Abstract
Two rapid diagnostic assays, utilizing two different Luminex flow cytometry methods, were developed for identification of clinically important ascomycetous yeast species. Direct hybridization and allele-specific primer extension methods were both successful in establishing a DNA-based assay that can rapidly and accurately identify Candida albicans, Candida krusei, Candida parapsilosis, Candida glabrata, and Candida tropicalis as well as other clinical species. The direct hybridization assay was designed to identify a total of 19 ascomycetous yeast species, and the allele-specific primer extension assay was designed to identify a total of 34 species. Probes were validated against 438 strains representing 303 species. From culture to identification, the allele-specific primer extension method takes 8 h and the direct hybridization method takes less than 5 h to complete. These assays represent comprehensive, rapid tests that are well suited for the clinical laboratory.
Opportunistic fungal infections are a major threat to immunocompromised patients (9, 15). Many factors compromise the immune system, including low birth weight, cancer, diabetes, AIDS, burns, and organ transplantation (2, 4, 8, 10, 16, 19, 21). Candida albicans and related species account for a significant portion of fungal infections as well as a large portion of the mortality rate associated with such infections. Accurate identification of the species causing infection is important for treatment, as not all species respond to the same treatment. This is evidenced by the increasing numbers of Candida glabrata strains showing resistance to azoles, one of the major treatment options (14, 23). Along with accurate identification, the ability to detect many different species has become critical as the emergence of non-C. albicans Candida species continues to be increasingly documented (2).
Current methods for identification of pathogenic yeasts require as little time as a day or as long as several weeks. Rapid, reliable tests that identify a majority of species known to cause infections would greatly improve patient treatment. Many of the recently developed molecular-based assays are real-time assays that provide rapid results, but they are often limited in the number of species that can be detected in a single assay (18). Standard PCR-based assays are also limited in the number of species that can be identified from a single amplification. DNA sequencing continues to be the most reliable method for species identification, but the cost of equipment and the need to compare the sequences from unknowns with a database that includes sequences from all known species may not be practical for many clinical laboratories.
Flow cytometry using Luminex technology allows a high level of multiplexing by sorting assays with uniquely colored microspheres so that up to 100 determinations can be made with either hybridization or extension DNA assays. A single-well assay that tests for numerous pathogenic yeast species can be developed, and several groups have already shown this technology to be practical. Most recently, this technology has been successfully used for detection of species in the yeast genus Trichosporon (3). Additionally, a comparison of methods suited to the Luminex format has been studied using single-nucleotide polymorphisms from plants (13) and application of the allele-specific primer extension (ASPE) assay to thrombophilia genotyping (1), as well as bacterial identification (5).
The nucleotide sequences of domains 1 and 2 (D1/D2) of the large subunit of the rRNA gene often have sufficient substitutions to resolve individual yeast species. A database is now available that includes D1/D2 sequences for all known ascomycetous and basidiomycetous yeasts (see references 6 and 11 and subsequent submissions to GenBank), thus providing the basis for rapid, accurate species identification. We report two microsphere-based methods for rapid molecular identification of yeast species that have been developed from the D1/D2 database. One method relies on species-specific DNA oligonucleotides that are attached to the microspheres (beads) and hybridized to D1/D2 amplicons, which are initiated with biotinylated primers. Only amplicons with sufficient sequence complementary to the species-specific primer will hybridize to the bead and be detected by the flow cytometer. In the second method, species-specific primers are hybridized to complementary sites on the D1/D2 amplicons and during the subsequent extension reaction incorporate biotin-dCTP for detection. All extension primers are hybridized to beads, but only extended products are biotinylated and detected by the Luminex flow cytometer. The first method takes less than 5 h to complete from culture to identification, whereas the second method takes 8 to 10 h. Both assays reliably identify Candida albicans, Candida parapsilosis, Candida tropicalis, Candida krusei, and C. glabrata, along with many other medically important yeast species.
MATERIALS AND METHODS
Growth of cultures and DNA isolation.
The growth of cultures and the DNA isolation techniques used were described by Kurtzman and Robnett (12). Species compared are listed in Table S1 in the supplemental material.
Direct hybridization method. (i) Attachment of probes to microspheres (beads).
Probes were purchased with a 5′ amino 12-carbon linker (Integrated DNA Technologies, Coralville, IA) and dissolved in water at a concentration of 1 mM. Probes were then diluted 10:1, and 2 μl of this concentration was used for the attachment reaction. Carboxylated beads (Luminex, Austin, TX) were prepared by placing 200 μl of stock suspension in a 1.5-ml microcentrifuge tube and centrifuging at 8,000 rpm in a bench top centrifuge for 1 min. The supernatant storage buffer was decanted, and the beads were resuspended in 50 μl of 0.1 M MES [2-(N-morpholino)ethanesulfonic acid] solution set to pH 4.5 by using 5 N NaOH. Care was taken to expose the beads to as little light as possible to prevent photobleaching, which hinders the Luminex detection system from registering the individual beads. Once resuspended, the beads were vortexed and 2 μl of 100 μM primer stock was added to the bead mixture. Preweighed aliquots of desiccated 1-ethyl-3-(dimethylaminopropyl)carbodimide HCl (EDC) powder (Pierce Biotechnology) were warmed to room temperature. A 10-mg EDC aliquot was combined with 1 ml of water, and 2.5 μl of this solution was immediately added to each bead-primer mixture for attachment of the amine-modified primers to the carboxylated beads. The bead mix was incubated in the dark at room temperature for 30 min. Following this incubation step, a second 10-mg aliquot of EDC was combined with 1 ml of water, and 2.5 μl was added to each bead preparation. The beads with primers were again incubated at room temperature in the dark for 30 min. After incubation with EDC, 1 ml of 0.02% polyoxyethylenesorbitan monolaurate (Tween 20) was added, and the beads were vortexed and then centrifuged at 8,000 rpm for 1 min in a table top centrifuge. The supernatant was aspirated, taking care not to disturb the pelleted beads. One milliliter of 0.1% sodium dodecyl sulfate was then added. The beads were vortexed and then centrifuged again at 8,000 rpm for 1 min in a table top centrifuge. The supernatant was once again aspirated, taking care not to disturb the pelleted beads. The beads were then resuspended in Tris-EDTA, and a dilution stock of 100:1 was made in order to count the number of beads per microliter with a hemacytometer. Bead sets with the newly attached primers were then stored at 4°C for future use.
(ii) Hybridization of bead sets/probes with PCR products.
The D1/D2 region of the rRNA gene is approximately 600 bp long and was amplified using the primers NL-1 (5′GCATATCAATAAGCGGAGGAAAAG3′) and NL-4 (5′GGTCCGTGTTTCAAGACGG3′). The 3′ primer, which in all cases was the NL-4 primer, was biotinylated. PCR for the D1/D2 region with NL-1 and biotinylated NL-4 as PCR primers was carried out using an initial 2-min 94°C step for denaturation of the DNA molecules followed by a 30-s 94°C denaturation step, a 30-s 55°C annealing step, and then a 1-min 72°C extension step. Steps 2 to 4 were repeated 40 additional times, with a final 8-min extension step before storage of the PCR products at either 4°C or −20°C. For the hybridization step, 5 μl of PCR product was used in a total volume of 17 μl of Tris-EDTA. Successful concentration ranges of PCR template for hybridization have previously been detailed (3). Each bead set with attached assay probe was diluted so that approximately 4,000 beads were used in each sample well. The bead sets were diluted using 1.5× tetramethylammonium chloride (TMAC) solution (Sigma, St. Louis, MO) containing 4.5 M TMAC, 0.15% Sarkosyl, 75 mM Tris-HCl pH 8.0, and 6 mM EDTA at pH 8.0, so that 33 μl of 1.5× TMAC containing the appropriate amount of beads was added to each sample well. The beads and PCR products were mixed together by being pipetted up and down five times and then incubated in the dark at an initial denaturation temperature of 96°C for 5 min, followed by 15 min of incubation in the dark at 55°C. This incubation step was followed by the addition of 25 μl of 1× TMAC solution containing 10 ng/μl streptavidin-R-phycoerythrin (Molecular Probes, Eugene, OR) per well and incubation for 10 min in the dark at 55°C. The preceding incubation steps were done in a thermocycler. Samples were then measured on the basis of fluorescence intensity in a Luminex100 flow cytometer.
(iii) Probe design.
Probes were designed by aligning ascomycetous yeast D1/D2 DNA sequences and searching for species-specific DNA variation. Alignment and searches for species-specific DNA sequences were aided by MacClade (Sinauer Associates, Inc., Sunderland, MA) and ARB (Technical University of Munich, Munich, Germany) software. Probe design was accomplished using ARB software as well as manual design. Probe sequences ranged from 15 bp to 25 bp in length. All probes were designed to place the sequence differences as close to the center of the probe as possible. Secondary structure and runs of three or more guanines (Gs) or cytosines (Cs) on the 3′ or 5′ ends of the probes were avoided where possible. Cross hybridization and probe interaction required the use of trial and error to redesign several probes so that they would work in a multiplex assay.
Allele-specific primer extension method. (i) Amplification and extension.
The D1/D2 region of the rRNA gene was amplified using primers NL-1 and NL-4. The initial PCR with 55°C annealing and 72°C extension temperatures was as stated above, with the exception that the 3′ primer was not biotinylated. Amplification and subsequent processing were performed in 96-well thermowell plates (Costar, Corning, NY). Following amplification, free nucleotides and excess amplification primers were removed by adding 3.0 μl of Exo-SAP-IT enzyme mix (USB, Cleveland, OH) to 7.5 μl of each PCR product. The samples were incubated at 37°C for 30 min and then at 80°C for 15 min. Alternatively, amplification products were purified by filtering on a Millipore (Billerica, MA) MultiScreen PCR plate to eliminate free nucleotides and excess primer. The purified samples (5 μl) were then used as templates for the extension reactions. The extension reactions were carried out in 20-μl volumes of ASPE buffer (2 mM Tris-HCl, pH 8.4; 5 mM KCl) containing 2.0 mM MgCl2; 0.75 U Tsp DNA polymerase (Invitrogen, Carlsbad, CA); 5 μM dATP, dTTP, and dGTP; 5 μM biotin-dCTP (Invitrogen), and 25 nM of each extension primer. All extension primers were modified at the 5′ end by 24-mer oligonucleotides that were reverse complements of the 24-mer oligonucleotides attached to the given bead sets used in the assay. Each bead set had its own unique 24-mer oligonucleotide. The extension reactions were performed with an initial 96°C step for 2 min, followed by 94°C for 30 s, 55°C for 1 min, and 72°C for 2 min. Steps 2 to 4 were repeated for 30 cycles, and samples were held at 4°C until hybridization with bead sets. Successful extension reactions incorporated biotin-dCTP into an extension product so that a positive result could be measured using the same streptavidin-R-phycoerythrin reagent as used in the direct hybridization method.
(ii) Hybridization.
The FlexMap system (Luminex, Austin, TX) consists of an array of 100 different bead sets, each of which is already coupled with a specific 24-mer oligonucleotide from the supplier. Extension primers were designed with 24-mer antitags so that a specific extension primer and product are hybridized to a specific bead set. The extension products were hybridized to the FlexMap bead sets by adding 2,500 beads of each bead set per sample well and 5 μl of extension product in a total volume of 50 μl in 1× Tm buffer (0.2 M NaCl, 0.1 M Tris, 0.08% Triton X-100). The samples were incubated at 96°C for 90 s and then at 37°C for 45 min. The beads were pelleted by centrifugation at 2,250 × g (3,700 rpm) for 3 min in an Eppendorf model 5804 centrifuge and washed with 70 μl of 1× Tm buffer per sample. A second centrifugation step at 2,250 × g for 3 min was followed by the addition of 70 μl of 1× Tm buffer containing 2 μg/ml of streptavidin-R-phycoerythrin. The samples were incubated at 37°C for 15 min and then analyzed with the Luminex100 flow cytometer. A lower hybridization temperature is sufficient for the ASPE method because the hybridization step is not the discriminatory step of the process. All extension primers hybridize to their respective bead set regardless of whether or not a successful extension reaction has taken place. Successful extensions are detected with streptavidin-R-phycoerythrin, which reacts with the biotin-dCTP incorporated into the newly extended DNA strand.
(iii) Probe design.
Probes were designed in the same manner as for direct hybridization, with the exception that single-nucleotide differences in sequences were placed on the 3′ end of the extension primer to disrupt extension in DNAs that did not match the sequence at that nucleotide. The 3′ nucleotide was chosen first to differentiate the species from its nearest related species and second to differentiate the species from the largest number of other species. In addition, probes were designed so that species that did not differ from other species at the 3′ nucleotide had three or more nucleotide differences from the other species distributed throughout the probe so that hybridization, and thus extension, would be unlikely to occur.
RESULTS
Direct hybridization.
Probes were designed to identify 19 ascomycetous yeast species either individually or as part of a small group of species. Initial probe development used a test group of 96 strains representing 94 species that included the target species as well as closely related species most likely to cross-react with the probes developed. The species-specific probes and their target species are shown in Table 1. Probes were designed with as many nucleotide differences as possible between the target species DNA and the DNA of the closest related species to achieve maximum discrimination. As recommended by Luminex protocols (5), we set a standard for probes that the positive signal must be at least 2 times greater than any signal from a nonpositive strain once background from a water blank negative control is subtracted. All probes were developed and tested as a multiplex, single-well assay. The complete multiplex assay was then applied to all strains.
TABLE 1.
Species-specific direct hybridization probes
| Probe | Species or strain identified | Probe sequence | Probe signal range (AFU)
|
Min ratio of signalsc | |
|---|---|---|---|---|---|
| Negativea | Positiveb | ||||
| Alb7 | Candida albicans | TATTTTGCATGCTGCTCTCTC | 0-58 | 299-1,268 | 5.2 |
| Dub5 | C. dubliniensis | TATTTTGCAAGTTACTCTTTC | 0-98 | 676-934 | 6.9 |
| Fari2 | Stephanoascus farinosus | TTGGTTTGTAACGATCAACT | 0-161 | 843 | 5.2 |
| Glab5 | C. glabrata | TTGCCTCTCGTGGGCTTGGGA | 0 | 466-743 | NAd |
| Hellen2 | Zygoascus hellenicus | AAGGGATCTAAATCAGACAT | 0-100 | 792 | 7.9 |
| HemC | C. haemulonii | AACGAGCAGTCGATGTAGTACA | 0 | 921-1,219 | NA |
| HemIIB | C. haemulonii type II | AAAGTGGGAGCTGATGTAGCAAC | 0-71 | 757-1,031 | 10.7 |
| KrusF | C. krusei | GAGGACTGCGCCGTGTAGG | 0-70 | 2,015-2,463 | 28.8 |
| Lusit2 | Clavispora lusitaniae | CGGGCCAGCGTCAAATAAAC | 0 | 269 | NA |
| Malt5 | C. maltosa | TATAGCCACTGTCGATACTG | 0-251 | 1,389 | 5.5 |
| Nee5 | C. neerlandica | AGGAGAATCGCTTGGGAACG | 0-201 | 2,058 | 10.2 |
| Pcifer3 | Pichia ciferrii | AAGATAATAGCAGTTAAATG | 0-521e | 2,775 | 5.3 |
| SojC | C. sojae | GCCTTCGTAGATACTGC | 0-611f | 1,279-1,419 | 2.1 |
| SP12 | Candida sp. strain NRRL Y-17456 | GGACAATTGCAAAGAAATGT | 0-50 | 1,301-1,373 | 26 |
| VisA | C. viswanathiig | GCGGCAGGACAATCGCGTGG | 0-100 | 791-1,271 | 7.9 |
| UniD | All species tested | GTGAAATTGTTGAAAGGGAA | NA | 782-5,571 | NA |
Range of adjusted fluorescence values for all negative strains after subtraction of background from a water blank negative.
Range of AFU for all positive strains after subtraction of background.
The min ratio is the lowest recorded positive value divided by the highest recorded negative value.
NA, adjusting the fluorescence values of negative strains by subtracting background resulted in no discernable signals above background for all negative strains for these probes, and min ratios therefore could not be calculated.
C. silvicultrix gave a signal of 521 AFU. All other strains recorded less than 97 AFU.
C. tropicalis and L. elongisporus strains cross-reacted with SojC at about 50% of the signal of C. sojae strains. See text for discussion. All other strains were recorded at less than 330 AFU.
C. viswanathii includes C. lodderae.
The hybridization probes from Table 1 were further validated with 342 additional strains representing 209 species (see Table S1 in the supplemental material). The test species were chosen because of their proximity to a clade containing a species for which a probe was designed, because they were members of clades containing a species for which a probe was designed, or because they represented additional strains of species for which probes were designed. Ninety-two of these strains were clinical isolates. Multiple strains of a species for which a probe was designed represented a rigorous positive test for the species-specific probe and established a range of positive values to determine the amount of variation that can be expected from this test. Likewise, hundreds of strains predicted to be negative for the designed probes represent an extremely robust negative test. Table 1 catalogues the values for negative and positive ranges from each probe tested. The min ratio was established as the ratio of the lowest recorded positive signal divided by the highest recorded negative signal. Analysis by standard deviation could not be applied uniformly, as several species were represented by a single strain. All 15 species-specific probes had min ratios above the standard 2.0. Since no measurable background signals were detected on species negative for Lusit2, Glab5, and HemC, the infinitely large min ratios were arbitrarily considered to be above 2.0. VisA identifies both Candida viswanathii and Candida lodderae, but it is considered species-specific because C. lodderae and C. viswanathii appear to be conspecific.
For two of the species-specific probes, SojC and Pcifer3, cross-reactive signals were over 500 adjusted fluorescence units (AFU). SojC reacted with Candida sojae, C. tropicalis, and Lodderomyces elongisporus, but C. tropicalis and L. elongisporus reacted at about 50% of the signal for C. sojae. In addition to this signal differential, C. tropicalis and L. elongisporus are identified by the Trop/Elong5B probe, which permits unambiguous identification of C. sojae. The Pcifer3 probe showed cross-reactivity with Candida silvicultrix, but the signal from this probe when hybridized to C. silvicultrix was only 19% of the signal obtained with Pichia ciferrii. No additional cross-reactivity from these 15 probes was encountered among the 438 strains tested.
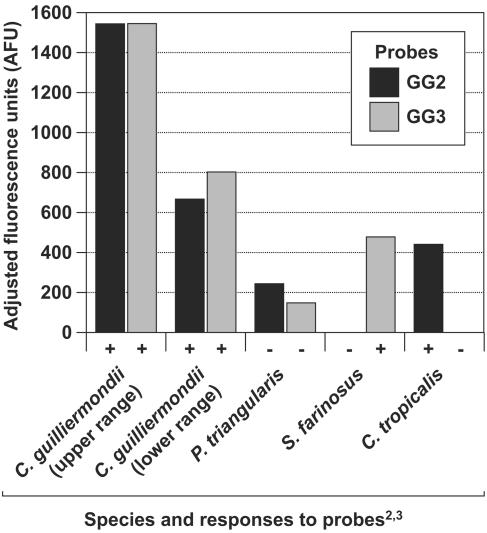
It was not possible to develop species-specific probes for some target species. For some of these species, a combination of probes will unambiguously identify them, but for others we developed a probe that would identify a group of species that included the target species. For example, all Candida parapsilosis strains were positive for Parap2B and LPW. Although both probes individually exhibited a high level of cross-reactivity with non-C. parapsilosis strains, no non-C. parapsilosis strain that was tested cross-reacted with both probes. Thus, the combination of probes gave us a reliable method for identifying C. parapsilosis even though a single species-specific probe could not be developed from the D1/D2 domain. In a similar fashion, we were able to identify other species with a series of multispecies probes as well (Table 2). Lodderomyces elongisporus and Candida tropicalis are both identified by the Trop/Elong5B probe, distinguishing these two species from all other species tested, whereas the LPW probe identifies C. parapsilosis, Candida sp. strain NRRL Y-17456, and L. elongisporus but not C. tropicalis. Therefore, the combination of the Trop/Elong5B probe and the LPW probe provides unambiguous identification of C. tropicalis and L. elongisporus. Likewise, the combination of the GG2 and GG3 probes identifies Candida guilliermondii, C. guilliermondii var. carpophila, and Candida fermentati. Each of the probes cross-reacts with several other species, but only species of the C. guilliermondii-C. fermentati complex are positive for both probes (Fig. 1). The positive reaction with both probes was consistent for 48 strains of the above species with no exception, while the remaining 389 strains tested, including the closely related Candida glucosophila, were negative for at least one of the two probes. One species tested gave slight signals for both probes, but as illustrated in Fig. 1, the signals are only 37% and 18%, respectively, of the signals for the lowest recorded positive sample. Overall, the complete assay identifies 18 individual ascomycetous yeast species and the C. guilliermondii clade. Table 3 details the requirements for positive identification of each of the target species. Multiple positive signals in a single well would indicate a mixed culture.
TABLE 2.
Multispecies direct hybridization probes
| Species identified | Probe(s) | Probe sequence | Cross-reactivitya | Positive rangeb | Cross-reactive nontarget speciesc |
|---|---|---|---|---|---|
| Candida fermentati, C. guilliermondii var. guilliermondii, C. guilliermondii var. carpophila | GG2 GG3 | ATCAGACTCGATATTTTGTG GTGACCCGCAGCTTATCGGG | 0-441 0-551 | 667-1544 878-1515 | 0 |
| C. parapsilosis | LPW Parap2B | TGCGGCTTCGGCCTAGGATG GGTAGGATAAGTGCAAAG | 0-2272 0-943 | 2696-3432 1376-1853 | 0 |
| C. tropicalis, Lodderomyces elongisporus | LPW Trop/Elong5B | TGCGGCTTCGGCCTAGGATG AGAATTGCGTTGGAATGT | 0-2272 0-521 | 2696-3432 915-1820 | 0 |
| All species tested | UniD | GTGAAATTGTTGAAAGGGAA | NAd | 782-5571 | NA |
Range of fluorescence values for strains cross-reactive with individual probes. See text and Fig. 1.
Range of fluorescence values for strains considered positive for the individual probe indicated.
Number of strains considered to be cross-reactive (greater than 40% of the lowest positive signal) with both multispecies probes of the probe sets.
NA, not applicable.
FIG. 1.
Detection of the C. guilliermondii clade by using two probes, illustrating that only clade species are positive for both probes. Measurement in fluorescence units was adjusted by subtracting the mean of values for water blank samples as background. All C. guilliermondii var. carpophila and C. fermentati strains gave signals for both probes within the range established by the two C. guilliermondii strains shown. Pichia triangularis gave the highest recorded signal of both probes for a nontarget species. The two signals were 37% and 18% of the lowest positive signal and therefore were considered negative for both probes.
TABLE 3.
Probes required for species identification using the direct hybridization assay
| Species or strain | Probe(s) required for identificationa |
|---|---|
| Candida albicans | Alb7 |
| C. dubliniensis | Dub5 |
| C. fermentati | GG2 + GG3 |
| C. glabrata | Glab5 |
| C. guilliermondii var. guilliermondii | GG2 + GG3 |
| C. guilliermondii var. carpophila | GG2 + GG3 |
| C. haemulonii | HemC |
| C. haemulonii type II | HemIIB |
| C. krusei | KrusF |
| C. lodderae | VisA |
| C. maltosa | Malt5 |
| C. neerlandica | Nee5 |
| C. parapsilosis | Parap2B + LPW |
| C. sojae | SojC |
| C. tropicalis | Trop/Elong5B |
| C. viswanathii | VisA |
| Candida sp. strain NRRL Y-17456 | SP12 + LPWb |
| Clavispora lusitaniae | Lusit2 |
| Lodderomyces elongisporus | LPW + Trop/Elong5B |
| Pichia ciferrii | Pcifer3 |
| Stephanoascus farinosus | Fari2 |
| Zygoascus hellenicus | Hellen2 |
Probes do not separate C. fermentati, C. guilliermondii var. guilliermondii and C. guilliermondii var. carpophila, which were shown to be separate species (22), from each other.
Although SP12 is specific to Candida sp. strain NRRL Y-17456, LPW is always positive for the strain as well and therefore is a requirement for positive identification.
To verify that successful D1/D2 amplifications were produced in each PCR, we incorporated a direct hybridization probe (UniD) complementary to a conserved D1/D2 region. By having an internal positive control, it is possible to measure the success of the PCR without the time-consuming step of testing all products by gel electrophoresis. We do not report a min ratio for this probe, as all species tested were positive for the probe.
ASPE.
Species-specific ASPE extension primers and their target species are given in Table 4. Sequence data from the D1/D2 domains of the 26S rRNA gene were used for development of these primers as described for development of direct hybridization probes. Overall, 27 species-specific primers were developed and tested simultaneously on the same 438 strains used for the direct hybridization assay. A positive control extension primer (UniB) was developed to serve as a measurement of PCR amplification and primer extension success for each sample.
TABLE 4.
ASPE species-specific primers
| Primer | Species or strain identified | Primer sequence | Probe signal range (AFU)
|
Min ratio of signalsc | |
|---|---|---|---|---|---|
| Negativea | Positiveb | ||||
| AlbA | Candida albicans | GATGAGATGACCCGGGTCTG | 0-118 | 926-2,991 | 7.9 |
| AnomA | Pichia anomala | TTGAATGTGGCTTCACTTCG | 0-236 | 2,117-2,609 | 9.0 |
| DelphinA | Kluyveromyces delphensis | ACTCCGGGAATGTAGCTGTC | 0-317 | 5,262 | 16.6 |
| DubA1 | C. dubliniensis | GATGAGATGGCCCGGGTCTA | 0-130 | 2,792-3,556 | 21.5 |
| FabiA1 | P. fabianii | CAAGATAATAGCTTGGGAAT | 0-715d | 2,979 | 4.2 |
| GlabC | C. glabrata | AGAGTACCACTTTGGGACTG | 0-46 | 1,026-1,545 | 22.3 |
| HaemA | C. haemulonii | GAGCAGTCGATGTAGTACAG | 0-52 | 239-909 | 4.6 |
| HaemIIA | C. haemulonii type II | GGAACTAAAAGTGGGAGCTG | 0-71 | 347-1,402 | 4.9 |
| HellenA | Zygoascus hellenicus | GTTTATGGTGAAGAACCCAC | 0-112 | 2,075 | 18.5 |
| JadB2 | P. jadinii | GGCAGTGTGTTTATAGCCCC | 0-660e | 1,356 | 2.1 |
| LelonA | Lodderomyces elongisporus | AGCGGTAGGAGAATTGCGTA | 0-227 | 2,804-6,270 | 12.4 |
| LipoA | Yarrowia lipolytica | GAAATGAGTGGAGAGTGGCC | 0-185 | 2,301 | 12.4 |
| LusitA | Clavispora lusitaniae | GGAAAGAATGTGGCGCGTGC | 0-130 | 2,949 | 22.7 |
| MaltA | C. maltosa | CCACTGTCGATACTGCCAGT | 0-265 | 4,911 | 18.5 |
| NeerA | C. neerlandica | TAGGAGAATCGCTTGGGAAC | 0-36 | 2,010 | 55.8 |
| OccidA | Issatchenkia occidentalis | GTTTTTGCCGCAGGAGAAGGC | 0-117 | 1,481-2,316 | 12.7 |
| OnychA | P. onychis | TCTGGTTGTGCCTGGCGCAG | 0-62 | 2,478-2,762 | 40.0 |
| ParapB50 | C. parapsilosis | AGTTTGAGCGGTAGGATAAG | 0-201 | 2,673-3,893 | 13.3 |
| PciferB2 | P. ciferrii | TATAGTTTCTGTTGATATTG | 0-364 | 1,298 | 3.6 |
| PeterA | P. petersonii | CCGTGTGGCGGGGAGCCCGGT | 0-43 | 1,455 | 33.8 |
| PseudoA | C. pseudointermedia | AGCCCCGTAGGTACCACAAC | 0-48 | 1,621 | 33.8 |
| SciferB | Stephanoascus ciferrii | CAAAAGAATGTGGCTCCGCT | 0-223 | 658 | 3.0 |
| SfariA | S. farinosus | CAACTTTGGAGACGGCCTTG | 0-68 | 2,624 | 38.6 |
| SojA | C. sojae | GGCGGTAGGAGAATTGCGA | 0-82 | 1,378-1,585 | 16.8 |
| SsmithA | S. smithiae | GCCTAGGTTGATACTACCTA | 0-58 | 1,068-1,628 | 18.4 |
| WojA | Candida sp. strain NRRL Y-17456 | TGGGCGGTAGGACAATTGCA | 0-198 | 2,829-3,063 | 14.3 |
| VisA2 | C. viswanathiif | GGCGGCAGGACAATCGCGT | 0-109 | 1544-2096 | 14.2 |
| UniB | All species | TCGAGTTGTTTGGGAATGCA | NAg | 1050-3760 | NA |
Range of adjusted fluorescence units for all strains negative for the given probe.
Range of AFU for all strains positive for the given probe.
The min ratio is the lowest recorded positive value divided by the highest recorded negative value.
P. veronae and C. blankii cross-reacted with FabiA1. All other strains tested gave less than 112 AFU.
C. lodderae NRRL Y-17317, Pichia sp. strain NRRL Y-11569, and C. viswanathii NRRL Y-27370 cross-reacted with JadB2. All other strains gave less than 109 AFU.
C. viswanathii includes C. lodderae.
NA, not applicable.
Table 4 shows the negative and positive ranges for all species-specific primers tested as well as their min ratios. The min ratios of 27 species-specific primers ranged from 2.1 to 55.8, with positive signals ranging from 239 to 6,270. Two of the species predicted to be negative for the FabiA1 primer, Pichia veronae and Candida blankii, gave signals that were 24% and 12%, respectively, of the signal given by Pichia fabianii. Additionally, three of the test strains predicted to be negative for the JadB2 primer, C. lodderae NRRL Y-17317, Pichia sp. strain NRRL Y-11569, and C. viswanathii NRRL Y-27370, gave signals that were 37%, 49%, and 35%, respectively, of the signal given by Pichia jadinii. Although high cross-reacting signals were recorded for these primers, they can be differentiated from a true positive by the difference in signal intensities as long as a positive control for the target species is included.
Although most target species were identified by a single probe, a few required a combination of multispecies probes for identification. Multispecies primers and their targets are given in Table 5. The multispecies primers had min values ranging from 2 to 37 and positive signals ranging from 461 to 5,189. There were few cross-reactive species even with the multispecies primers. The Trop/ElongA primer cross-reacted with nine strains of both C. guilliermondii and C. fermentati, but since all nine strains are clearly identified by the GuilA primer, no attempt was made to redesign this otherwise discriminatory primer. Similarly, the HansenA primer cross-reacted with Candida sake, but the signal for C. sake was 50% or less than those for the four species (Debaryomyces hansenii var. hansenii, D. hansenii var. fabryi, Debaryomyces nepalensis, and Candida psychrophila) identified by the HansenA primer. Other than these two exceptions, all extension primers for the ASPE assay gave clear discrimination of species.
TABLE 5.
ASPE multispecies primers
| Primer | Species identified | Primer sequence | Cross-reactivitya | Positive rangeb | No. of species cross-reacting |
|---|---|---|---|---|---|
| AtellA | Kazachstania telluris, K. bovina, K. heterogenica, K. pintolopesii | TGTAATTTGGAGAGTGGATC | 0-151 | 523-1,080 | 0 |
| GuilA | C. fermentati, C. guilliermondii var. guilliermondii, C. guilliermondii var. carpophila | CAGACTCGATATTTTGTGAG | 0-226 | 1,569-5,145 | 0 |
| HansenA | Debaryomyces hansenii var. hansenii, D. hansenii var. fabryi, D. nepalensis, C. psychrophila | GACTAAGGAATGTGGCTCTA | 0-719 | 1,414-4,414 | 1c |
| KrusD | C. krusei, C. pseudointermedia | CTTGGAACAGGGCGCCCAG | 0-79 | 2,923-4,724 | 0 |
| Trop/ElongA | C. tropicalis, Lodderomyces elongisporus | GGCGGTAGGAGAATTGCGT | 0-1,049 | 2,150-5,189 | 1d |
| ZeylaG | C. zeylanoides, C. santamariae var. santamariae, C. santamariae var. membranifaciens | GACATTGGAATGTAGCTTTA | 0-121 | 461-1,412 | 0 |
Range of fluorescence values for all strains that cross-react with the given probe. Values less than 40% of the lowest positive value are not considered to be cross-reactive.
Range of fluorescence values for all strains positive for the given probe.
Cross-reacts with C. sake.
Cross-reacts with eight strains of C. guilliermondii and one strain of C. fermentati.
Overall, the ASPE extension assay that was developed is capable of uniquely identifying 30 ascomycetous yeast species as well as the C. guilliermondii, D. hansenii, Candida zeylanoides, and Kazachstania (Arxiozyma) telluris clades. Table 6 details the requirements for species identification with this assay. We were able to unambiguously identify all Candida tropicalis strains with the dual Trop/ElongA extension primer and by subtraction of the strains positive for the LelonA species-specific primer. Similarly, all C. krusei strains could be identified with the combination of the KrusD and PseudoA primers. The KrusD primer identified all C. krusei strains as well as Candida pseudointermedia. The C. pseudointermedia-specific primer, PseudoA, allowed identification of C. krusei by subtraction. The ZeylaG group primer identified a group consisting of C. zeylanoides, Candida santamariae var. santamariae, and C. santamariae var. membranifaciens. Group primer AtellA identified K. telluris, Kazachstania bovina, Kazachstania pintolopesii, and Kazachstania hetergenica and reliably differentiated this group of four species from all other species tested. The HansenA group primer identified a group consisting of Debaryomyces hansenii var. hansenii, D. hansenii var. fabryi, Candida psychrophila, and C. nepalensis. As with the direct hybridization assay, multiple positive signals in a single well would indicate a mixed culture.
TABLE 6.
Probes for species identification using the ASPE assay
| Species or strain | Probe(s) required for identification |
|---|---|
| Candida albicans | AlbA |
| C. dubliniensis | DubA1 |
| C. fermentati | GuilA |
| C. glabrata | GlabC |
| C. guilliermondii var. guilliermondii | GuilA |
| C. guilliermondii var. carpophila | GuilA |
| C. haemulonii | HaemA |
| C. haemulonii type II | HaemIIA |
| C. krusei | KrusDa |
| C. lodderae | VisA2 |
| C. maltosa | MaltA |
| C. neerlandica | NeerA |
| C. parapsilosis | ParapB50 |
| C. pseudointermedia | PseudoA + KrusD |
| C. psychrophila | HansenA |
| C. santamariae var. santamariae | ZeylaG |
| C. santamariae var. membranifaciens | ZeylaG |
| C. sojae | SojA |
| C. tropicalis | Trop/ElongA |
| C. viswanathii | VisA2 |
| C. zeylanoides | ZeylaG |
| Candida sp. strain NRRL Y-17456 | WojA |
| Clavispora lusitaniae | LusitA |
| Debaryomyces hansenii var. hansenii | HansenA |
| D. hansenii var. fabryi | HansenA |
| D. nepalensis | HansenA |
| Issatchenkia occidentalis | OccidA |
| Kazachstania telluris | AtellA |
| K. bovina | AtellA |
| K. heterogenica | AtellA |
| K. pintolopesii | AtellA |
| Kluveryomyces delphensis | DelphenA |
| Lodderomyces elongisporus | LelonA + Trop/ElongAb |
| Pichia anomala | AnomA |
| P. ciferrii | PciferB2 |
| P. fabianii | FabiA1 |
| P. jadinii | JadB2 |
| P. onychis | OnychA |
| P. petersonii | PeterA |
| Stephanoascus ciferrii | SciferB |
| S. farinosus | SfariA |
| S. smithiae | SSmithA |
| Yarrowia lipolytica | LipoA |
| Zygoascus hellenicus | HellenA |
Candida krusei identification requires a positive signal from KrusD along with absence of a signal from PseudoA.
Although LelonA is specific for Lodderomyces elongisporus, the Trop/ElongA primer is also always positive for the species and is therefore required for positive identification.
DISCUSSION
We have developed two assays utilizing two different methods, both of which are capable of identifying clinically important ascomycetous yeast species in a single-well test within one working day when starting from culture. In the development process, it became clear that each method had its benefits and drawbacks. When designing probes for the direct hybridization method, there were few consistent rules that appeared to apply to the hybridization reactions. With some probes, a single nucleotide difference was sufficient to distinguish species, while other probes might have up to three or more nucleotides of mismatch and still not distinguish the two species. Other ambiguities became apparent when resolving C. tropicalis, C. sojae, and L. elongisporus, which have relatively few nucleotide differences in the D1/D2 region. We designed probes with single nucleotide differences for each of the species, and while this was sufficient to distinguish C. sojae and C. tropicalis from one another, it was not sufficient to differentiate C. tropicalis from L. elongisporus. Altering the probe in length and shifting the position of the mismatch did not result in a successful probe. Intentional mismatches were designed for both the C. tropicalis and the C. sojae probes, but none of the mismatched probes gave sufficient signal for discrimination. For this reason, we instead designed a group probe to recognize L. elongisporus, C. parapsilosis, and Candida sp. strain NRRL Y-17456. The combination of probes resulted in differentiation of the species. Once we had working probes for a particular species, the probes were reliable over multiple trials and different PCR amplifications.
By contrast, when working with ASPE, we encountered very few primers that did not work well upon first design. In most cases, a single nucleotide difference on the 3′ end was sufficient for differentiation of the species. The ease of design allowed us to develop additional species-specific primers, which resulted in a more comprehensive assay for the ASPE method. One of the drawbacks of the extension assay was developing group primers. It was difficult to develop a group primer that was conserved across all members of the group but yet retained a unique 3′ nucleotide relative to all species outside of the group. Group probes or primers provide a valuable tool for identifying new species or variant strains that species-specific probes do not detect due to variation that might occur within the probe sequence. The direct hybridization assay appears better suited for group probe development, as it assays a region of DNA sequence rather than a single nucleotide. The ability of the ASPE assay to recognize a single nucleotide, however, lends itself to other applications, such as epidemiology. With variants of the same species differing in sequence by one or only a few nucleotides in a given region, a combination of species-specific and strain-specific primers could easily track specific haplotypes with the ASPE method. Previous studies have shown the practical importance of epidemiology, especially in regard to Candida albicans (7, 17, 20).
The availability of a large database of sequences for the D1/D2 region was essential for successful probe design. Originally, the LPW probe was designed to be specific for Lodderomyces elongisporus, Candida parapsilosis, and Candida sp. strain NRRL Y-17456, and in testing it on the 96 strains used for development, this specificity was confirmed. Once the probes were tested on the remaining 341 strains, however, several strains gave a positive signal for the LPW probe. While many of these strains had a single nucleotide difference in the middle of the probe region, some strains that gave a positive signal had 3 or more differences distributed throughout the probe sequence. For example, Candida caseinolytica had nucleotide substitutions at the 1st and 19th positions of the probe, as well as two deletions at the 5th and 13th positions, yet C. caseinolytica still gave a positive signal for the LPW probe. These results nullified the effectiveness of conducting a BLAST search for verifying the specificity of probes. Instead, by having the full D1/D2 sequences of over 900 strains in our database and using nearly half of these strains in testing the probes, we were in a unique position to design many effective probes from a single DNA region.
While the approach of using a single DNA region was largely successful for these assays, these results do not discount the value of other DNA regions for species identification. The choice of the D1/D2 large-subunit region of the rRNA gene was based on the amount of sequence information available relative to other DNA regions. Sequence databases of the internal transcribed spacer, the intergenic spacer, and other genes are growing and may be suitable for similar assay development. In some cases, a more substituted gene region may be more suitable to differentiate closely related species such as those of the C. guilliermondii complex. Testing of new regions will still require an extensive database to minimize the likelihood of false positives.
Our main goal was to develop a DNA-based identification assay that was rapid and reliable. To that end, a comparison between the two methods showed a slight advantage for the direct hybridization method as a more rapid assay. A rapid-cycling thermocycler could reduce the time for both methods by as much as 30%. Both methods are rapid in comparison to current methods of identification, and using a rapid-cycling thermocycler would make both methods even more attractive for the clinical laboratory. The ASPE assay was more discriminatory and therefore provided a slightly more comprehensive assay; however, both methods were very reliable when validated against 438 ascomycetous yeast strains.
In the present work, we have rapidly and reliably identified 18 ascomycetous yeast species as well as the C. guilliermondii clade, using the direct hybridization method. The ASPE assay rapidly and reliably identified 31 ascomycetous yeast species and four groups of species containing one or more opportunistic pathogens. The prospect for introducing these assays into a clinical setting is very good because the equipment and methodology can easily be applied to other fungal pathogens as well as to bacterial pathogens and thereby form a comprehensive, single-platform, diagnostic assay system.
Supplementary Material
Acknowledgments
We thank Christie Robnett for DNA samples and Dave Labeda for advice on the use of ARB software for sequence comparisons. Jack Fell and Mara Diaz kindly provided advice on direct hybridization protocols, and we are grateful to Allen Ward and Sherry Dunbar (Luminex Corp.) for providing technical input on Luminex technology. Todd Ward and Tom Usgaard graciously shared technical knowledge for primer extension protocols. Don Fraser assisted in figure preparation.
This research was funded in part by National Institutes of Health grant 1-OU1 AI53879-01.
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bortolin, S., M. Black, H. Modi, I. Boszko, D. Kobler, D. Fieldhouse, E. Lopes, J. M. Lacroix, R. Grimwood, P. Wells, R. Janeczko, and R. Zastawny. 2004. Analytical validation of the tag-it high-throughput microsphere-based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia-associated single-nucleotide polymorphisms. Clin. Chem. 50:2028-2036. [DOI] [PubMed] [Google Scholar]
- 2.Clark, T. A., and R. A. Hajjeh. 2002. Recent trends in the epidemiology of invasive mycoses. Curr. Opin. Infect. Dis. 6:569-574. [DOI] [PubMed] [Google Scholar]
- 3.Diaz, M. R., and J. W. Fell. 2004. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J. Clin. Microbiol. 42:3696-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doctor, B. A., N. Newman, N. M. Minich, H. G. Taylor, A. A. Fanaroff, and M. Hack. 2001. Clinical outcomes of neonatal meningitis in very-low birthweight infants. Clin. Pediatr. 40:473-480. [DOI] [PubMed] [Google Scholar]
- 5.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMap system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 6.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 7.Forche, A., G. Schonian, Y. Graser, R. Vigalys, and T. G. Mitchell. 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28:107-125. [DOI] [PubMed] [Google Scholar]
- 8.Garbino, J., L. Kolarova, P. Rohner, D. Lew, P. Pichna, and D. Pittet. 2002. Secular trends of candidemia over 12 years in adult patients at a tertiary care hospital. Medicine (Baltimore) 81:425-433. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins, C., and D. Armstrong. 1984. Fungal infections in the immunocompromised host. Clin. Haematol. 13:599-630. [PubMed] [Google Scholar]
- 10.Kossoff, E. H., E. S. Buescher, and M. G. Karlowicz. 1998. Candidemia in neonatal intensive care unit: trends during fifteen years and clinical features of 111 cases. Pediatr. Infect. Dis. J. 17:504-508. [DOI] [PubMed] [Google Scholar]
- 11.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzman, C. P., and C. J. Robnett. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 3:417-432. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. H., D. R. Walker, P. B. Cregan, and H. R. Boerma. 2004. Comparison of four flow cytometric SNP detection assays and their use in plant improvement. Theor. Appl. Genet. 110:167-174. [DOI] [PubMed] [Google Scholar]
- 14.Li, X., N. Brown, A. S. Chau, J. L. Lopez-Ribot, M. T. Ruesga, G. Quindos, C. A. Mendrick, R. S. Hare, D. Loebenberg, B. DiDomenico, and P. M. McNicholas. 2004. Changes in the susceptibility to posaconazole in clinical isolates of Candida albicans. J. Antimicrob. Chemother. 53:74-80. [DOI] [PubMed] [Google Scholar]
- 15.Meunier-Carpentier, F., T. E. Kiehn, and D. Armstrong. 1981. Fungemia in the immunocompromised host. Changing patterns, antigenemia, high mortality. Am. J. Med. 71:363-370. [DOI] [PubMed] [Google Scholar]
- 16.Patterson, J. E. 1999. Epidemiology of fungal infections in solid organ transplant patients. Transplant. Infect. Dis. 1:229. [DOI] [PubMed] [Google Scholar]
- 17.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvarangan, R., U. Bui, A. P. Limaye, and B. T. Cookson. 2003. Rapid identification of Candida species directly from blood culture bottles. J. Clin. Microbiol. 41:5660-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith, D. J., and P. D. Thomson. 1992. Changing flora in burn and trauma units: historical perspective experience in the United States. J. Burn Care Rehabil. 13:276-280. [DOI] [PubMed] [Google Scholar]
- 20.Soll, D. R., and C. Pujol. 2003. Candida albicans clades. FEMS Immunol. Med. Microbiol. 39:1-7. [DOI] [PubMed] [Google Scholar]
- 21.Tortorano, A. M., J. Peman, H. Bernhardt, L. Klingspor, C. C. Kibbler, O. Faure, E. Biraghi, E. Canton, K. Zimmermann, S. Seaton, R. Grillot, and the ECMM Working Group on Candidaemia. 2004. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23:317-322. [DOI] [PubMed] [Google Scholar]
- 22.Vaughan-Martini, A., C. P. Kurtzman, S. A. Meyer, and E. B. O'Neill. 2005. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5:463-469. [DOI] [PubMed] [Google Scholar]
- 23.Vermitsky, J. P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.