Abstract
The increasing demand for molecular diagnostics in clinical microbiology laboratories necessitates automated sample processing. In the present study, we evaluated the performance of the MagNA Pure LC total nucleic acid isolation kit (M extraction) in comparison with the manual method (Si extraction) according to Boom et al. (R. Boom, C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa, J. Clin. Microbiol. 28:495-503, 1990) for the detection of viral DNA by competitive quantitative PCR. Reconstruction experiments with HindIII-digested phage lambda DNA and HaeIII-digested φX174 DNA showed that the recovery of DNA from phosphate-buffered saline, cerebrospinal fluid, EDTA-anticoagulated plasma, and EDTA-anticoagulated whole blood by M extraction is, on average, 6.6-fold lower compared to Si extraction. PCR signals of spiked PCR control DNAs for Epstein-Barr virus and varicella-zoster virus were also between 1.9- and 14.2-fold lower after M extraction compared to Si extraction, also suggesting impaired DNA recovery. M extraction of spiked cytomegalovirus strain AD 169 in whole blood showed a 5- to 10-fold reduction in PCR sensitivity compared to Si extraction. This reduction of PCR sensitivity was also observed when clinical whole blood samples were processed by M extraction. Before implementing M extraction, the clinical consequences of the reduced recovery should first be considered, especially when maximal sensitivity is required.
In recent years, molecular diagnostic testing has become an essential part of the routine work flow in clinical microbiology laboratories. To comply with the demands for sensitivity and reliability, highly qualitative methods for extraction, amplification, and detection of nucleic acids (NA) are required. Most amplification-detection systems today, like PCR-hybridization and real-time PCR, have the analytical power to detect and identify a single target molecule (5, 7, 8, 9, 22). When combined with the extraction procedure developed by Boom et al. (Si extraction) (3), acknowledged for its potency in removing inhibitors from clinical samples and widely used for the purification of NA from a variety of clinical samples (2, 6, 24), optimal clinical sensitivity and reliability are achieved. However, with the increasing number of molecular assays becoming available to the clinical laboratory, the total throughput of samples also increases and makes automation of the extraction procedure mandatory. The use of an automated commercial NA extraction method also has other potential benefits, like a high degree of standardization and transferring part of the quality control from the clinical microbiology laboratory to the manufacturer.
Several robotic platforms have recently become available for sample preparation in molecular diagnostics. For low-throughput settings, the BioRobot EZ1 (QIAGEN) and MagNA Pure Compact (Roche) systems are suitable platforms, whereas for medium- to high-throughput settings the MagNA Pure LC instrument (Roche) and the BioRobot M48/9604 system (QIAGEN) are available. For most clinical microbiology laboratories performing several DNA- and RNA-based assays on a wide range of clinical specimens, medium- to high-throughput extractors combined with generic extraction chemistry will provide an efficient solution for sample preparation, although for smaller laboratories low-throughput extractors can also be sufficient.
The MagNA Pure LC total NA isolation kit (M extraction) on the MagNA Pure LC instrument provides generic extraction chemistry on a medium- to high-throughput extraction platform. Several reports have described the application of M extraction in the sample preparation of cytomegalovirus (CMV), enterovirus, hepatitis B virus, hepatitis C virus (HCV), herpes simplex virus type 1 and 2, human immunodeficiency virus type 1, and broad-range bacterial rRNA genes (10, 11, 13, 14, 15, 16, 18, 20, 21). Most of these reported assays perform with satisfactory analytical and clinical sensitivities compared to reference procedures. However, there is some evidence for reduced extraction efficiency with M extraction. For instance, HCV RNA recovery seems to be less efficient with M extraction and analysis by the (COBAS) AMPLICOR HCV 2.0 test (10, 11, 14) compared to other manual (Roche) and automated (NucliSens) extraction procedures (1, 19). Furthermore, for enterovirus RNA, Knepp et al. (16) have shown that M extraction was less sensitive than the automated BioRobot viral RNA M48 and manual QiaAmp viral RNA isolation kits. Also, data presented by Mohammadi et al. suggest less efficient recovery of the Escherichia coli 16S rRNA gene by M extraction in comparison with Si extraction (manual NucliSens extraction) (21). Finally, Burghoorn-Maas et al. (C. Burghoorn-Maas, P. van Deursen, M. Jacobs, and H. G. M. Niesters, Abstr., 2nd Eur. Congr. Virol., p. 7-10, 2004) have shown that M extraction was 0.5 to 1.0 log10 less sensitive compared to the NucliSens miniMAG platform.
Because the lower recovery of M extraction is currently supported by a limited number of small-scale experiments, it has to date not been recognized as a possible issue of concern. Furthermore, in most reports the extraction methods used are difficult to compare directly, due to differences in input/output ratios and therefore the equivalent of sample volume tested by the amplification-detection system. In this respect, a more extensive study of the extraction efficiency in a direct comparison, with equivalent sample volumes, could provide more insights into the possible drawbacks of M extraction. Here we report the evaluation of M extraction in comparison with the Si extraction reported by Boom et al. (3), which is currently widely used throughout the field of molecular diagnostic testing (2, 6).
MATERIALS AND METHODS
Clinical specimens.
EDTA-anticoagulated whole blood (whole blood) was obtained from healthy volunteers. EDTA-anticoagulated plasma (plasma) was obtained by separation from the whole blood after centrifugation for 10 min at 1,750 × g. Clinical whole blood specimens were obtained from patients for whom CMV and/or EBV PCR was requested. Cerebrospinal fluid (CSF), stored at −20°C, was obtained from a previous study (24).
Human CMV.
Sucrose density gradient-purified human CMV strain AD 169 (lot no. 80-165-1; 5.38 × 109 viral particles/ml of virus dilution buffer [10 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5], as determined electron microscopically by direct particle counting, which discriminates between full and empty particles) was obtained from Advanced Biotechnologies Inc. (Colombia, Md.). The virus titer was corrected to 1.61 × 1010 viral particles/ml as described previously (5).
DNA purification.
DNA was purified from 200 μl of phosphate-buffered saline (PBS), plasma, or CSF or 50 μl of whole blood by manual Si extraction or automated M extraction. Specimens were spiked with either HindIII-digested phage lambda (λ-HindIII) DNA (Gibco BRL, Breda, The Netherlands), HaeIII-digested φX174 DNA (molecular weight marker IX; Roche Diagnostics Nederland BV, Almere, The Netherlands), or internal control (IC) DNA before extraction.
Manual Si extraction was carried out as described previously (3), with 20 μl of size-fractionated silica particles (SC). Elution was in 100 μl of TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]).
Automated M extraction on the MagNA Pure LC instrument (Roche Diagnostics Nederland BV) was carried out with the MagNA Pure LC total NA isolation kit (Roche Diagnostics Nederland BV) according to the manufacturer's instructions, using the protocol total NA external lysis, with the elution volume set at 100 μl.
Recovery of lost DNA.
DNA potentially lost during washing steps of the purification procedures was retrieved by re-extraction of the discarded washing solutions. For M extraction, 200 μl of washing solution was processed by Si extraction and eluted in 50 μl of TE buffer. For Si extraction, the discarded lysis buffer L6 was added to 20 μl of fresh SC and subsequently extracted by Si extraction with elution in 50 μl of TE buffer. For the washing stages (L2, 70% ethanol and acetone), 200 μl of the discarded washing solution was re-extracted by Si extraction and eluted in 50 μl of TE buffer. Noneluted DNA from the silica matrices of both methods (magnetic glass particles or SC) was recovered by elution in 50 μl of TE buffer for 1 h at 56°C.
Agarose gel electrophoresis.
DNA was electrophoresed through 1% agarose (Agarose MP; Roche Diagnostics Nederland BV) gels containing 1 μg ethidium bromide per ml of 0.5× Tris-borate-EDTA buffer. Appropriate recovery markers were included in all gels for estimation of the DNA recovery after image capturing under UV illumination. The Gel Doc 1000 system (Bio-Rad, Veenendaal, The Netherlands) was used for image capturing, and the graphic files were exported as 8-bit TIFF images. DNA band intensities were calculated from the peak surface areas using Scion Image Release Beta 4.0.2 (Scion Corporation, Frederick, Md.). Recoveries were calculated in comparison to the appropriate recovery markers.
Primers and probes.
The primers and probes used (high-performance liquid chromatography purified; Applied Biosystems, Nieuwererk a/d IJssel, The Netherlands) are listed in Table 1.
TABLE 1.
Primers and probes used in this study
| Name | Function | Sequencea | Reference |
|---|---|---|---|
| CMV-531 | Forward PCR primer | 5′-ACA AGG TGC TCA CGC ACA TTG ATC-3′ | 5 |
| bio-CMV-1107 | Reverse PCR primer | 5′-biotin-CAC TGG CTC AGA CTT GAC AGA CAC-3′ | 5 |
| TBR-CMV-1 | CMV-specific probe | 5′-TBR-TGA AGG TTG CCC AGT ACA TTC T-3′ | 5 |
| TBR-CMV-2 | CMV-IC-specific probe | 5′-TBR-CCC TTT ACA TCT TTC TGA AGT AGG G-3′ | 5 |
| VZV-3 | Forward PCR primer | 5′-TCT TTC ACG GAG GCA AAC AC-3′ | 8 |
| bio-VZV-4 | Reverse PCR primer | 5′-biotin-TCC AAG GCG GGT GCA TAT CT-3′ | 8 |
| TBR-VZV-1 | VZV-specific probe | 5′-TBR-AAC GGT TTG GGT TTT CAC GCT GCC-3′ | 8 |
| TBR-VZV-2 | VZV-IC-specific probe | 5′-TBR-ACC TGT CGG ACT CGT AGT TGC TGT-3′ | 8 |
| bio-EBV-1 | Forward PCR primer | 5′-biotin-TGG GTC GCC GGT GTG TTC GTA TA-3′ | |
| EBV-2 | Reverse PCR primer | 5′-CTA AAC GGA GGG ACC AAA GGT GG-3′ | |
| TBR-EBV-1 | EBV-specific probe | 5′-TBR-GGC CAT TCC AAA GGG GAG ACG-3′ | |
| TBR-EBV-2 | EBV-IC-specific probe | 5′-TBR-GAG CAG TCA GGA TCC GAG AGC-3′ |
TBR, tris-(2,2′-bipyridine)-ruthenium (II) chelate.
IC DNA.
Construction of the IC DNAs for CMV and VZV has been described previously (5, 8). Construction of the IC DNA for EBV will be described elsewhere (V. Bekker et al., submitted for publication). The design of the EBV-IC DNA is essentially the same as for CMV and VZV.
CMV, VZV, and EBV PCRs.
CMV and VZV PCRs were carried out as described previously (5, 8). EBV PCR was carried out in the same format as the CMV and VZV PCRs and in general has performance characteristics similar to both the CMV and VZV PCRs. The validation of the EBV PCR assay will be described elsewhere (Bekker et al., submitted). Briefly, the reaction mixtures (50 μl) consisted of 10 mM Tris HCl (pH 8.3); 50 mM KCl; 3 (CMV), 4 (VZV), or 4.5 (EBV) mM MgCl2; dATP, dGTP, and dCTP at a concentration of 200 μM each; 400 μM dUTP; 2.5 U of AmpliTaq DNA polymerase (CMV and VZV) or AmpliTaq Gold DNA polymerase (EBV); 0.5 U of Amperase (uracil-N-glycosylase); 200 ng each of the forward and reverse primers (Table 1); 20 μg of alpha-casein (C 6780; Sigma-Aldrich Chemie, Zwijndrecht, The Netherlands); 5 μg of bovine serum albumin (Roche Diagnostics Nederland BV); and 25 μl of DNA. Amplifications were done in a PE 9600 thermocycler (Applied Biosystems) under the following conditions: 2 min at 50°C, 5 min (CMV and VZV) or 10 min (EBV) at 95°C, followed by 35 cycles each consisting of 20 s at 95°C, 20 s at 63°C, and 1 min at 72°C, followed by 5 min at 72°C.
Removal of excess primers, hybridization, and electrochemiluminescence measurement.
Excess primers were removed as described previously (5) by protocol Delta Y-A. Hybridization and electrochemiluminescence detection were also performed as described previously (4, 5, 8).
Algorithm for quantitation.
For quantitation of CMV and EBV, the following algorithms were used. After correcting for the background, the ratio of the virus DNA-specific signal to the IC DNA-specific signal (R) was determined and the number of copies of viral DNA per milliliter of whole blood was calculated by multiplying R by factor F. Factor F was derived by multiplying the factors ICextraction × D. The factor ICextraction represents the number of IC DNA molecules present in the DNA extraction, and factor D is required to reach the copy number per milliliter, which is set at 20 for whole blood.
RESULTS
Recovery of HindIII-digested phage λ DNA.
Four micrograms of spiked HindIII-digested phage lambda DNA was extracted from PBS, CSF, and plasma with four to eight replicates each by both M extraction and Si extraction. Table 2 shows the summarized data for all specimen types tested.
TABLE 2.
Recovery of DNAa
| Method and specimen typeb | DNA typec | Mean % DNA recoveryd
|
% of DNA which was not recovered | |||
|---|---|---|---|---|---|---|
| By method | From binding step | From washing steps | From silica matrix | |||
| M extraction | ||||||
| PBS | HMW | 10 | 10 | 5 | 15 | 60 |
| LMW | 5 | 15 | 20 | 10 | 50 | |
| CSF | HMW | 15 | 20 | 0 | 5 | 60 |
| LMW | 15 | 25 | 0 | 5 | 55 | |
| Plasma | HMW | 25 | 25 | 0 | 5 | 45 |
| LMW | 25 | 25 | 0 | 5 | 45 | |
| Whole blood | HMWf | 20 | 20 | 0 | 60 | NAg |
| LMW | 25 | 40 | 0 | 10 | 25 | |
| Si extraction | ||||||
| PBS | HMW | 100 | 0 | NDe | ND | 0 |
| LMW | 100 | 0 | ND | ND | 0 | |
| CSF | HMW | 80 | 5 | ND | 5 | 10 |
| LMW | 80 | 5 | ND | 5 | 10 | |
| Plasma | HMW | 90 | 5 | ND | 5 | 0 |
| LMW | 70 | 5 | ND | 5 | 20 | |
| Whole blood | HMWf | 70 | 5 | 0 | 25 | NA |
| LMW | 65 | 15 | 0 | 5 | 15 | |
For PBS, CSF, and plasma, HindIII-digested phage λ DNA was used. For whole blood, HaeIII-digested φX174 DNA was used.
Plasma, EDTA-anticoagulated plasma; whole blood, EDTA-anticoagulated whole blood.
HMW DNA, 23.1 kb, 9.4 kb, 6.6 kb, and 4.4 kb (λ-HindIII) and chromosomal DNA (marker IX); LMW DNA, 2.3 kb and 2.0 kb (λ-HindIII) and 1.4 kb, 1.1 kb, 0.9 kb, and 0.6 kb (marker IX).
Calculated percentages were rounded to the nearest multiple of 5% for simplified representation.
ND, not determined.
Due to the fact that the exact input of chromosomal DNA was unknown, the percentages reflect the relative amounts of DNA that could be detected.
NA, not applicable (could not be determined because the exact input was unknown).
Table 2 clearly shows that M extraction had a substantially lower recovery compared to Si extraction when λ-HindIII DNA was extracted from PBS, CSF, and plasma, with, on average, 7.8-fold (range, 2.8-fold to 20-fold) lower recoveries. To identify purification steps where DNA was possibly lost in the procedure, DNA was extracted from the fluids in which binding to the silica matrix takes place for both procedures. Approximately 20% of the missing DNA is not bound by the magnetic glass particles in M extraction, whereas approximately 3% of the λ-HindIII DNA remains unbound in Si extraction. In a second elution of the silica matrix, about 7.5% and 5% of the missing DNA could be retrieved for M and Si extractions, respectively. Since a large portion (on average, 53%) of the λ-HindIII DNA was still missing with M extraction, the other washing solutions were also investigated for potentially lost DNA. However, within the detection limit of agarose gel electrophoresis, no DNA could be retrieved from discarded wash buffers I, II, and III, thus leaving about 53% of the input of λ-HindIII DNA irretrievable.
Recovery of DNA from whole blood.
For whole blood, HaeIII-digested φX174 DNA was used to monitor low-molecular-weight (LMW) DNA recovery. For recovery of high-molecular-weight (HMW) DNA, the human chromosomal DNA present in the blood was used, but no estimation of irretrievable DNA was possible for HMW DNA, since the exact amount of chromosomal DNA per milliliter was unknown.
With whole blood, a similar trend in recovery was observed with M extraction, with, on average, 3.1-fold lower recovery compared to Si extraction (Table 2). Again part of the missing DNA could be retrieved from the binding step (30% with M extraction and 10% with Si extraction), and no DNA was lost during the washing procedures. For both extraction procedures, more HMW DNA could be retrieved from the silica matrix compared to PBS, CSF, and plasma, whereas the amount of LMW DNA retrieved by a second elution was the same as for the other specimen types.
Influence of DNA recovery on PCR.
To assess the influence of the impaired DNA recovery of M extraction on PCR assays, we spiked PCR-negative whole blood samples with low (50 copies [LPC]) and high (250 copies [HPC]) positive control DNAs for EBV or VZV and the appropriate IC DNA (200 copies of EBV-IC or 50 copies of VZV-IC). Both M and Si extractions were performed in five replicates for each sample. After correcting for the background signal level, the signals obtained by M extraction were between 7.6- and 14.2-fold lower for both EBV wild-type and IC DNAs compared to the signals obtained after Si extraction, as can be seen in Table 3. For VZV, similar results were obtained with signals between 1.9- and 7.9-fold lower after M extraction (Table 3).
TABLE 3.
Summarized data for EBV and VZV LPC and HPC control DNAs, CMV strain AD 169, and EBV and CMV clinical samples after Si extraction and M extraction from EDTA-treated whole blood
| Specimen typea | Quantityb | Average Si extraction/M extraction signal ratioc
|
|
|---|---|---|---|
| Virus | IC | ||
| EBV | |||
| LPC | 3 | 9.0 (2.8 to 20.3; n = 4) | 14.2 (4.4 to 37.7; n = 5) |
| HPC | 3.7 | 7.6 (4.1 to 13.1; n = 5) | 7.7 (3.9 to 10.2; n = 5) |
| VZV | |||
| LPC | 3 | 7.9 (3.5 to 12.8; n = 4) | 5.1 (2.4 to 8.2; n = 5) |
| HPC | 3.7 | 2.1 (0.7 to 3.3; n = 4) | 1.9 (0.2 to 3.5; n = 3) |
| CMV strain AD169 | 1.5 | NAd | 2.2 (0.1 to 7.2; n = 8) |
| 2.5 | 1.8 (0.8 to 2.9; n = 7) | 3.5 (2.5 to 7.3; n = 8) | |
| 3.5 | 1.1 (0.7 to 1.6; n = 8) | 2.2 (1.1 to 3.2; n = 8) | |
| 4.5 | 1.1 (0.9 to 1.4; n = 8) | 3.2 (2.3 to 4.2; n = 8) | |
| EBV(clinical) | Unknown | 9.4 (1.0 to 33.5; n = 16) | 7.2 (0.9 to 26.9; n = 23) |
| CMV (clinical) | Unknown | 4.8 (2.2 to 6.2; n = 3) | 3.6 (1.7 to 8.8; n = 16) |
LPC, 1,000 copies/ml, 50 copies/extraction; HPC, 5,000 copies/ml, 250 copies/extraction.
Log10 number of viral DNA copies/ml.
Ratio calculated from background-corrected luminosity signals. Values in parentheses reflect the range of the measurements and the number of samples.
NA, not applicable.
Extraction and PCR of human CMV strain AD 169 from whole blood.
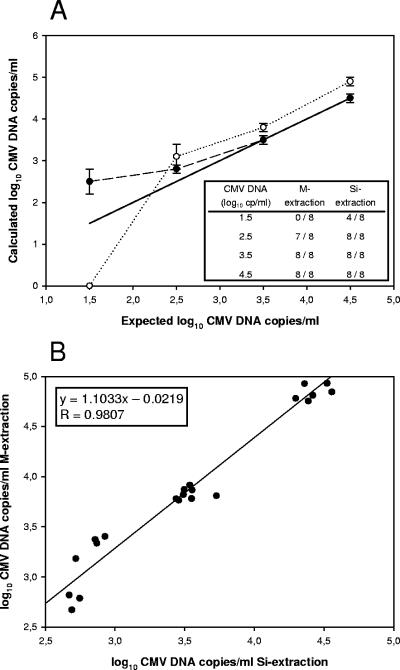
In order to assess the influence of the impaired DNA recovery of M extraction with complete virus, another reconstruction experiment was performed with human CMV strain AD 169. A PCR-negative whole blood sample was spiked with 10-fold serial dilutions of CMV ranging from 1.5 to 4.5 log10 copies/ml, and each dilution was extracted in eight replicates by both procedures in the presence of 140 copies of CMV-IC DNA. Figure 1A shows the quantitative results of both procedures after CMV PCR. At 1.5 log10 copies/ml, all eight replicates isolated by M extraction were PCR negative, whereas only 50% (4/8) of the replicates from Si extraction were negative. All replicates at subsequent dilutions for both extraction procedures were positive, except 1/8 replicates (13%) for M extraction at 2.5 log10 copies/ml. Also the luminosity signals were lower after M extraction compared to Si extraction, especially for CMV-IC. For CMV and for CMV-IC, the signals were between 1.1- to 1.8-fold and 2.2- to 3.5-fold lower, respectively, as can be seen in Table 3.
FIG. 1.
(A) Quantitative results obtained for human CMV strain AD 169 by CMV PCR after extraction from EDTA-anticoagulated whole blood by Si extraction (closed symbols) and M extraction (open symbols). Error bars indicate the standard deviation of the log-transformed quantitative results. The table shows the number of positives for the eight replicates tested with the different dilutions of CMV. (B) Correlation between the quantitative results obtained by Si extraction and M extraction.
The correlation between quantitative results was in excellent accordance for both extraction procedures, as illustrated in Fig. 1B, with r = 0.9807, a slope of 1.1033, and an intercept of −0.0219. At 2.5, 3.5, and 4.5 log10 CMV copies/ml, the mean difference from the expected value, 0.1 (range, −0.2 to 0.4) log10 copy/ml, was well within the accepted range (±0.5 log10 copy/ml) for Si extraction. For M extraction, the mean difference was greater, with approximately 0.4 (range, 0.2 to 0.9) log10 copy/ml, but still acceptable. At the lowest input of 1.5 log10 copies/ml, Si extraction overestimated the expected CMV DNA load by 1.0 log10 copy/ml, which is significantly outside the acceptable range of ±0.5 log10 copy/ml. This overestimation is most likely caused by the Poisson distribution of CMV near the limit of detection of the assay, resulting in inconsistent positive and negative reactions and therefore overestimation of the CMV DNA load in the portion of positive reaction mixtures.
Effect of impaired DNA recovery in clinical whole blood samples.
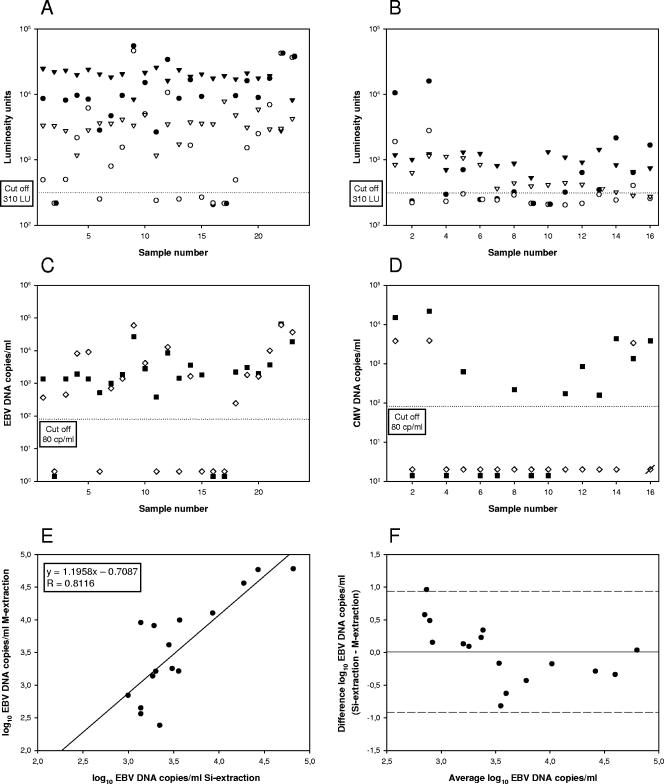
To assess the effect of impaired DNA recovery of M extraction in clinical samples, whole blood samples were analyzed by PCR in parallel for EBV (n = 23) and CMV (n = 16) after M extraction and Si extraction in the presence of 200 EBV-IC or 70 CMV-IC molecules. PCR and quantitation results are shown in Fig. 2. Figure 2A shows that for 17 (Fig. 2A, samples 1, 3 to 8, 10 to 15, and 18 to 21) of the 23 samples (74%), both EBV and EBV-IC signals were lower after M extraction compared to Si extraction. Another five samples (Fig. 2A, samples 2, 9, 16, 17, and 23) (22%) had comparable EBV signals but lower EBV-IC signals. Only one sample (Fig. 2A, sample 22) (4%) showed comparable results in both EBV and EBV-IC signals with both extraction methods. For CMV, 8 of the 16 samples (Fig. 2B, samples 1, 5, and 11 to 16) (50%) showed lower signals for both CMV and CMV-IC after M extraction (Fig. 2B). Another seven samples (Fig. 2B, samples 2, 4, and 6 to 10) (44%) showed comparable signals for CMV but lower signals for CMV-IC. One sample (Fig. 2B, sample 3) (6%) showed a comparable CMV-IC signal but yielded a lower CMV signal after M extraction. In addition, M extraction yielded four and six false-negative results for EBV- and CMV-positive specimens with viral DNA loads ranging from 2.4 to 3.4 log10 copies/ml, respectively (Fig. 2C, samples 6, 11, 13, and 15, and D, samples 5, 8, and 11 to 14). The EBV and CMV status of these samples had been confirmed by a reference PCR that was conducted a day earlier (data not shown). One weakly CMV-positive whole blood sample with a viral DNA load of approximately 2.0 log10 copies/ml, as judged by the reference PCR, yielded negative results with both methods (Fig. 2D, sample 4). This specimen's viral DNA load was near the limit of detection for the assay (1.9 log10 copies/ml). For the EBV- and CMV-positive specimens, the luminosity signals obtained after M extraction were, on average, 9.4-fold (range, 1.0-fold to 33.5-fold) and 4.8-fold (range, 2.2-fold to 6.2-fold) lower compared to the signals obtained after Si extraction (Table 3). For EBV-IC and CMV-IC, respectively, the luminosity signals were 7.2-fold (range, 0.9-fold to 26.9-fold) and 3.6-fold (range, 1.7-fold to 8.8-fold) lower after M extraction compared to Si extraction (Table 3). Although M extraction resulted in lower luminosity signals, this did not seem to significantly influence the viral load calculation for EBV, resulting in a high correlation between the quantitation results for EBV by both methods. This is shown in Fig. 2E with an overall correlation of 0.8116, a slope of 1.1958, and an intercept of −0.7087. The agreement between viral load values for the specimens processed by both methods was measured by determining the differences in log10 DNA loads for each sample (i.e., the loads obtained by Si extraction minus the loads obtained by M extraction) and calculating the mean and standard deviation of the differences. The values were in good agreement, with a mean log10 difference of 0.01 and a standard deviation of 0.463. The 95% confidence interval (±2 standard deviations) for the mean difference was 0.94 and −0.92 log10. A plot of the difference versus the average log10 viral load for each sample is shown in Fig. 2F. Furthermore, for the majority (75%) of the EBV-positive specimens, the difference between the two methods was within the ±0.5 log10 copy/ml range (Fig. 2F). For CMV, only three samples were available for comparison of the quantitation results between the two extraction procedures. These three samples, Fig. 2D, samples 1, 3, and 15, showed differences of 0.60, 0.75, and −0.43 log10 copy/ml, respectively.
FIG. 2.
PCR signals in luminosity units obtained for wild-type (circles) and IC (triangles) DNAs after processing with Si extraction (closed symbols) and M extraction (open symbols) for EBV (A) and CMV (B). The cutoff value of 310 luminosity units (LU) is indicated by the horizontal dotted line. Quantitative results for EBV (C) and CMV (D) after Si extraction (closed squares) or M extraction (open diamonds). A diagonal rod through the diamond symbol shows one invalid result for M extraction. The correlation (E) and mean difference (F) between quantitative results for EBV after Si extraction and M extraction are also shown. The solid line (F) represents the mean difference (0.01 log10 copy of EBV DNA/ml) between the quantitative results for Si extraction and M extraction. The dashed lines (F) represent the upper and lower 95% agreement lines (mean ± 2 standard deviations [0.94 and −0.92 log10 copy of EBV DNA/ml]).
DISCUSSION
We have evaluated the performance of M extraction as an automated NA extraction system, aiming to replace the manual Si extraction procedure that we currently use for several applications. DNA recovery experiments with HindIII-digested phage lambda DNA and HaeIII-digested φX174 DNA showed substantially lower DNA yields for M extraction compared to Si extraction, with, on average, 6.6-fold lower recoveries for M extraction. A major problem identified with M extraction was the retrieval of DNA from the magnetic glass particles, where up to 60% of the DNA could not be retrieved. PCR signals of spiked PCR control DNAs for EBV and VZV were also between 1.9- and 14.2-fold lower after M extraction compared to Si extraction, also suggesting impaired DNA recovery. This resulted in a 5- to 10-fold reduction in PCR sensitivity when spiked CMV strain AD 169 was extracted from whole blood by M extraction. Furthermore, it also resulted in false-negative results with 10 (4 EBV, 6 CMV) of the 39 clinical whole blood specimens tested. For manual Si extraction, high yields were observed with minimal loss of DNA during the procedure, which is in accordance with previous studies (2, 3, 5, 6, 8).
Although the experiments presented here do not exclude the contribution of PCR inhibition to the lower signals obtained after M extraction, no evidence for PCR inhibition was found in several experiments where eluates extracted from whole blood and TE buffer with M extraction were spiked with EBV-IC DNA and analyzed by EBV PCR. The signals obtained with the spiked eluates were in excellent accordance with the signals obtained from Si-extracted samples with the corresponding amount of EBV-IC DNA, showing that PCR inhibition did not contribute to the lower signals after M extraction (data not shown).
Several reports have described less efficient recovery of small RNA transcripts used as IC or quantification standard (QS) samples for (COBAS) AMPLICOR HCV 2.0 and HIV-1 MONITOR v1.5 tests (10, 11, 14, 20). Therefore, the amount of these transcripts per specimen needed to be increased when applying M extraction to allow reliable quantification. Our data suggest that relatively small DNA molecules, like the ICs (3- to 4-kb plasmids) used in this study are not susceptible to the lower extraction efficiency that has been described for the small RNA transcripts. If the ICs used in this study had been extracted with lower efficiency compared to the larger viral chromosomal DNAs (125 to 230 kb) by M extraction, overestimation of the viral DNA loads would have occurred. However, quantification did not seem to be significantly influenced by the lower recovery of DNA by M extraction, as was shown by high correlations between quantitative results from the two methods. Furthermore, the differences in quantitative results for the majority of specimens were within the acceptable range of ±0.5 log10 copy/ml. This is important when monitoring disease progression in solid organ and stem cell transplant recipients, where both primary and reactivated CMV infections need to be detected and quantified quickly and reliably in order to start preemptive therapy (15, 18). For managing EBV-associated posttransplantation lymphoproliferative disease, it has also been suggested that EBV load measurements can contribute to preemptive therapy and therefore reliable quantification is necessary (17).
We consider that the problems we have identified in our study are structural for M extraction and not the result of a malfunctioning instrument or one specific lot of M extraction reagents. This conclusion is based on the observation that we obtained results similar to those presented here with multiple lots of M extraction reagents on two different MagNA Pure LC instruments at two different locations over a 4-year time period (data not shown). Furthermore, the impaired recovery of DNA seems to be a specific M extraction problem, since we did not observe substantially lower recoveries in preliminary experiments with other DNA extraction kits available for the MagNA Pure LC instrument (DNA I Isolation Kit, DNA III Isolation Kit Bacteria Fungi, Microbiology Kit MGRADE). However, the use of DNA- and RNA-specific isolation kits is a disadvantage when both DNA- and RNA-targeted assays have to be performed and limited material is available.
The clinical impact of the lower efficiency of M extraction does not seem to be widespread, as is shown by other investigators who were satisfied with the performance of M extraction (10, 11, 13, 14, 15, 16, 18, 20, 21). However, there are some applications were the reduced recovery might have clinical implications. For instance, molecular diagnosis of herpes simplex virus (HSV) encephalitis, with its severe morbidity and high mortality, requires optimal methods for demonstrating the presence of HSV DNA near the lower range of the observed concentrations (200 to 4 × 107 copies/ml) of HSV DNA in primary CSF specimens from HSV encephalitis patients (23). High sensitivity is also required for detecting bacterial meningitis, where bacterial counts of less than 1,000 CFU/ml have been observed in 15% of the cases in children (17). Furthermore, Schuurman et al. have also shown that a very sensitive assay (100 to 200 CFU/ml) utilizing Si extraction for sample preparation did not detect 14% of the culture-proven cases of bacterial meningitis due to the small numbers of bacteria present in these samples (24). Finally, bacterial loads have been reported as low as 1 to 10 CFU/ml in bacteremia (12) and therefore requiring maximal sensitivity when molecular detection methods are used.
To what extent the reduced recovery of M extraction may prove detrimental to an application is largely dependent upon the extraction method that it needs to replace. For instance, the manual extraction method provided with the (COBAS) AMPLICOR HCV, and HIV tests can readily be replaced with M extraction (10, 11, 14, 20), due to the fact that the NA are concentrated by M extraction compared to the manual procedure. This concentrating of the NA counteracts the effects of the reduced recovery and results in a sensitivity similar to the manual extraction method. However, compared with Si extraction, this advantage is largely redundant because the sample input/output ratios are identical for the two methods.
In conclusion, M extraction seems to have a structurally impaired DNA recovery, resulting in a loss of sensitivity in molecular diagnostic testing. Whether this reduced recovery is detrimental to the application in which M extraction is used is largely dependent on the method that is intended to be replaced by M extraction and the clinical application. We therefore recommend that the clinical consequences of the reduced recovery, especially when maximal sensitivity is required, should be considered before implementing M extraction in the routine work flow.
REFERENCES
- 1.Beld, M., M. R. Habibuw, S. P. H. Rebers, R. Boom, and H. W. Reesink. 2000. Evaluation of automated RNA extraction technology and a qualitative HCV assay for sensitivity and detection of HCV RNA in pool-screening systems. Transfusion 40:575-579. [DOI] [PubMed] [Google Scholar]
- 2.Boom, R., C. Sol, M. Beld, J. Weel, J. Goudsmit, and P. Wertheim-van Dillen. 1999. Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J. Clin. Microbiol. 37:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. A. Sol, T. Schuurman, A. van Breda, J. F. L. Weel, M. Beld, I. J. M. ten Berge, P. M. E. Wertheim-van Dillen, and M. D. de Jong. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J. Clin. Microbiol. 40:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom, R., C. Sol, J. Weel, Y. Gerrits, M de Boer, and P. Wertheim-van Dillen. 1999. A highly sensitive assay for the detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence. J. Clin. Microbiol. 37:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. Sol, J. Weel, K. Lettinga, Y. Gerrits, A. van Breda, and P. Wertheim-Van Dillen. 2000. Detection and quantitation of human cytomegalovirus DNA in faeces. J. Virol. Methods 84:1-14. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., G. Martinez, and A. Mulchandani. 2000. Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal. Biochem. 280:166-172. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, M. D., J. F. Weel, T. Schuurman, P. M. Wertheim-van Dillen, and R. Boom. 2000. Quantitation of varicella-zoster virus DNA in whole blood, plasma, and serum by PCR and electrochemiluminescence. J. Clin. Microbiol. 38:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhar, A. K., M. M. Roux, and K. R. Klimpel. 2001. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR Green chemistry. J. Clin. Microbiol. 39:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiebelkorn, K. R., B. G. Lee, C. E. Hill, A. M. Caliendo, and F. S. Nolte. 2002. Clinical evaluation of an automated nucleic isolation system. Clin. Chem. 48:1613-1615. [PubMed] [Google Scholar]
- 11.Germer, J. J., M. M. Lins, M. E. Jensen, W. S. Harmsen, D. M. Ilstrup, P. S. Mitchell, F. R. Cockerill III, and R. Patel. 2003. Evaluation of the MagNA Pure LC instrument for extraction of hepatitis C virus RNA for the COBAS AMPLICOR hepatitis C virus test, version 2.0. J. Clin. Microbiol. 41:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry, N. L., C. A. McLimans, A. J. Wright, R. L. Thompson, W. R. Wilson, and J. A. Washington II. 1983. Microbiological and clinical evaluation of the isolator lysis-centrifugation blood culture tube. J. Clin. Microbiol. 17:864-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalpoe, J. S., A. C. M. Kroes, M. D. de Jong, J. Schinkel, C. S. de Brouwer, M. F. C. Beersma, and E. C. J. Claas. 2004. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J. Clin. Microbiol. 42:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessler, H. H., A. M. K. Clarici, E. Stelzl, G. Mühlbauer, E. Daghofer, B. I. Santner, E. Marth, and R. E. Stauber. 2002. Fully automated detection of hepatitis C virus RNA in serum and whole-blood samples. Clin. Diagn. Lab. Immunol. 9:1385-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessler, H. H., G. Mühlbauer, E. Stelzl, E. Daghofer, B. I. Santner, and E. Marth. 2001. Fully automated nucleic acid extraction: MagNA Pure LC. Clin. Chem. 47:1124-1126. [PubMed] [Google Scholar]
- 16.Knepp, J. H., M. A. Geahr, M. S. Forman, and A. Valsmakis. 2003. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J. Clin. Microbiol. 41:3532-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Scolea, L. J., Jr., and D. Dryja. 1984. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J. Clin. Microbiol. 19:187-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leb, V., M. Stöcher, E. Valentine-Thon, G. Hölzl, H. Kessler, H. Stekel, and J. Berg. 2004. Fully automated internally controlled quantification of hepatitis B virus DNA by real-time PCR by use of the MagNA Pure LC and LightCycler instruments. J. Clin. Microbiol. 42:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Guteknust, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, B. G., K. R. Fiebelkorn, A. M. Caliendo, and F. S. Nolte. 2003. Development and verification of an automated sample processing protocol for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:2062-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammadi, T., H. W. Reesink, C.M.J.E. Vandenbroucke-Grauls, and P. H. M. Savelkoul. 2003. Optimization of real-time PCR assay for rapid and sensitive detection of eubacterial 16S ribosomal DNA in platelet concentrates. J. Clin. Microbiol. 41:4796-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, S., M. Zuckerman, and M. Smith. 2003. Real-time quantitative PCR of Epstein-Barr virus BZLF1 DNA using the LightCycler. J. Virol. Methods 109:227-233. [DOI] [PubMed] [Google Scholar]
- 23.Schloss, L., A. M. van Loon, P. Cinque, G. Cleator, J.-M. Echevarria, K. I. Falk, P. Klapper, J. Schirm, B. F. Vestergaard, H. Niesters, T. Popow-Kraupp, W. Quint, and A. Linde. 2003. An international external quality assessment of nucleic acid amplification of herpes simplex virus. J. Clin. Virol. 28:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Schuurman, T., R. F. de Boer, A. M. D. Kooistra-Smid, and A. A. van Zwet. 2004. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for the diagnosis of bacterial meningitis in a clinical setting. J. Clin. Microbiol. 42:734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]