Abstract
In this study, 61 drug-resistant Streptococcus pneumoniae strains were characterized by multilocus sequence typing (MLST). These strains were representatives of 26 major clones (defined using pulsed-field gel electrophoresis) accounting for 93% of the 1,285 drug-resistant Streptococcus pneumoniae isolates recovered from the nasopharynges of healthy children attending day-care centers in Lisbon during 2001 to 2003. Using MLST, 13 of the 26 clones were found to be identical or closely related to 11 Pneumococcal Molecular Epidemiology Network (PMEN) clones, 4 clones were found to be unique as there were no identical or highly related allelic profiles deposited in the MLST database, and the remaining 9 clones had sequence types that matched or differed at a single or double locus from allelic profiles available in the MLST database. These nine clones were of serotypes 33F, 10A, 19A, 19F, 6A, 20, 24F, and 3, one was nontypeable, and, by MLST, they were found to be identical or highly related to isolates from disease origin that were dispersed internationally. Since the majority of these clones had serotypes that are not included in the 7-valent conjugate pneumococcal vaccine, monitoring of these clones is important for surveying their possible spread in the future. We propose the inclusion of these novel international clones in the PMEN.
Streptococcus pneumoniae (pneumococcus) is a major cause of invasive diseases, such as bacteremia, septicemia, and meningitis, and also causes less severe conditions, such as middle-ear infections, sinusitis, and recurrent bronchitis (48). It causes annually over one million deaths worldwide, mainly in children younger than 5 years of age, and the highest incidence is registered in developing countries (48).
The normal habitat of pneumococci is the nasopharynx, and pneumococcal nasopharyngeal colonization is the starting point for pathogenesis and horizontal spread of strains. The highest colonization rates are found in populations attending crowded places such as day-care centers. Children are frequently colonized by pneumococcus without having disease symptoms, as the mucosal membranes often confer immunity (15). However, when the host condition is altered due to internal and/or external factors, the existent balance is disturbed and disease may occur (15, 25).
For these reasons, it is important to enhance the knowledge of the dynamics of the colonization process (3), including surveillance and characterization of the nasopharyngeal pneumococci.
The continuous emergence of pneumococcal strains resistant to antimicrobial agents is a major cause of concern, narrowing the therapeutic alternatives for pneumococcal disease treatment. Respiratory tract infections, often caused by pneumococci, are the most common reason for antibiotic prescription in children (20). The emergence of penicillin-resistant pneumococci has led to the use of alternative antimicrobials, and macrolides in particular provide a good alternative for treatment of respiratory tract infections (21, 22). However, in the last decade, the prevalence of both penicillin- and macrolide-resistant pneumococci has rapidly increased worldwide (1, 9, 24, 42).
Although only a few drug-resistant pneumococcus (DRPn) clones have successfully achieved worldwide spread (23), with the introduction of the conjugate pneumococcal vaccine the emergence of novel pandemic drug-resistant clones expressing capsular serotypes not covered by the vaccine is a strong possibility.
The prevalence of DRPn clones may vary according to geographic area and also over time. Therefore, surveillance is important for detecting local epidemics, monitoring patterns of resistant bacteria, and helping in the choice of appropriate antimicrobial agents (35).
Typing techniques such as antimicrobial susceptibility testing, serotyping, and pulsed-field gel electrophoresis (PFGE) are commonly used for local surveillance of pathogens as pneumococci. For global epidemiology, multilocus sequence typing (MLST) (10) has become the most valuable tool (16).
In this study 61 DRPn strains were characterized by MLST in order to identify and relate their genetic backgrounds with the ones of isolates recovered in other geographic areas. These strains were representatives of the 26 most frequent clonal types that, together, accounted for 93% of the DRPn isolates recovered from the nasopharynges of healthy children attending day-care centers in the Lisbon and Oeiras regions during 2001 to 2003.
MATERIALS AND METHODS
Pneumococcal carriage isolates.
From 2001 to 2003, 3,539 pneumococcal isolates were collected from the nasopharynges of healthy children (aged 6 months to 6 years) attending 13 day-care centers in Lisbon and Oeiras. These strains were collected during the European Resistance Intervention Study Reducing Resistance in Respiratory Tract Pathogens in Children project.
Antimicrobial susceptibility testing, according to the NCCLS guidelines (26) identified 1,285 DRPn. Oxacillin, clindamycin, erythromycin, tetracycline, chloramphenicol, levofloxacin, and sulfamethoxazole-trimethoprim were tested by the disk diffusion method. Penicillin and ceftriaxone MICs were determined with the Etest (AB Biodisk, Solna, Sweden) according to the manufacturers' instructions. Serotyping (43) and PFGE (38) were performed for all DRPn, and 80 clonal types were defined based on PFGE typing.
From this collection 61 pneumococci were selected for MLST according to the following criteria: two representative strains of clonal types with more than five isolates. These strains had the most common PFGE subtypes within the clone group. In addition, representatives of possible capsular switch events (strains sharing the same PFGE clonal type but with different capsular types) were also selected.
Pneumococcal reference strains.
The 26 strains currently accepted by the Pneumococcal Molecular Epidemiology Network (PMEN) were used as reference strains (23; http://www.sph.emory.edu/PMEN/).
PFGE clonal types.
Clonal types were defined using PFGE (7). Briefly, PFGE patterns were analyzed with Bionumerics software (version 3.0; Applied Maths, Ghent, Belgium), and relatedness among the PFGE profiles was evaluated, using S. pneumoniae strain R6 as a molecular marker.
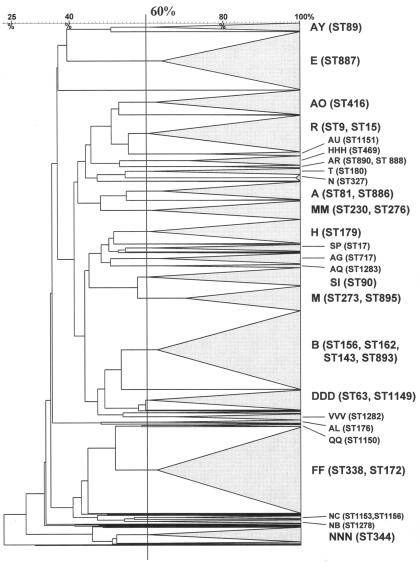
The dendrogram was generated from a similarity matrix calculated with the Jaccard coefficient, and patterns were clustered by the unweighted pair group method with arithmetic mean, using an optimization of 0% and a tolerance of 1.4%. By comparing the clusters generated by Bionumerics with the clusters obtained by visual classification using Tenover's criteria (44), a similarity of 60% was found to be the most appropriate cutoff above which isolates would belong to a common clone.
PFGE patterns obtained in this study were compared with those obtained for strains isolated between 1996 and 1999 in the same settings to determine their prevalence through time (27, 36-38) and with PFGE patterns of the 26 reference strains from PMEN.
The nomenclature of the PFGE patterns was given arbitrarily and is the same used in previous studies. Two or three identical capital letters were assigned to patterns found during 1996 to 1998. Two different capital letters were assigned to new PFGE profiles that were found since 1999.
MLST.
MLST was performed as described previously (10). The primers used for PCR amplification were the ones originally described (10), with the exception of two primers: rec2-dn, GTT CCA TTT TCA ACC AAG GC, and spi2-up, AGA GTG GGG ATT ATT CCT CC. These two new primers were used to overcome difficult PCR amplifications observed for some isolates which DNA did not anneal adequately with the original primers, rec-dn and spi-up. Sequencing was performed either at The Rockefeller University (New York, NY) or at Macrogen, Inc. (Seoul, Korea). In the interpretation of results, single locus variants (SLVs) and double locus variants (DLVs) of a specific clone were assumed to be genetically related (4, 36).
eBURST.
Correlation between the sequence types (ST) of novel S. pneumoniae clones with all ST existent in the database was performed, using the eBURST software available at the MLST website (www.mlst.net).
RESULTS
The 1,285 DRPn strains were characterized by PFGE typing, antimicrobial susceptibility testing, and serotyping. Eighty clones were identified by PFGE typing, of which 26 clones had five or more isolates and accounted for 1,195 (93%) DRPn isolates (Fig. 1). Sixty-one strains representative of the 26 DRPn clones were selected for MLST according to the criteria described in Materials and Methods.
FIG. 1.
Unweighted pair group method with arithmetic mean dendrogram of the 1,285 DRPn PFGE patterns using the Jaccard coefficient. Bionumerics comparison settings were 0% optimization and 1.4% tolerance. Each cluster tree was collapsed. The 60% similarity cutoff value is shown. Capital letters refer to PFGE nomenclature of the major clonal types. In parentheses is the MLST determined for selected isolates within a PFGE clone group.
MLST.
Data obtained by MLST and comparison of the results with the MLST database led us to classify the 26 clonal types into three groups: PMEN clones, unique clones, and novel international clones. PMEN clones are clones whose representatives have identical allelic profiles or are SLVs or DLVs of resistant international clones included in the PMEN; novel international clones are clones recovered in other countries (besides Portugal) and that are currently not included in the PMEN; unique clones are clones that appear to have been detected for the first time in this study.
PMEN clones.
This group comprised 13 clonal types (based on PFGE classification) corresponding to 945 (74%) DRPn isolates. MLST results confirmed that 10 PFGE clonal types matched the following 10 PMEN clones: Spain 23F-1, Spain 9V-3, England 14-9, Colombia 23F-26, Sweden 15A-25, Poland 6B-20, Portugal 19F-21, Greece 6B-22, Spain 6B-2, and Spain 14-5 (Table 1). One additional clone PFGE pattern, QQ, was highly related (DLV) to Colombia 23F-26 (FF). The remaining two PFGE patterns, HHH and AL, were different DLVs of Hungary 19A-6, although expressing a different serotype (Table 1).
TABLE 1.
Properties of representatives of Pneumococcal Molecular Epidemiology Network clones detected in this study
| Strain code | PG MIC (μg/ml) | Antibiotic resistancea | Serotype | PFGE patternb | ST | Description |
|---|---|---|---|---|---|---|
| 542 | 1.5 | C, TE, SXT, TX | 23F | A | 886 | SLV of Spain 23F-1 |
| 4450 | 0.75 | C, TE, SXT | 23F | A | 81 | Spain 23F-1 |
| 2574 | 0.5 | C, TE, SXT | 19A | A | 81 | Spain 23F-1 |
| 1570 | 0.75 | SXT | 14 | B | 156 | Spain 9V-3 |
| 2807 | 2 | E, DA, TE, SXT, TX | 14 | B | 893 | DLV of Spain 9V-3 |
| 2737 | 1.5 | E, DA | 14 | B | 143 | DLV of Spain 9V-3 |
| 4140 | 0.047 | E, DA, TE, SXT | 9V | B | 162 | SLV of Spain 9V-3 |
| 1683 | 1.5 | SXT | NTc | B | 156 | Spain 9V-3 |
| 952 | 0.023 | E | 14 | R | 9 | England 14-9 |
| 3451 | 0.5 | E, DA, SXT | 14 | R | 15 | SLV of England 14-9 |
| 1309 | 0.094 | 23F | FF | 338 | Colombia 23F-26 | |
| 2844 | 0.094 | 23F | FF | 338 | Colombia 23F-26 | |
| 2901 | 0.094 | 15B | FF | 172 | SLV of Colombia 23F-26 | |
| 1304 | 0.094 | 6A | 1150 | DLV of Colombia 23F-26 | ||
| 3831 | 0.094 | 6A | 1150 | DLV of Colombia 23F-26 | ||
| 1730 | 0.19 | E, DA, TE | 15A | DDD | 63 | Sweden 15A-25 |
| 464 | 0.125 | E, DA, TE | 19F | DDD | 1149 | SLV of Sweden 15A-25 |
| 1331 | 0.023 | E, DA, TE | 19A | DDD | 63 | Sweden 15A-25 |
| 3919 | 0.094 | E, DA, TE | 6B | E | 887 | SLV of Poland 6B-20 |
| 1605 | 0.094 | E, DA, TE | 6B | E | 887 | SLV of Poland 6B-20 |
| 1282 | 0.016 | E, DA, TE | 19F | H | 179 | SLV of Portugal 19F-21 |
| 4099 | 0.047 | E, DA, TE | 19F | H | 179 | SLV of Portugal 19F-21 |
| 3026 | 0.012 | C, E, DA, TE, SXT | 6B | M | 895 | SLV of Greece 6B-22 |
| 3536 | 0.016 | C, E, DA, TE, SXT | 6B | M | 273 | Greece 6B-22 |
| 1175 | 0.023 | C, E, DA, TE, SXT | 6A | M | 273 | Greece 6B-22 |
| 3104 | 1 | C, E, DA, TE, SXT | 6B | SI | 90 | Spain 6B-2 |
| 2236 | 0.75 | C, E, DA, TE, SXT | 6B | SI | 90 | Spain 6B-2 |
| 4034 | 1 | C, E, DA, TE, SXT | 14 | SP | 17 | SLV of Spain 14-5 |
| 2667 | 1.5 | E, DA, TE, SXT | 14 | SP | 17 | SLV of Spain 14-5 |
| 2703 | 0.023 | E | 6B | HHH | 469 | DLV of Hungary 19A-6 |
| 2743 | 0.016 | E | 6B | HHH | 469 | |
| 3572 | 0.023 | E, DA, TE | 6B | AL | 176 | DLV of Hungary 19A-6 |
| 4058 | 0.023 | E, DA, TE | 6B | AL | 176 | DLV of Hungary 19A-6 |
Abbreviations: C, chloramphenicol; E, erythromycin; DA, clindamycin; TE, tetracycline; SXT, sulfamethoxazole-trimethoprim; TX, ceftriaxone; PG, penicillin. SLV, single-locus variants; DLV, double-locus variants.
Numbers of isolates with each PFGE clonal type were as follows: A, 42; B, 184; R, 78; FF, 200; QQ, 5; DDD, 46; E, 135; H, 54; M, 119; SI, 44; SP, 10; HHH, 22; and AL, 6.
NT, nontypeable.
Different SmaI PFGE patterns with the same or highly related ST are probably the result of rapid microevolution attributed to S. pneumoniae (12), resulting in alteration of the band pattern (changing band weights by changing restriction sites) though not changing the sequence of the housekeeping genes included in the MLST scheme.
Capsular switch evidence was observed for strains belonging to the following six PMEN clones: Spain 23F-1, expressing serotype 19A; Colombia 23F-26, expressing serotypes 15B and 6A; Sweden 15A-25, expressing serotypes 19F and 19A; Hungary 19A-6, expressing serotype 6B; Greece 6B-22, expressing serotype 6A; and Spain 9V-3, expressing serotypes 14 and 9A. A nontypeable isolate of Spain 9V-3 was also found.
Interestingly, a single isolate of serotype 6A with PFGE pattern H—characteristic of clone Portugal 19F-21—had ST 1152, which shares only three of seven alleles with the typical ST of this clone. This result was confirmed and, in our study, it is the single case where PFGE was a less discriminatory typing technique than MLST.
Unique clones.
Four clones (PFGE patterns AR, AU, NB, and NC)—two of serotype 19A and two nontypeable—were assigned new ST (890, 1151, 1153, 1156, and 1278) (Table 2). No isolates with the same ST, neither SLVs nor DLVs, were available in the MLST database, with the exception of one strain with PFGE pattern AR (ST 888), which was formerly determined for one isolate from our previous studies (strain PT1804b).
TABLE 2.
Properties of representatives of unique and novel international clone groups
| Strain code | PG MIC (μg/ml) | Antibiotic resistancea | Serotype | PFGE patternb | Allele
|
ST | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | rec | spi | xpt | ddl | ||||||
| Unique clones | ||||||||||||
| 2808 | 0.023 | TE, SXT | 19A | AR | 8 | 74 | 19 | 15 | 6 | 40 | 26 | 888 |
| 2143 | 0.023 | TE, SXT | 19A | AR | 8 | 74 | 4 | 15 | 6 | 40 | 27 | 890 |
| 3171 | 0.094 | 19A | AU | 7 | 60 | 9 | 8 | 6 | 3 | 29 | 1151 | |
| 1620 | 0.094 | 19A | AU | 7 | 60 | 9 | 8 | 6 | 3 | 29 | 1151 | |
| 1817 | 0.125 | SXT | NTc | NB | 2 | 6 | 4 | 29 | 91 | 19 | 147 | 1278 |
| 4812 | 0.19 | SXT | NT | NB | 2 | 6 | 4 | 29 | 91 | 19 | 147 | 1278 |
| 3201 | 4 | E, DA, TE, SXT, TX | NT | NC | 2 | 13 | 2 | 29 | 91 | 19 | 141 | 1153 |
| 5002 | 0.047 | E, DA, TE | NT | NC | 2 | 13 | 2 | 29 | 91 | 19 | 59 | 1156 |
| Novel international clones | ||||||||||||
| 2655 | 0.012 | E, DA | 33F | AG | 5 | 35 | 29 | 1 | 45 | 39 | 18 | 717 |
| 2673 | 0.016 | E, DA | 33F | AG | 5 | 35 | 29 | 1 | 45 | 39 | 18 | 717 |
| 2757 | 0.016 | E, TE | 19A | AO | 1 | 13 | 14 | 4 | 17 | 51 | 14 | 416 |
| 2752 | 0.016 | E, DA, TE | 19A | AO | 1 | 13 | 14 | 4 | 17 | 51 | 14 | 416 |
| 1809 | 0.023 | E, DA, TE | 19F | AQ | 15 | 16 | 96 | 5 | 6 | 1 | 26 | 1283 |
| 1750 | 0.023 | E, DA, TE | 19F | AQ | 15 | 16 | 96 | 5 | 6 | 1 | 26 | 1283 |
| 4313 | 0.38 | C, E, DA, TE, SXT | 19F | AY | 5 | 5 | 7 | 7 | 8 | 5 | 1 | 89 |
| 3815 | 0.38 | C, E, DA, TE, SXT | 19F | AY | 5 | 5 | 7 | 7 | 8 | 5 | 1 | 89 |
| 4914 | 0.023 | E, DA, TE | 3 | T | 7 | 15 | 2 | 10 | 6 | 1 | 22 | 180 |
| 4076 | 0.023 | E, DA, TE | 3 | T | 7 | 15 | 2 | 10 | 6 | 1 | 22 | 180 |
| 2436 | 0.5 | E, DA, TE | 19A | MM | 2 | 19 | 2 | 17 | 6 | 22 | 14 | 276 |
| 2679 | 0.38 | E, DA, TE | 19A | MM | 12 | 19 | 2 | 17 | 6 | 22 | 14 | 230 |
| 4815 | 0.125 | E, DA, TE | 19F | MM | 12 | 19 | 2 | 17 | 6 | 22 | 14 | 230 |
| 2942 | 0.75 | E, DA, TE | 24F | MM | 12 | 19 | 2 | 17 | 6 | 22 | 14 | 230 |
| 5137 | 0.38 | E, DA, TE | 20 | MM | 12 | 19 | 2 | 17 | 6 | 22 | 14 | 230 |
| 320 | 0.023 | E, SXT | 6A | N | 1 | 5 | 7 | 12 | 10 | 1 | 14 | 327 |
| 3651 | 0.023 | E, SXT | 6A | N | 1 | 5 | 7 | 12 | 10 | 1 | 14 | 327 |
| 780 | 0.125 | E, DA, TE, SXT | NT | NNN | 8 | 37 | 9 | 29 | 2 | 12 | 53 | 344 |
| 944 | 0.125 | E, TE, SXT | NT | NNN | 8 | 37 | 9 | 29 | 2 | 12 | 53 | 344 |
| 4737 | 0.016 | E, DA | 10A | VVV | 7 | 7 | 4 | 2 | 10 | 1 | 27 | 1282 |
| 3700 | 0.016 | E, DA | 10A | VVV | 7 | 7 | 4 | 2 | 10 | 1 | 27 | 1282 |
Abbreviations: C, chloramphenicol; E, erythromycin; DA, clindamycin; TE, tetracycline; SXT, sulfamethoxazole-trimethoprim; TX, ceftriaxone; PG, penicillin.
Numbers of isolates with each PFGE clonal type were as follows: AR, 7; AU, 9; NB, 10; NC, 7; AG, 26; AO, 53; AQ, 8; AY, 20; T, 16; MM, 44; N, 15; NNN, 21; and VVV, 14.
NT, nontypeable.
Novel international clones.
Nine clonal types were found to be distributed internationally through comparison of our results with those available in the MLST database. None was related to the 26 PMEN clones (approved up to December 2004 [http://www.sph.emory.edu/PMEN/]). Seven of these clonal types (AG, AQ, MM, N, NNN, T, and VVV) had been found in previous studies by our group (1996 to 1999) (27, 36-38), while the remaining two (AO and AY) were found for the first time in Portuguese day-care centers during this study (Table 3).
TABLE 3.
Number of isolates of novel international clones recovered from 1996 to 2003
| PFGE pattern | 1996 | 1997 | 1998 | 1999 | 2001 | 2002 | 2003 |
|---|---|---|---|---|---|---|---|
| AG | 5 | 7 | 16 | 3 | |||
| AO | 27 | 16 | 10 | ||||
| AQ | 1 | 7 | 1 | ||||
| AY | 5 | 9 | 6 | ||||
| T | 4 | 1 | 1 | 10 | 5 | ||
| MM | 2 | 3 | 7 | 25 | 12 | ||
| N | 4 | 3 | 13 | 2 | |||
| NNN | 1 | 3 | 6 | 15 | 3 | 3 | |
| VVV | 21a | 2 | 12 |
Nineteen strains were drug susceptible.
Clonal type AG (serotype 33F): ST 717.
This clonal type was first detected in 1999 in the Lisbon area (27). This clone represents 2.0% of the total DRPn detected between 2001 and 2003. According to the MLST database, strains of the same ST were detected in Scotland in 2003 and found to be causative of invasive disease. A nontypeable DLV isolate, also of invasive origin, was found in the United States. SLVs were recovered during our studies carried out in 1999 (27).
Clonal type AO (serotype 19A): ST 416.
Clonal type AO had not been detected in previous studies. This was the most frequent clonal type within the group of novel international clones, accounting for 4.1% of all DRPn. Identical allelic profiles and serotypes were detected in strains of carriage and invasive origin from the United Kingdom in 1999 to 2000. An SLV with the same capsular type was isolated in Greece. Several DLVs were found in the MLST database. Their origins were The Netherlands, Iceland (39), England, Scotland, Hungary, and the United States, and isolation dates ranged from 1987 through 2003. The majority of the isolates were from serotype 19A, but serotype 15B and serogroups 6 and 19 were also identified.
Clonal type AQ (serotype 19F): ST 1283.
This clonal type was detected in 1999 in Lisbon day-care centers (27). It was attributed a new ST and was found to be a DLV of isolates having the same serotype, recovered in Malaysia and Brazil in 2000, both of which were associated with invasive disease.
Clonal type AY (serotype 19F): ST 89.
Clonal type AY was not identified by our group in previous studies. The ST of this clonal type was detected in 1997 in Spain. It is genetically related to isolates recovered in Denmark, Italy (8), Spain, and Portugal (27). It is an SLV of a minor multiresistant Spain 19F clone (ST 88). Isolation dates of these foreign isolates ranged from 1987 to 1999, with the majority of the isolates being from invasive origin. All isolates were of serotype 19F.
Clonal type MM (serotype 19A): ST 230 and 276.
This clonal type has been isolated from Portuguese day-care centers attendees since 1997 (38). This clone represented 3.4% of all DRPn, being the second most frequent clonal type among the novel international clones. Evidence of diverse capsular switch was observed, since strains of serotypes 19F, 20, and 24F were detected. Isolates with the same ST were recovered in Denmark, Italy (29), and Sweden (40). These also had different serotypes: 14, 24F, and 19F, respectively. When searching the MLST database for SLVs and DLVs of this ST, a variety of isolates from The Netherlands, Poland, Norway, Sri Lanka, Sweden, India, the United States, Kenya, Finland, Australia, and Turkey was detected. Serotype diversity was also present in these latter isolates, where capsular types 23F, 19A, 19F, 14, and 3 were detected. The majority of these isolates were of invasive origin.
Clonal type NNN (nontypeable): ST 344.
Clonal type NNN was first detected in 1997 as previously reported (38). Isolates having the same ST were also recovered in Norway, Australia, and Poland, from invasive origin and carriage. An SLV was recovered in the United States from a noninvasive disease, and DLVs were found in Finland, England, and Portugal in our previous studies (27). These isolates were also nontypeable and were all from carriage.
Clonal type N (serotype 6A): ST 327.
This clonal type was first detected in 1996 in day-care centers of Lisbon (7). Isolates with identical MLST allelic profiles were recovered in England and Brazil from carriage and invasive sources, respectively. Invasive SLVs were recovered during 1982 in The Netherlands and during 1998 in the United States. Carriage SLV and DLV strains were found in England, and one invasive DLV was recovered in Scotland. The isolates were of serotype 6A or had only the indication of serogroup 6.
Clonal type VVV (serotype 10A): ST 1282.
This PFGE pattern was first detected during 1998 in Lisbon day-care centers where the majority of the strains found were drug susceptible (7). A new ST was attributed to this clone. Invasive SLV isolates from Spain, Scotland, and Portugal were found and had the same capsular type, 10. DLV of this ST were recovered in the United Kingdom, Finland, USA and Portugal, being the majority from invasive origin. The serotypes of these isolates were 10A and 6A.
Clonal type T (serotype 3): ST 180.
This clonal type was identified in 1998 in Portuguese day-care centers (38). Isolates with the same ST were found in The Netherlands, the United Kingdom, Denmark, Spain (31), Canada, and Taiwan. SLVs of this clone were found in the United Kingdom, Denmark, Taiwan, the United States, and Portugal. A unique DLV was recovered in Finland. This clone was first recovered in 1984 in The Netherlands, and the vast majority of the foreign isolates were of invasive origin. All isolates were of serotype 3. This clone is presently being proposed to PMEN for acceptance by P. Hermans and B. Spratt (L. McGee, Minutes of the 8th Meeting of the PMEN, Helsinki, Finland, 12 May 2004).
Antibiotype of novel international clones.
The novel international clones comprised 216 DRPn strains (16.8% of the DRPn), the majority being disseminated in several day-care centers (Table 4). Nearly all strains were resistant to erythromycin (99.1%), which was often associated with resistance to clindamycin (90.3%), tetracycline (76.4%), and occasionally sulfamethoxazole-trimethoprim (32.4%) and chloramphenicol (9.3%). Only three clones (MM, AY, and NNN) out of nine had intermediate resistance to penicillin, representing 40% of the novel international isolates. The remaining clones were susceptible to penicillin, with the exception of single isolates of clones AQ and T (Table 4).
TABLE 4.
Antimicrobial resistance patterns of novel international clones and dissemination in day-care centersa
| PFGE pattern | Serotype(s) | No. of isolates | Antibiotype
|
No. of DCC where clone was present (out of 13) | |
|---|---|---|---|---|---|
| MIC of penicillin (μg/ml) | Other resistance markers | ||||
| MM | 19A (29), 19F (3), 20 (3), 24F (9) | 44 | 0.125-0.75 | C, E (44), DA (44), TE (44), SXT (14) | 6 |
| N | 6A | 15 | Susceptible | E (15), SXT (15) | 1 |
| NNN | NT | 21 | 0.094-1 | E (21), DA (17), TE (21), SXT (21), TX | 7 |
| VVV | 10A | 14 | Susceptible | E (14), DA (14), TE (4) | 1 |
| AG | 33F | 26 | Susceptible | E (26), DA (26), TE (2) | 8 |
| AO | 19A | 53 | Susceptible | E (53), DA (53), TE (53) | 2 |
| AQ | 19F | 1 | 0.25 | SXT | 1 |
| 7 | Susceptible | E (7), DA (7), TE (7) | 1 | ||
| AY | 19F | 19 | 0.094-0.5 | C (19), E (18), DA (18), TE (18), SXT (18), TX | 5 |
| T | 3 | 1 | 0.38 | E, DA, TE, SXT | 1 |
| 15 | Susceptible | E (15), DA (15), TE (15) | 3 | ||
Abbreviations: C, chloramphenicol; E, erythromycin; DA, clindamycin; TE, tetracycline; SXT, sulfamethoxazole-trimethoprim; TX, ceftriaxone; NT, nontypeable; DCC, day-care centers. Numbers between parentheses indicate numbers of isolates.
eBURST.
To assess the distribution of the ST of the novel clones within the entire database of MLST, we performed an eBURST analysis (11). In eBURST, the correlation of ST is based on the allelic profiles, where the ancestor within a group is the ST with more SLVs. eBURST analysis was performed for our clones together with the entire S. pneumoniae ST database, and isolates with related ST (5/7 shared alleles) were grouped. Eighty-six groups were obtained in the MLST database, where group 1 was the major one with 1,037 isolates; the following eight groups ranged between 114 and 26 isolates. We observed that seven of the nine novel international clones were dispersed into seven groups. This was in sharp contrast with the 11 PMEN clones identified in our study: they were all clustered in group 1. Two of the novel international clones (ST 180 and ST 416) were also in group 1. Interestingly, the ST of two of the four unique clones were singletons. One of the remaining unique clones belonged to group 2 (the same as clone AQ, ST 1283), and the other one (clone NC) and its two ST (ST 1153 and 1156) formed a new group.
DISCUSSION
Children constitute the main reservoir of pneumococci and are probably the vector for spread of the bacterium to adults (36, 37). Characterization of the nasopharyngeal pneumococci in children is important for monitoring trends in antimicrobial resistance and for a better understanding of the population biology of this pathogen.
PMEN clones were the most frequently isolated group in our study, similarly to what has been reported in previous studies in the same settings (36, 37). This group accounted for 79.1% of the 1,195 DRPn considered in this study (Table 1). By PFGE, serotyping, and antimicrobial susceptibility testing it was possible to assign 76.3% of the isolates to representatives of PMEN clones. MLST not only confirmed these results but also identified three additional PFGE clones as having an ST highly related to those of these international clones. The assignment of different PFGE patterns to the same MLST highlights the genetic evolution of these clones: their genotypes are changing, leading to an increasing number of PFGE subtypes and, eventually over time, to the emergence and assignment of a distinct PFGE pattern. This phenomenon reflects the high genomic plasticity of this pathogen (5). For instance, acquisition or loss of resistance genes (for example, macrolide and tetracycline resistance determinants) associated with integrative mobile elements, such as transposons, may alter the original PFGE pattern of an isolate by originating new band profiles.
By comparing the allelic profiles of our isolates with the ones included in the MLST database, we have found that several of the clones we characterized in the Lisbon area were internationally disseminated and thus were named as novel international clones. Besides this information, data on the clinical sources and capsular types of those identical/highly related isolates (SLVs and DLVs) were also retrieved.
A common property of these novel international drug-resistant pneumococcal clones was resistance to erythromycin often associated with resistance to clindamycin and tetracycline (Table 4). Interestingly, only three of these clones had intermediate resistance to penicillin, corroborating previous findings that erythromycin-resistant and penicillin-susceptible clones are emerging worldwide (37, 41). This is probably related to the increase of macrolide-resistant strains observed in recent years (18, 19, 32).
The introduction of the 7-valent pneumococcal conjugate vaccine may also contribute to the emergence of new or less frequent drug-resistant pneumococci expressing capsular serotypes not covered by the vaccine (14, 33), since this vaccine was conceived to target seven of the most common serotypes responsible for invasive disease (4, 6B, 9V, 14, 18C, 19F, and 23F), particularly in the United States. Recent studies reported its high efficacy in reducing invasive disease (2, 47) and there is evidence that it also reduces the nasopharyngeal carriage of vaccine-type strains (6, 13, 14, 17). However, an increasing frequency in carriage of nonvaccine-type strains has been observed by some and this may lead to the emergence of nonvaccine-type drug-resistant clones (14, 28, 46). The behavior and the disease potential of these nonvaccine types is still unknown, but it is possible that diseases caused by nonvaccine serotypes will increase over time (30). In fact, in one study the number of otitis media cases caused by nonvaccine types increased after introduction of the vaccine (34).
In this study the majority of the novel international clones have nonvaccine serotypes unusual in carriage: serotypes 3 (clonal type T), 33F (clonal type AG), 10A (clonal type VVV), 20 and 24F (clonal type MM), and nontypeable (NNN), all of which are identical or highly related to invasive isolates according to the data provided by the MLST database. Only two of the nine clones (AY and AQ) had a vaccine serotype, 19F (Table 2). Serotypes 19A and 6A were also present in this group, and although these serotypes may be considered to be covered by the vaccine due to cross-reactivity with serotypes 19F and 6B, respectively (45, 49), a recent study did not find conclusive evidence that serotype 19A was adequately covered by the 7-valent conjugate vaccine (47).
eBURST performed on novel international clone ST illustrate the diversity of these clones as the nine novel international clones were dispersed through eight distinct groups, suggesting that these clones have distinct evolutionary origins.
Our results reinforce the idea that careful monitoring is becoming increasingly important, especially regarding the emergence of nonvaccine-type DRPn strains with unusual genetic backgrounds.
We propose that the novel international clones should be considered for inclusion in the PMEN (Table 5). The increasing frequency of these clones (27, 36-38) with disease potential might be an indication of the invasive multidrug-resistant international clones of the near future.
TABLE 5.
Clones to be proposed for Pneumococcal Molecular Epidemiology Network inclusion
| ST | Serotype | Antimicrobial resistancesa | PFGE pattern |
|---|---|---|---|
| 327 | 6A | E, SXT | N |
| 230 | 19A | PGI, E, DA, TE | MM |
| 416 | 19A | E, DA, TE | AO |
| 1282 | 10A | E, DA | VVV |
| 717 | 33F | E, DA | AG |
| 344 | NTb | PGI, E, DA, TE, SXT | NNN |
| 1283 | 19F | E, DA, TE | AQ |
| 89 | 19F | PGI, C, E, DA, TE, SXT | AY |
Abbreviations: PGI, penicillin intermediate resistance (0.1 μg/ml ≤ MIC < 1.5 μg/ml); C, chloramphenicol; E, erythromycin; DA, clindamycin; TE, tetracycline; SXT, sulfamethoxazole-trimethoprim.
NT, nontypeable.
Acknowledgments
This work was supported by EURIS (the European Resistance Intervention Study) (contract QLK2-CT-2000-01020) and PREVIS (Pneumococcal Resistance, Epidemicity and Virulence: an International Study) (contract LSHM-CT-2003-503413) projects of the European Community. R.S.-L., M.I.C., and J.A.C. were supported by Fundação para a Ciência e Tecnologia (SFRH/BPD/14596/2003, SFRH/BD/5205/2001, and SFRH/BD/3123/2000, respectively). C.S. and N.F. were supported by grants from Instituto de Biologia Experimental e Technológica (project WLP, no. 31, CEM/NET, and reference 28/12/02 CB, respectively).
We thank Alexander Tomasz for providing the opportunity to perform part of this work at the Rockefeller University, NY, with a grant from The Lounsbery Foundation to A. Tomasz. We are grateful to António Brito Avô and Joana Saldanha for their valuable contribution during the project, nurses Anabela Gonçalves and Paula Poção for the collection of the samples, and Lesley McGee for providing the PMEN strains. We acknowledge the use of the pneumococcal MLST database, which is located at the Imperial College in London and is funded by the Wellcome Trust.
REFERENCES
- 1.Albrich, W. C., D. L. Monnet, and S. Harbarth. 2004. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg. Infect. Dis. 10:514-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, S., H. Shinefield, R. Baxter, R. Austrian, L. Bracken, J. Hansen, E. Lewis, and B. Fireman. 2004. Postlicensure surveillance for pneumococcal invasive disease after use of heptavalent pneumococcal conjugate vaccine in Northern California Kaiser Permanente. Pediatr. Infect. Dis. J. 23:485-489. [DOI] [PubMed] [Google Scholar]
- 3.Bogaert, D., R. De Groot, and P. W. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144-154. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 6.Dagan, R., R. Melamed, M. Muallem, L. Piglansky, D. Greenberg, O. Abramson, P. M. Mendelman, N. Bohidar, and P. Yagupsky. 1996. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J. Infect. Dis. 174:1271-1278. [DOI] [PubMed] [Google Scholar]
- 7.De Lencastre, H., K. G. Kristinsson, A. Brito-Avô, I. S. Sanches, R. Sá-Leão, J. Saldanha, E. Sigvaldadottir, S. Karlsson, D. Oliveira, R. Mato, M. Aires de Sousa, and A. Tomasz. 1999. Carriage of respiratory tract pathogens and molecular epidemiology of Streptococcus pneumoniae colonization in healthy children attending day care centers in Lisbon, Portugal. Microb. Drug Resist. 5:19-29. [DOI] [PubMed] [Google Scholar]
- 8.Dicuonzo, G., G. Gherardi, R. E. Gertz, F. D'Ambrosio, A. Goglio, G. Lorino, S. Recchia, A. Pantosti, and B. Beall. 2002. Genotypes of invasive pneumococcal isolates recently recovered from Italian patients. J. Clin. Microbiol. 40:3660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doern, G. V., and S. D. Brown. 2004. Antimicrobial susceptibility among community-acquired respiratory tract pathogens in the USA: data from PROTEKT US 2000-01. J. Infect. 48:56-65. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein, J. A., S. S. Huang, J. Daniel, S. L. Rifas-Shiman, K. Kleinman, D. Goldmann, S. I. Pelton, A. DeMaria, and R. Platt. 2003. Antibiotic-resistant Streptococcus pneumoniae in the heptavalent pneumococcal conjugate vaccine era: predictors of carriage in a multicommunity sample. Pediatrics 112:862-869. [DOI] [PubMed] [Google Scholar]
- 14.Frazão, N., A. Brito-Avô, C. Simas, J. Saldanha, R. Mato, S. Nunes, N. G. Sousa, J. A. Carriço, J. S. Almeida, I. Santos-Sanches, and H. de Lencastre. 2005. Effect of the seven-valent conjugate pneumococcal vaccine on carriage and drug resistance of Streptococcus pneumoniae in healthy children attending day-care centers in Lisbon. Pediatr. Infect. Dis. J. 24:243-252. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Rodriguez, J. A., and M. J. Fresnadillo Martinez. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 50:S59-S73. [DOI] [PubMed] [Google Scholar]
- 16.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghaffar, F., T. Barton, J. Lozano, L. S. Muniz, P. Hicks, V. Gan, N. Ahmad, and G. H. McCracken, Jr. 2004. Effect of the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clin. Infect. Dis. 39:930-938. [DOI] [PubMed] [Google Scholar]
- 18.Hyde, T. B., K. Gay, D. S. Stephens, D. J. Vugia, M. Pass, S. Johnson, N. L. Barrett, W. Schaffner, P. R. Cieslak, P. S. Maupin, E. R. Zell, J. H. Jorgensen, R. R. Facklam, and C. G. Whitney. 2001. Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 286:1857-1862. [DOI] [PubMed] [Google Scholar]
- 19.Inoue, M., N. Y. Lee, S. W. Hong, K. Lee, and D. Felmingham. 2004. PROTEKT 1999-2000: a multicentre study of the antibiotic susceptibility of respiratory tract pathogens in Hong Kong, Japan and South Korea. Int. J. Antimicrob. Agents 23:44-51. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, M. R. 2003. Worldwide trends in antimicrobial resistance among common respiratory tract pathogens in children. Pediatr. Infect. Dis. J. 22:S109-S119. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, M. R., and C. E. Johnson. 2003. Macrolide resistance: an increasing concern for treatment failure in children. Pediatr. Infect. Dis. J. 22:S131-S138. [DOI] [PubMed] [Google Scholar]
- 22.Lonks, J. R., J. Garau, and A. A. Medeiros. 2002. Implications of antimicrobial resistance in the empirical treatment of community-acquired respiratory tract infections: the case of macrolides. J. Antimicrob. Chemother. 50:S87-S92. [DOI] [PubMed] [Google Scholar]
- 23.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melo-Cristino, J., M. Ramirez, N. Serrano, and T. Hanscheid. 2003. Macrolide resistance in Streptococcus pneumoniae isolated from patients with community-acquired lower respiratory tract infections in Portugal: results of a 3-year (1999-2001) multicenter surveillance study. Microb. Drug Resist. 9:73-80. [DOI] [PubMed] [Google Scholar]
- 25.Mulholland, K. 1999. Strategies for the control of pneumococcal diseases. Vaccine 17:S79-S84. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. Approved standard M2-A8, M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Nunes, S., R. Sá-Leão, J. Carriço, C. R. Alves, R. Mato, A. B. Avô, J. Saldanha, J. S. Almeida, I. S. Sanches, and H. de Lencastre. 2005. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J. Clin. Microbiol. 43:1285-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obaro, S. K., R. A. Adegbola, W. A. Banya, and B. M. Greenwood. 1996. Carriage of pneumococci after pneumococcal vaccination. Lancet 348:271-272. [DOI] [PubMed] [Google Scholar]
- 29.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 30.Pelton, S. I., R. Dagan, B. M. Gaines, K. P. Klugman, D. Laufer, K. O'Brien, and H. J. Schmitt. 2003. Pneumococcal conjugate vaccines: proceedings from an interactive symposium at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy. Vaccine 21:1562-1571. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Trallero, E., J. M. Marimon, L. Iglesias, and J. Larruskain. 2003. Fluoroquinolone and macrolide treatment failure in pneumococcal pneumonia and selection of multidrug-resistant isolates. Emerg. Infect. Dis. 9:1159-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pihlajamaki, M., P. Kotilainen, T. Kaurila, T. Klaukka, E. Palva, and P. Huovinen. 2001. Macrolide-resistant Streptococcus pneumoniae and use of antimicrobial agents. Clin. Infect. Dis. 33:483-488. [DOI] [PubMed] [Google Scholar]
- 33.Porat, N., A. Arguedas, B. G. Spratt, R. Trefler, E. Brilla, C. Loaiza, D. Godoy, N. Bilek, and R. Dagan. 2004. Emergence of penicillin-nonsusceptible Streptococcus pneumoniae clones expressing serotypes not present in the antipneumococcal conjugate vaccine. J. Infect. Dis. 190:2154-2161. [DOI] [PubMed] [Google Scholar]
- 34.Porat, N., G. Barkai, M. R. Jacobs, R. Trefler, and R. Dagan. 2004. Four antibiotic-resistant Streptococcus pneumoniae clones unrelated to the pneumococcal conjugate vaccine serotypes, including 2 new serotypes, causing acute otitis media in southern Israel. J. Infect. Dis. 189:385-392. [DOI] [PubMed] [Google Scholar]
- 35.Sahm, D. F., J. A. Karlowsky, L. J. Kelly, I. A. Critchley, M. E. Jones, C. Thornsberry, Y. Mauriz, and J. Kahn. 2001. Need for annual surveillance of antimicrobial resistance in Streptococcus pneumoniae in the United States: 2-year longitudinal analysis. Antimicrob. Agents Chemother. 45:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sá-Leão, R., A. Tomasz, and H. de Lencastre. 2001. Multilocus sequence typing of Streptococcus pneumoniae clones with unusual drug resistance patterns: genetic backgrounds and relatedness to other epidemic clones. J. Infect. Dis. 184:1206-1210. [DOI] [PubMed] [Google Scholar]
- 37.Sá-Leão, R., A. Tomasz, I. S. Sanches, A. Brito-Avô, S. E. Vilhelmsson, K. G. Kristinsson, and H. de Lencastre. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 182:1153-1160. [DOI] [PubMed] [Google Scholar]
- 38.Sá-Leão, R., A. Tomasz, I. S. Sanches, S. Nunes, C. R. Alves, A. B. Avô, J. Saldanha, K. G. Kristinsson, and H. de Lencastre. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 38:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sá-Leão, R., S. E. Vilhelmsson, H. de Lencastre, K. G. Kristinsson, and A. Tomasz. 2002. Diversity of penicillin-nonsusceptible Streptococcus pneumoniae circulating in Iceland after the introduction of penicillin-resistant clone Spain 6B-2. J. Infect. Dis. 186:966-975. [DOI] [PubMed] [Google Scholar]
- 40.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 41.Schito, G. C., E. A. Debbia, and A. Marchese. 2000. The evolving threat of antibiotic resistance in Europe: new data from the Alexander Project. J. Antimicrob. Chemother. 46S:3-9. [DOI] [PubMed] [Google Scholar]
- 42.Song, J. H., S. I. Jung, K. S. Ko, N. Y. Kim, J. S. Son, H. H. Chang, H. K. Ki, W. S. Oh, J. Y. Suh, K. R. Peck, N. Y. Lee, Y. Yang, Q. Lu, A. Chongthaleong, C. H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumarasinghe, F. Jamal, A. Kamarulzaman, N. Parasakthi, P. H. Van, C. Carlos, T. So, T. K. Ng, and A. Shibl. 2004. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob. Agents Chemother. 48:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorensen, U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vakevainen, M., C. Eklund, J. Eskola, and H. Kayhty. 2001. Cross-reactivity of antibodies to type 6B and 6A polysaccharides of Streptococcus pneumoniae, evoked by pneumococcal conjugate vaccines, in infants. J. Infect. Dis. 184:789-793. [DOI] [PubMed] [Google Scholar]
- 46.Veenhoven, R., D. Bogaert, C. Uiterwaal, C. Brouwer, H. Kiezebrink, J. Bruin, I. J. E., P. Hermans, R. de Groot, B. Zegers, W. Kuis, G. Rijkers, A. Schilder, and E. Sanders. 2003. Effect of conjugate pneumococcal vaccine followed by polysaccharide pneumococcal vaccine on recurrent acute otitis media: a randomised study. Lancet 361:2189-2195. [DOI] [PubMed] [Google Scholar]
- 47.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. 2003. State of the art of new vaccines: research and development. Initiative for vaccine research. World Health Organization, Geneva, Switzerland.
- 49.Yu, X., B. Gray, S. Chang, J. I. Ward, K. M. Edwards, and M. H. Nahm. 1999. Immunity to cross-reactive serotypes induced by pneumococcal conjugate vaccines in infants. J. Infect. Dis. 180:1569-1576. [DOI] [PubMed] [Google Scholar]