and mtp-40 amplification, we reported the presence of Mycobacterium tuberculosis complex strains in a notable percentage (71.7%) of Greek patients with sarcoidosis (4).
Nevertheless, the PCR techniques used did not provide a detailed differentiation among isolates of the M. tuberculosis complex. Since the discovery of polymorphic DNA in M. tuberculosis complex strains, strain differentiation has become a valuable tool in the study of the epidemiology of tuberculosis and perhaps of sarcoidosis as well. Spoligotyping is a typing method that is based on DNA polymorphism in the M. tuberculosis complex. The chromosomal region consisting of identical 36-bp direct repeats alternating with 35- to 41-bp unique spacer sequences is the target of the spoligotyping technique (5). Since spoligotyping is PCR-based, it can be performed directly with M. tuberculosis complex organisms, including those that are nonviable or found in tissues in paraffin-embedded blocks or in archeological samples (13).
The objective of this study was to determine the molecular epidemiology of sarcoidosis in Greece by spoligotyping. We wanted to determine whether M. tuberculosis complex isolates from patients with sarcoidosis have spoligotypes that are common in the world and similar to those isolated from Greek patients with tuberculosis or if they have specific spoligotypes.
A total of 33 formalin-fixed paraffin-embedded tissues (lung and lymph node tissues) and 7 bronchoalveolar lavage samples, previously determined to be IS6110 positive (4), from Greek patients suffering from sarcoidosis (S1 to S40) were subjected to molecular analysis. In addition, formalin-fixed paraffin-embedded material (lung tissue) from 32 Greek patients with tuberculosis (T1 to T32) was analyzed in parallel as controls. DNA extraction was performed as previously described (4). To confirm the integrity of the DNAs, a 430-bp sequence of the human glyceraldehyde-3-phosphate dehydrogenase gene was amplified.
The DNAs were subjected to direct repeat-specific PCR and hybridization with membrane-bound spacer oligonucleotides and a spoligotyping kit (ISOGEN Bioscience, Maarsen, The Netherlands) according to the manufacturer's instructions. The spoligotyping results were entered in a binary format as Excel spreadsheets and were compared to the World Spoligotype Database spoIDB3.0 (11). At the time of the matching analysis, spoIDB3.0 contained 13,008 patterns distributed into 813 shared types and 1,300 orphan patterns from >90 countries (11). An isolate was assigned a shared type if the same spoligotype was found for isolates obtained from two or more patients in the world. If no matching spoligotype was identified in the database, the isolate was defined as unique. All of the identified spoligotypes have been submitted to the World Spoligotype Database (http://www.pasteur-guadeloupe.fr/tb/spoldb3). A dendrogram was generated using Bionumerics V4.0 (Applied Maths, Sint-Martin-Latem, Belgium) with the UPGMA Dice comparison setting, an optimization of 2%, and a position tolerance of 1%.
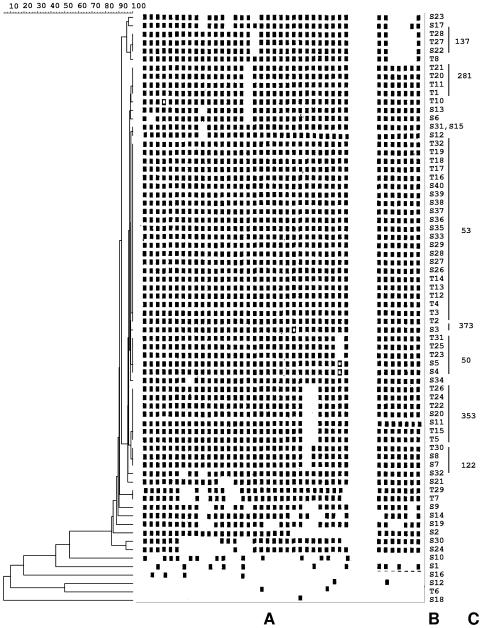
Thirty-eight samples from patients with sarcoidosis and 31 samples from patients with tuberculosis had intact DNA for spoligotyping. A total of 24 distinct spoligotypes were obtained from the 38 samples from sarcoidosis patients, and 9 were obtained from the 31 patients with tuberculosis (Fig. 1). Nineteen (50%) isolates from patients with sarcoidosis were represented by unique patterns, whereas 19 (50%) isolates belonged to six shared types already defined in spoIDB3.0. Five (16.13%) isolates from patients with tuberculosis were represented by unique patterns, whereas 26 (83.87%) isolates were clustered in six shared types. The spoligotypes specific to Greece and the shared type designations of the isolates that matched with worldwide strains, together with their geographic distribution, are indicated in Table 1. All of the patients were epidemiologically unrelated and were born and live in Greece. As indicated in Fig. 1, 18 isolates from patients with sarcoidosis exhibited similar patterns to those of certain strains from patients with tuberculosis. Only one major spoligotype was observed (type 53), which accounted for 28.95% of the total isolates from sarcoidosis patients and for 35.48% of the total isolates from patients with tuberculosis. Additionally, it is noteworthy that no association between specific spoligotypes and particular tissue origins was observed.
FIG. 1.
Dendrogram built from 69 M. tuberculosis complex samples from Greece based on spoligotyping results. (A) Spoligotype description; (B) sample number; (C) shared type identification number.
TABLE 1.
Shared types designations of the isolates, and geographic distribution
| Shared typea | Isolates with shared type | Geographic distributionb |
|---|---|---|
| 53 | T2, T3, T4, T12, T13, T14, T16, T17, T18, T19, T32, S26, S27, S28, S29, S33, S35, S36, S37, S38, S39, S40 | Ubiquitous |
| 281 | T1, T11, T20, T21 | AUS, FXX, NLD, PAK, IND, GRC |
| 122 | T30, S7, S8 | FXX, ITA, IND, GRC |
| 353 | T5, T15, T22, T24, T26, S11, S20 | GBR, AUS, NLD, GRC |
| 137 | T27, T28, S22 | Ubiquitous |
| 373 | S3 | AUS, FXX, NLD, USA, GRC |
| 50 | T23, T25, T31, S4, S5 | Ubiquitous |
| Unique | T6, T7, T8, T10, T29, S1, S2, S6, S9, S10, S12, S13, S14, S15, S16, S17, S18, S19, S23, S24, S30, S31, S32, S34 | GRC |
Designation of the spoligotype in the World Spoligotype Database (spoIDB3.0).
Ubiquitous, spoligotype present in all seven geographic areas; AUS, Australia; FXX, metropolitan France; GBR, Great Britain; ITA, Italy; NLD, The Netherlands; PAK, Pakistan; IND, India; USA, United States; GRC, Greece.
This is the first report describing the molecular characterization of M. tuberculosis complex isolates from patients with sarcoidosis and tuberculosis in Greece. Almost 34.78% of the spoligotypes were found to be specific to Greece, and thus 50% of the sarcoidosis patients and 16.13% of the patients with tuberculosis were infected with an M. tuberculosis complex isolate having a spoligotype that had not been previously submitted to the global database.
Approximately half of the spoligotypes obtained from the Greek samples belong to previously identified shared spoligotypes. A significant proportion (31.88%) of the total isolates belong to shared type 53. Shared type 53, along with shared type 50, representing 11.8% of the M. tuberculosis complex isolates in the World Spoligotype Database, seems to have been prevalent in Europe for several centuries (3; H. D. Donoghue, H. Fletcher, I. Pap, and M. Spigelman, presented at the 22nd Annual Congress of the European Society of Mycobacteriology, Berlin, Germany, 2001).
Focusing on the M. tuberculosis complex isolates from the patients with sarcoidosis, we observed that half of them had different and unique spoligotypes. Interestingly, 50% of the isolates from sarcoidosis patients did not resemble the spoligotypes of the isolates from patients with tuberculosis, most of which pertained to shared spoligotypes. These results support the hypothesis that members of the M. tuberculosis complex may contribute to the development of sarcoidosis. However, further study is needed to determine whether these strains are specific to sarcoidosis, specific to Greece, or more widespread in the Greek population. We are, indeed, far from resolving whether these results also suggest that sarcoidosis is related to unique spoligotypes.
One of the most widespread groups of M. tuberculosis strains is the Beijing type, which has been associated with outbreaks in a number of communities throughout the world (1, 6, 7, 9, 10, 12). Although during the last few years Greece has accepted immigrants from Russia, China, Estonia, and other Balkan countries, the Beijing genotype of M. tuberculosis was not observed. Furthermore, spoligotypes localized to Australia, France, Great Britain, Italy, The Netherlands, Pakistan, India, and the United States were found in low proportions.
Collectively, our results point to M. tuberculosis complex isolate spoligotypes among Greek patients with sarcoidosis and tuberculosis. Unfortunately, no information is available from Greece, and there is limited information from neighboring countries, regarding the molecular epidemiology of tuberculosis and especially sarcoidosis to aid in explaining and classifying the spread of these spoligotypes. Since a significant number of the samples from patients with sarcoidosis are distributed in different and unique spoligotypes, the analysis of a greater number of samples could reveal spoligotypes specifically associated with sarcoidosis. Through that effort, the hypothesis of the involvement of some types of mycobacteria in the etiopathology of the disease could be tested more robustly. Similarly, the description of the genetic diversity of M. tuberculosis complex isolates from patients with sarcoidosis could be improved.
REFERENCES
- 1.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. I. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 2.duBois, R. M., N. Goh, D. McGrath, and P. Cullinan. 2003. Is there a role for microorganisms in the pathogenesis of sarcoidosis? J. Int. Med. 253:4-17. [DOI] [PubMed] [Google Scholar]
- 3.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. Duc Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, H. Minh Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Morkousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. Ngo Niobe-Eyangoh, J. W. Pape, V. Rasolfo-Razanamparany, M. Ridell, M. Lucia Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Ragosti. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazouli, M., J. Ikonomopoulos, R. Triggidou, M. Foteinou, C. Kittas, and V. Gorgoulis. 2002. Assessment of mycobacterial, propionibacterial, and human herpesvirus 8 DNA in tissues of Greek patients with sarcoidosis. J. Clin. Microbiol. 40:3060-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruuner, A., S. E. Hoffner, H. Sillastu, M. Danilovits, K. Levina, S. B. Svenson, S. Ghebremichael, T. Koivula, and G. Kallenius. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 39:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in katG is predominant in genetically heterogenous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGrath, D. S., N. Goh, P. J. Foley, and R. M. duBois. 2001. Sarcoidosis genes and microbes: soil or seed. Sarcoidosis Vasc. Diffuse Lung Dis. 18:149-164. [PubMed] [Google Scholar]
- 9.Niemann, S., S. Rusch-Gerdes, and E. Richter. 1997. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J. Clin. Microbiol. 35:3015-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian, L., J. D. Van Embden, A. G. Van Der Zanden, E. F. Weltevreden, H. Duanmu, and J. T. Douglas. 1999. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J. Clin. Microbiol. 37:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola, C., I. Filliol, C. Guttierez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographical distribution of shared types and epidemiological and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 13.Zink, A. R., and A. G. Nerlich. 2004. Molecular strain identification of the Mycobacterium tuberculosis complex in archival tissue samples. J. Clin. Pathol. 57:1185-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]