Abstract
We compared a rapid slide latex agglutination test (LAT; Oxoid, Basingstoke, United Kingdom) that detects penicillin binding protein 2a (PBP2a) with MicroScan conventional panels (Dade Behring, West Sacramento, CA) for detection of oxacillin resistance in Staphylococcus aureus. The PBP2a LAT demonstrated 99% agreement with MicroScan oxacillin MIC results for 388 isolates of S. aureus. All 249 oxacillin-resistant isolates gave strong positive reactions in the LAT (100% sensitivity). Three of the 139 oxacillin-susceptible isolates were also strongly positive and one was weakly positive in the LAT (97.1% specificity). The three oxacillin-susceptible isolates with strongly positive reactions were further characterized. The mecA gene was detected in all three by PCR; one isolate was determined to be resistant to oxacillin by reference broth microdilution testing (MIC, 8 μg/ml), one isolate was inducibly resistant to oxacillin (MIC of 16 μg/ml after overnight induction), and one isolate remained susceptible regardless of the method used for testing. Sequence analysis of a 2.1-kb gene fragment of the mecA gene from the susceptible isolate revealed a one-base substitution at nucleotide position 1449 which results in a Met-to-Ile change for amino acid residue 483. This amino acid substitution has not been previously reported and may be associated with a change in the function of PBP2a resulting in oxacillin susceptibility. An additional 487 isolates were tested in parallel with the both the LAT and MicroScan panels using criteria in which only strong (3 to 4+) or repeatedly weak (1 to 2+) LAT reactions were considered positive, and the results showed 99.4% agreement. The PBP2a LAT provided rapid and reliable detection of oxacillin resistance and proved a useful adjunct to the phenotypic method. Both methods provided reliable detection of oxacillin-resistant S. aureus and facilitated the discovery of a novel, functionally impaired form of PBP2a.
Infections due to Staphylococcus aureus are associated with significant morbidity and mortality and represent a large burden on the healthcare system. Nosocomial and increasingly community-acquired strains of S. aureus often exhibit resistance to oxacillin. Infections with oxacillin-resistant S. aureus (ORSA) have poorer outcomes, longer hospitalizations, and increased costs compared to infections with oxacillin-susceptible S. aureus (3, 9, 12, 13, 16). In large part, this may be due to inappropriate or inadequate treatment, while other factors such as underlying patient condition and virulence of the organism may also contribute to the poor outcomes. Rapid detection of oxacillin resistance can facilitate early targeted treatment and offers an opportunity for improving patient outcomes. Also, many centers are interested in rapidly identifying patients infected or colonized with ORSA for isolation or selected presurgical decontamination.
Phenotypic oxacillin resistance can be detected by various techniques, no one of which is completely reliable, especially for heterogeneously resistant strains of S. aureus (18). Oxacillin resistance is primarily mediated by the production of penicillin binding protein 2a (PBP2a), which is encoded by the mecA gene. Because its production is under complex control by regulatory genes and rare mecA-negative resistance mechanisms exist, phenotypic or even genotypic evaluation is not completely straightforward (2, 7). Furthermore, genetic testing requires significant technical and financial resources that may be lacking in many clinical laboratories.
Rapid latex agglutination tests (LATs) for the mecA gene product, PBP2a, have been developed, modified, and tested in numerous studies (11, 14, 18, 22). They have generally shown excellent performance characteristics for the detection of oxacillin resistance compared with other reference methods. Here we report our experience with a commercial LAT in comparison with MicroScan conventional susceptibility panels and how it led to the discovery of a novel PBP2a mutation.
MATERIALS AND METHODS
Oxacillin MICs.
The MICs of oxacillin for all isolates of S. aureus were determined with MicroScan PC20 panels using the turbidity standard technique for preparation of inocula. The panels were read after 24 h of incubation with a WalkAway 96 SI instrument. Inocula for selected isolates were also prepared using the Prompt system (Dade Behring) according to the manufacturer's instructions. Clinical and Laboratory Standards Institute reference broth microdilution MICs were determined as part of the discrepancy analysis with and without oxacillin induction (15). Oxacillin induction studies were used to determine whether prior exposure to oxacillin could significantly increase the oxacillin MIC. A significant increase in the oxacillin MIC was an increase of greater than 2 doubling dilutions and an increase from the Clinical and Laboratory Standards Institute susceptible category (≤2 μg/ml) to the resistant category (≥4 μg/ml). These studies were done by using growth from around an oxacillin disk to prepare the inoculum for broth microdilution testing.
PBP2a LAT.
The LAT was performed according to the manufacturer's instructions on 18- to 24-h-old cultures of S. aureus grown on 5% sheep blood agar plates. A 1-μl inoculating loop was used to harvest a heaped loopful of organisms from the plate, and this growth was suspended in 4 drops of the extraction reagent, boiled for 3 min, and centrifuged after adding buffer reagent. Agglutination with the supernatant was assessed after 3 min of rocking and scored as follows: 1+, fine granulation against a milky background; 2+, small but definite clumps against a slightly milky background; 3+, large and small clumps against a clear background; 4+, large clumps against a very clear background. Any level of agglutination seen with the test but not the control latex particles was considered a positive test. All tests of samples with weakly positive (1 to 2+) agglutination reactions were repeated.
mecA PCR and DNA sequencing.
Genomic DNA was prepared using the QIAGEN DNeasy Tissue Kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions, with one modification, the addition of lysostaphin (Sigma-Aldrich, St. Louis, MO) to the cell lysis buffer at a final concentration of 30 μg/ml. The mecA gene fragment was amplified using a PCR with forward primer 5′CATATCGTGAGCAATGAACTGA3′ and reverse primer 5′AGCAACCATCGTTACGGATT3′. The PCR was carried out in a GeneAmp PCR system 9700 (PE Applied Biosystems, Foster City, CA) with thermal cycle parameters as follows: an initial denaturing step of 10 min at 95°C; 30 cycles of 15 s at 95°C, 30 s at 55°C, and 2 min 30 s at 72°C; and a final elongation step of 72°C for 7 min. The PCR products were purified prior to DNA sequencing using the QIAquick PCR purification kit (QIAGEN). The sequence of the 2.1-kb mecA gene fragment was determined using the CEQ DTCS Quick Start kit and the CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA). The following primers were used for DNA sequencing: MecAF1, 5′CATATCGTGAGCAATGAACTGA3′; MecAF2, 5′GAAGTTAGATTGGGATCATAG3′; MecAF3, 5′GAAGATGGCTATCGTGTCAC3′; MecAF4, 5′GGTAATATCGACTTAAAACAAG3′; MecAF5, 5′ACAAGATGATACCTTCGTTCCACTT3′; MecAR1, 5′AGCAACCATCGTTACGGATT3′; MecAR2, 5′GTCCGTAACCTGAATCAGC3′; MecAR3, 5′GCAGTACCTGAGCCATAATC3′; MecAR4, 5′GATACATTCTTTGGAACGATG3′.
Sequence data were compiled and edited using the DNASIS Max software (Hitachi Software Engineering Co., South San Francisco, CA).
Nucleotide sequence accession number.
The sequence determined in this study was deposited in the GenBank database and assigned accession no. AY786579.
RESULTS
A total of 388 consecutive clinical isolates of S. aureus collected between 10/7/03 and 4/30/04 were tested with both the PBP2a LAT and the MicroScan PC20 panel for resistance to oxacillin (Table 1). Two hundred forty-nine (64%) isolates were resistant to oxacillin based on the MIC results. All of the isolates for which the oxacillin MICs were >2 μg/ml gave strongly positive (3+ to 4+) reactions in the LAT. Three of the 139 isolates that were susceptible to oxacillin were also strongly positive, and 1 was repeatedly weakly positive (1+) in the LAT. An additional five oxacillin-susceptible isolates had weakly positive reactions (1+ to 2+), which upon repeat testing were negative. These isolates were considered LAT negative in the data analysis. The LAT and MicroScan MIC results demonstrated 99% agreement, and the LAT had a sensitivity of 100% and a specificity of 97.1% for detection of ORSA.
TABLE 1.
Verification of the PBP2a LAT for detection of oxacillin resistance in 388 isolates of S. aureus
MicroScan PC20 panels were inoculated using the turbidity standard method.
Strong (3 to 4+) or repeatedly weak (1 to 2+) reactions were considered positive; weak positives that failed to repeat were considered negative.
The three oxacillin-susceptible isolates that were strongly positive in the LAT were further characterized by retesting in MicroScan panels inoculated with the Prompt system, the mecA PCR, and reference broth microdilution testing with and without overnight induction. The results of the discrepant analysis are shown in Table 2. The mecA gene was present in all three isolates. Isolate 1 was phenotypically resistant to oxacillin when tested by the reference broth microdilution method with and without induction and by the MicroScan method when the Prompt system was used to prepare the inoculum. Isolate 2 required prior oxacillin induction or use of the Prompt system for inoculation of the MicroScan panel to show resistance to oxacillin. Isolate 3 remained susceptible to oxacillin regardless of the method used for susceptibility testing.
TABLE 2.
Characterization of S. aureus isolates with discrepant MicroScan oxacillin MIC and PBP2a LAT results
| Isolate no. | MicroScan MIC (μg/ml)
|
Result of:
|
MIC (μg/ml)
|
|||
|---|---|---|---|---|---|---|
| 1a | 2b | PBP2a LAT | mecA PCR | Reference | Induced | |
| 1 | 2 | >2 | 4+ | + | 8 | 32 |
| 2 | 1 | >2 | 3+ | + | 1 | 16 |
| 3 | ≤0.25 | 0.5 | 4+ | + | 0.25 | 0.5 |
Inoculum prepared by the turbidity standard method.
Inoculum prepared with the Prompt system.
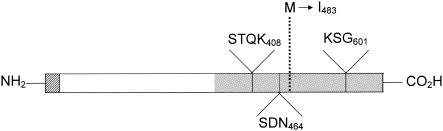
Both strands of a 2.1-kb mecA PCR amplification product from isolate 3 were sequenced. A one-base substitution was found at nucleotide position 1449 which results in a Met-to-Ile change for amino acid residue 483 (Fig. 1). This amino acid substitution in PBP2a has not been previously reported.
FIG. 1.
Diagram of the PBP2a protein indicating the amino acids in the active site of the transpeptidase domain and the amino acid substitution suspected of conferring oxacillin susceptibility. The hatched, white, and gray areas represent the transmembrane anchor, the non-penicillin binding domain, and the transpeptidase domain (penicillin binding domain), respectively. The solid lines indicate positions of conserved amino acids in the active site of the transpeptidase domain, and the dashed line indicates the position of the amino acid substitution found in the mecA-positive, oxacillin-susceptible isolate.
Based on the data presented above, we started using the PBP2a LAT routinely in our clinical microbiology laboratory for rapid detection of ORSA and asked the medical technologists performing the test to record the strength of the agglutination reactions on a scale of 1+ to 4+. We also changed from the turbidity standard to the Prompt method for inoculation of the MicroScan panels after our initial evaluation of the PBP2a LAT. We then monitored the agreement between the LAT and oxacillin MIC results for an additional 487 consecutive isolates tested from 5/1/04 to 9/30/04.
The in-use agreement between the test results was 97.9%, with a sensitivity of 100% and a specificity of 93.9% when any level of agglutination was considered positive. The results of the two tests showed 99.4% agreement when only 3 to 4+ or repeatedly 1 to 2+ agglutination reactions were called positive, with a sensitivity and a specificity of 99.1% and 100%, respectively (Table 3).
TABLE 3.
In-use correlation of PBP2a LAT and MicroScan oxacillin MIC results with different criteria for interpreting the PBP2a LAT results for 487 isolates of S. aureus
| Oxacillin MICa (μg/ml) | No. of PBP2a LAT-1b results
|
No. of PBP2a LAT-2c results
|
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| >2 | 324 | 0 | 321 | 3 |
| ≤2 | 10 | 153 | 0 | 163 |
Inoculum prepared with the Prompt system.
Any agglutination of test latex considered positive.
Only strong (3 to 4+) or repeatedly weak (1 to 2+) agglutination of test latex was considered positive.
DISCUSSION
Previous studies have shown that no phenotypic system, including reference broth microdilution, is completely reliable for detection of oxacillin resistance in S. aureus. This is particularly true for those strains that are heteroresistant (17, 18). Despite widespread use in clinical laboratories and regular modifications of the system, relatively little data on the performance of MicroScan panels for detection of oxacillin resistance in S. aureus have been published in the past decade. Farrell (4) reported that conventional and rapid MicroScan panels both had a sensitivity of 100% and a specificity of 92% for detection of oxacillin resistance in 335 isolates of S. aureus compared with a mecA gene PCR test. More recently, Swenson et al. (18) showed that MicroScan conventional panels had a sensitivity of 74% and a specificity of 100% for detection of oxacillin resistance in a challenge set of 55 S. aureus isolates enriched for difficult-to-detect heteroresistant strains. The rapid MicroScan panels were more sensitive (100%) but less specific (89%) for detection of oxacillin resistance than the conventional panels.
There have been numerous evaluations of the PBP2a LAT, with most using the presence of mecA to define ORSA (1, 8, 11, 19-22). All investigators have reported similar findings, with sensitivities of ≥97% and few, if any, false-positive LATs. The PBP2a LAT is a sensitive and specific method for detection of the mecA product.
In our evaluation of the PBP2a LAT, we found an agreement between the LAT and MicroScan panel oxacillin results of 99%. The LAT had a sensitivity of 100% and a specificity of 97% when a MicroScan oxacillin MIC of ≥2 μg/ml was used to define ORSA. The values are similar to the values reported in the previous evaluations.
The manufacturer recommends that any agglutination of the test reagent in the absence of agglutination of the control reagent should be considered a positive test. However, the package insert states that true positive results generally have strong reactions while false-positive reactions are usually limited to weak agglutination. We examined our data based on different criteria for interpretation of positive LATs. In our experience, the best combination of sensitivity and specificity for the LAT is obtained when only strong reactions (3 to 4+ agglutination) or repeatedly reactive weak reactions (1 to 2+ agglutination) are considered positive.
The sensitivity of the LAT for detection of ORSA in our study was very high (99.1 to 100%). Other evaluations used oxacillin induction (21), extended agglutination times (20, 22), or a larger inoculum (11) to increase the sensitivity of the LAT without sacrificing specificity. In our experience, such measures were not required to achieve high sensitivity.
Apparent false-positive LAT results were found with four isolates. Three of the four isolates were investigated further. All contained mecA, one was phenotypically resistant by reference broth microdilution methods, one required induction with oxacillin to express resistance, and one remained phenotypically susceptible regardless of the MIC testing method used. The first two isolates were most likely heterogeneously resistant to oxacillin since the larger inoculum delivered by the Prompt system enhanced the expression of resistance in the MicroScan system and oxacillin induction significantly increased the MICs determined by the reference broth microdilution method.
The third isolate was positive in the LAT, suggesting that mecA is expressed, the PBP2a protein is incorporated into the cell wall, and the monoclonal antibody binding sites are intact. The mecA gene of this isolate was sequenced in an attempt to provide an explanation for its phenotypic susceptibility to oxacillin. We found a previously undescribed M483I amino acid substitution in PBP2a.
S. aureus typically has four penicillin binding proteins (5). These proteins are enzymes that catalyze a transpeptidase reaction which is necessary for cell wall synthesis. The antimicrobial activity of oxacillin is based upon the ability of the drug to form a covalent bond with penicillin binding proteins at the active site and thereby inhibit the transpeptidase reaction. Wild-type PBP2a confers resistance to oxacillin through decreased binding efficiency, so the protein can still catalyze the transpeptidase reaction and compensate for inactivation of the other proteins (2). The M483I change in PBP2a could result in oxacillin susceptibility if the amino acid substitution resulted in either increased binding to oxacillin or elimination of PBP2a transpeptidase activity.
The methionine residue at position 483 is located in the transpeptidase domain of PBP2a (amino acid residues 342 to 668) (10). This domain contains three amino acid motifs that are conserved in all penicillin binding proteins (Fig. 1) (6), and the conserved amino acids are found in the enzymatically active site of PBP2a. Although Met 483 is not located in the active site, it is possible that this amino acid is important for the protein's secondary or tertiary structure and affects the protein's conformation at the active site. Additional studies are necessary to determine the effect M483I has on PBP2a structure and function.
We have not proven that the M483I amino acid substitution is responsible for the oxacillin-susceptible phenotype of the isolate described here. However, an amino acid substitution that either changes or eliminates the function of PBP2a could result in oxacillin susceptibility.
In conclusion, we demonstrated excellent agreement between the results of the PBP2a LAT and phenotypic expression of oxacillin resistance as detected by MicroScan conventional panels. We also identified further refinements for both tests to provide better discrimination between oxacillin-sensitive S. aureus and ORSA. Our discovery of an oxacillin-susceptible isolate with the mecA gene and expressed gene product has obvious ramifications for clinical laboratories performing mecA PCR and PBP2a testing and serves to underscore the observation that no one testing strategy is completely reliable for detection of ORSA. Further studies are under way to define the effects of the M483I amino acid substitution on PBP2a structure and function.
REFERENCES
- 1.Cavassini, M., A. Wenger, K. Jaton, D. S. Blanc, and J. Bille. 1999. Evaluation of MRSA-screen, a simple anti-PBP 2a slide latex agglutination kit, for rapid detection and methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:1591-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, D. J. 1997. The reliability of MicroScan conventional and rapid panels to identify Staphylococcus aureus and detect methicillin resistance: an evaluation using the tube coagulase test and mecA PCR. Pathology 29:406-410. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou, N. H., B. A. Dix, and Y. R. Mauriz. 1986. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob. Agents Chemother. 29:333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microb. Mol. Biol. Rev. 66:702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackbarth, C., C. Miick, and H. F. Chambers. 1994. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2568-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jafri, A. K., B. S. Reisner, and G. L. Woods. 2000. Evaluation of a latex agglutination assay for rapid detection of oxacillin resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 36:57-59. [DOI] [PubMed] [Google Scholar]
- 9.Kopp, B. J., D. E. Nix, and E. P. Armstrong. 2004. Clinical and economic analysis of methicillin-susceptible and -resistant Staphylococcus aureus infections. Ann. Pharmacother. 38:1377-1382. [DOI] [PubMed] [Google Scholar]
- 10.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 11.Louie, L., S. O. Matsumura, E. Choi, M. Louie, and A. E. Simor. 2000. Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 38:2170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mekontso-Dessap, A., M. Kirsch, C. Brun-Buisson, and D. Loisance. 2001. Poststernotomy mediastinitis due to Staphylococcus aureus: comparison of methicillin-resistant and methicillin-susceptible cases. Clin. Infect. Dis. 32:877-883. [DOI] [PubMed] [Google Scholar]
- 13.Melzer, M., S. J. Eykyn, W. R. Gransden, and S. Chinn. 2003. Is methicillin-resistant Staphylococcus aureus more virulent than methicillin-susceptible S. aureus? A comparative cohort study of British patients with nosocomial infection and bacteremia. Clin. Infect. Dis. 37:1453-1460. [DOI] [PubMed] [Google Scholar]
- 14.Nakatomi, Y., and J. Sugiyama. 1998. A rapid latex agglutination assay for the detection of penicillin-binding protein 2′. Microbiol. Immunol. 42:739-743. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests of bacteria that grow aerobically; approved standard-sixth edition. NCCLS document M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Selvey, L. A., M. Whitby, and B. Johnson. 2000. Nosocomial methicillin-resistant Staphylococcus aureus bacteremia: is it any worse than nosocomial methicillin-sensitive Staphylococcus aureus bacteremia? Infect. Control Hosp. Epidemiol. 21:645-648. [DOI] [PubMed] [Google Scholar]
- 17.Swenson, J. M., J. Spargo, F. C. Tenover, and M. J. Ferraro. 2001. Optimal inoculation methods and quality control for the NCCLS oxacillin agar screen test for detection of oxacillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 39:3781-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swenson, J. M., P. P. Williams, G. Killgore, C. M. O'Hara, and F. C. Tenover. 2001. Performance of eight methods, including two new rapid methods, for detection of oxacillin resistance in a challenge set of Staphylococcus aureus organisms. J. Clin. Microbiol. 39:3785-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udo, E. E., E. M. Mokadas, A. Al-Haddad, B. Mathew, L. E. Jacob, and S. C. Sanyal. 2000. Rapid detection of methicillin resistance in staphylococci using a slide latex agglutination kit. Int. J. Antimicrob. Agents 15:19-24. [DOI] [PubMed] [Google Scholar]
- 20.van Griethuysen, A., M. Pouw, N. van Leeuwen, M. Heck, P. Willemse, A. Buiting, and J. Kluytmans. 1999. Rapid slide latex agglutination test for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leeuwen, W. B., C. van Pelt, A. Luijendijk, H. A. Verbrugh, and W. H. Goessens. 1999. Rapid detection of methicillin resistance in Staphylococcus aureus isolates by the MRSA-screen latex agglutination test. J. Clin. Microbiol. 37:3029-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazumi, T., S. A. Marshall, W. W. Wilke, D. J. Diekema, M. A. Pfaller, and R. N. Jones. 2001. Comparison of the Vitek gram-positive susceptibility 106 card and the MRSA-screen latex agglutination test for determining oxacillin resistance in clinical bloodstream isolates of Staphylococcus aureus. J. Clin. Microbiol. 39:53-56. [DOI] [PMC free article] [PubMed] [Google Scholar]