Abstract
The phenotypic and genotypic characteristics of 25 Streptococcus porcinus isolates recovered from human sources were investigated and compared to the characteristics of 17 reference strains obtained from nonhuman sources. All of the S. porcinus isolates were beta-hemolytic (wide zones), susceptible to vancomycin, gave positive results for the leucine aminopeptidase and l-pyrrolidonylarylamidase tests, and produced acids from mannitol and sorbitol. Most of them were positive for the CAMP test and resistant to bacitracin. The isolates were susceptible to most of the 14 antimicrobials tested, except for tetracycline, for which 80% of the human isolates and 35.2% of the nonhuman strains were resistant. The tet(M) and the tet(O) genes were detected in 23 (88.5%) and 8 (30.8%) of the 26 tetracycline-resistant isolates, respectively. Analysis of whole-cell protein profiles obtained after sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a high similarity among the profiles. Chromosomal DNA was analyzed by pulsed-field gel electrophoresis (PFGE) after digestion with SmaI and by random(ly) amplified polymorphic DNA (RAPD)-PCR using primer 1254. Analysis of SmaI-restricted genomic DNA revealed the substantial genetic diversity among S. porcinus isolates from nonhuman sources, which were also serologically more diverse. Most of the human isolates belonged to serogroup NG1 and shared highly related PFGE profiles that were distinct from profiles of isolates from nonhuman sources. These results were in agreement with those obtained by analysis of amplicons after RAPD-PCR, indicating the potential ability of these techniques for typing S. porcinus and suggesting the occurrence of a few clonal groups of S. porcinus strains adapted to the human host.
The species Streptococcus porcinus was proposed in 1984 (4) to accommodate physiologically related beta-hemolytic streptococci belonging to Lancefield serological groups E, P, U, and V and three additional experimental antigenic groups (C1, 5916T, and 7155A). Comparative analysis of 16S rRNA sequences of members of the genus Streptococcus revealed the close relationship among S. porcinus and other beta-hemolytic streptococci, such as those belonging to groups A, B, and C, allowing the location of these species in the pyogenic group (2, 14).
Members of this species are commonly associated with pyogenic infections in swine, such as cervical lymphadenitis, leading to edematous lymph nodes and hemorrhagic abscesses, pneumonia, and sepsis (4, 29). The frequency of these infections ranges from 3 to 6% among swine hosts submitted to slaughtering (12). In addition, S. porcinus strains have also been isolated from other pathologic conditions of swine, in association with abortion and endocarditis cases (10, 13, 21), as well as from healthy swine and, more rarely, from other hosts (29).
Isolation of S. porcinus strains from human sources is rarely documented, and infections in the human host have mostly been related to genitourinary tract, especially women in the reproductive age (6). However, the involvement of these microorganisms as a cause of infections in humans has possibly been hindered by misidentification of S. porcinus strains, mainly due to the biochemical similarities and serological cross-reactivity with Streptococcus agalactiae (Lancefield group B) strains, which are commonly associated with female genitourinary tract infections (6, 24).
In 1995, a study involving the identification of S. porcinus associated with human infections was published by Facklam et al. (6) reporting the phenotypic characteristics of 13 isolates obtained from clinical specimens, mostly from the genitourinary tracts of individuals living in the United States and Canada. The molecular properties of such isolates were not examined. In the following 3 years, a similar number of isolates from human sources was sent to the Centers for Disease Control and Prevention (CDC) for confirmation of the identification, possibly due to an increasing attention given to the potential role of S. porcinus as an etiologic agent of human infections.
The aim of the present study was to investigate the phenotypic and molecular characteristics of 25 S. porcinus isolates obtained from human sources and received for identification by the CDC, including those studied by Facklam et al. (6). Isolates obtained from nonhuman sources, mostly from swine, as well as the type strain of S. porcinus, were also included for comparative purposes.
MATERIALS AND METHODS
Bacterial strains.
Twenty-five S. porcinus isolates recovered from human sources were studied. The type strain of S. porcinus (SS-1029, ATCC 43138, NTCC 10999) and 16 other S. porcinus reference strains obtained from nonhuman sources, mostly from swines, were also included (Tables 1 and 2). The strains were taken from the culture collection of the Streptococcus Laboratory (CDC) or received from the health department laboratories in different locations. Cultures were pure, and the coisolation of other pathogens was not documented. Five strains of S. agalactiae (SS-617, SS-620, SS-700, SS-1073, and SS-1240) and two strains of S. pyogenes (SS-103 and SS-745) were included as controls for evaluation of several tests and methods.
TABLE 1.
S. porcinus strains from human sources included in this study
| Strain | Yr of isolation/location | Clinical source | Age (yr) | Sex | Serogroup | Tetr genea |
|---|---|---|---|---|---|---|
| 1499-84 | 1984/New York | Blood | NBb | Female | NG1 (C1) | tet(M) |
| 628-86 | 1986/Oregon | Wound | 20 | Male | P | tet(O) |
| 1217-87 | 1987/Louisiana | Placenta | 27 | Female | NG1 (C1) | tet(M) |
| 1451-92 | 1992/New York | Placenta | NRc | Female | NG1 (C1) | tet(M) |
| 638-93 | 1993/Canada | Cervix | 26 | Female | NG1 (C1) | S |
| 2144-93 | 1993/Maryland | Cervix | NR | Female | P | S |
| 219-94 | 1994/Georgia | Vagina | 16 | Female | NG1 (C1) | tet(M) |
| 369-94 | 1994/South Carolina | Placenta | 20 | Female | NG1 (C1) | tet(M) |
| 834-94 | 1994/Canada | Wound | 47 | Female | P | tet(M) |
| 1013-94 | 1994/Georgia | Urine | 34 | Female | NG1 (C1) | tet(M) |
| 1014-94 | 1994/Maryland | Genital tissue | NR | Female | NG1 (C1) | tet(M) |
| 345-95 | 1995/Florida | Placenta | 22 | Female | NG1 (C1) | tet(M) |
| 1256-95 | 1995/Florida | Wound | 41 | Male | NG1 (C1) | tet(M) |
| 1533-95 | 1995/Florida | Cervix | 22 | Female | NG1 (C1) | tet(O) |
| 2885-95 | 1995/New Jersey | Blood | 39 | Female | NG1 (C1) | tet(M) |
| 3676-95 | 1995/New York | Wound | NR | NR | NG1 (C1) | S |
| 4188-95 | 1995/Georgia | NR | NR | NR | NG1 (C1) | S |
| 4189-95 | 1995/Georgia | NR | NR | NR | NG1 (C1) | S |
| 663-96 | 1996/New Jersey | NR | 30 | Female | NG1 (C1) | tet(M) |
| 4524-96 | 1996/Georgia | Placenta | NR | Female | NG1 (C1) | tet(M) |
| 3123-97 | 1997/Philadelphia | Cervix | 25 | Female | NG1 (C1) | tet(M) |
| 3176-97 | 1997/Georgia | Umbilicus | NR | NR | NG1 (C1) | tet(M)/tet(O) |
| 790-98 | 1998/Alberta | Placenta | 33 | Female | NG1 (C1) | tet(M)/tet(O) |
| 3657-98 | 1998/London | Vagina | NR | Female | NG1 (C1) | tet(M)/tet(O) |
| 3658-98 | 1998/London | Placenta | NR | Female | NG1 (C1) | tet(M)/tet(O) |
That is, the tetracycline resistance gene. S, isolate susceptible to tetracycline.
NB, newborn.
NR, not reported.
TABLE 2.
S. porcinus strains from nonhuman sources included in this study
| Straina | Sender and location [identification code(s)] | Serogroup | Tetr geneb |
|---|---|---|---|
| SS-9 | R. Lancefield, Rockfeller University, United States (K129) | E | S |
| SS-558 | R. E. D. Williams, Colindale, England (NTCC 9824; 54/851) | P | S |
| SS-607 | R. Deibel, Cornell University, Ithaca, New York (NTCC 600) | E | S |
| SS-662 | J. Rotta, Czech Republic (42/59; K131) | E | S |
| SS-670 | J. Rotta, Czech Republic (27/58; 54/8) | P | S |
| SS-740 | T. Thal, Stockholm, Sweden (U10) | U | S |
| SS-841 | T. Thal, Stockholm, Sweden (U87) | U | tet(M) |
| SS-857 | Statens Seruminstitut, Capenhagen Denmark (1969/66; Kφd 70) | P | tet(M) |
| SS-995 | R. Shuman, Ames, Iowa (A1) | NG1 | S |
| SS-996 | R. Shuman, Ames, Iowa (C1) | NG1 | S |
| SS-997 | R. Shuman, Ames, Iowa (5916T) | NG2 | tet(M) |
| SS-998 | R. Shuman, Ames, Iowa (8923T) | NG2 | tet(M) |
| SS-1024 | R. Shuman, Ames, Iowa (A127) | V | tet(M)/tet(O) |
| SS-1029c | J. Jelínkova, Czech Republic (176) | V | tet(O) |
| SS-1057 | R. Shuman, Ames, Iowa (7155A) | NG3 | S |
| SS-1058 | R. Shuman, Ames, Iowa (8016A) | NG3 | S |
| SS-1124 | J. Jelínkova, Czech Republic (101-897) | V | S |
SS, standard strain.
See Table 1, footnote a.
Type strain (corresponds to ATCC 43138 and NCTC 10999).
Physiological characterization.
The isolates were tested for their phenotypic characteristics by conventional physiological tests as described previously (7, 19). The physiological characterization was also performed by using the Rapid ID 32 Strep system (bioMérieux, Marcy l'Étoile, France) according to the manufacturer's instructions. Tests were read after incubation at 37°C for 4 h. Serogrouping was performed by capillary precipitation tests or double-diffusion tests in agarose gels using antigen extracts obtained by the Lancefield hot-acid extraction procedure (7, 8) and antisera for groups E, P, U, and V and new group 1 (NG1 [also known as C1]), NG2, and NG3 prepared in our laboratories according to previously described procedures (9).
Antimicrobial susceptibility testing.
MICs were determined by a broth microdilution assay using PML panels (PML Microbiologicals, Wilsonville, Oreg.) according to the manufacturer's instructions. The following 14 antimicrobials were tested: amoxicillin, cefotaxime, cefuroxime, chloramphenicol, clindamycin, erythromycin, levofloxacin, meropenem, penicillin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, trovafloxacin, and vancomycin. The results were interpreted according to Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratories Standards) recommendations for Streptococcus other than Streptococcus pneumoniae (18). The MIC was defined as the lowest concentration of a given drug that inhibited growth as observed by the unaided eye.
Detection of tetracycline resistance genetic determinants.
Tetracycline-resistant isolates were screened for the presence of genes tet(K), tet(L), tet(M), and tet(O), according to the method of Trzcinski et al. (25). Briefly, each reaction was carried out in 50 μl of a mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 0.2 mM concentrations of each nucleotide, 0.5 μM concentrations of each primer, 2.5 U of Taq DNA polymerase, and 5 μl of DNA template. The primers used to amplify the tet determinants were tet(K) (5′-TAT TTT GGC TTT GTA TTC TTT CAT-3′ and 5′-GCT ATA CCT GTT CCC TCT GAT AA-3′), tet(L) (5′-ATA AAT TGT TTC GGG TCG GTA AT-3′ and 5′-AAC CAG CCA ACT AAT GAC AAT GAT-3′), tet(M) (5′-AGT TTT AGC TCA TGT TGA TG-3′ and 5′-TCC GCA TAT TTA GAC GAC GG-3′), and tet(O) (5′-AGC GTC AAA GGG GAA TCA CTA TCC-3′ and 5′-CGG CGG GGT TGG CAA ATA-3′). The PCR consisted of 35 cycles of 1 min at 95°C, 1 min at 50°C, and 1 min 30 s at 72°C, followed by a final 5 min at 72°C, except for tet(O), for which the annealing temperature was 55°C. PCR products were resolved by electrophoresis on 1.2% agarose gels in 0.5× TBE buffer (1 mM Tris, 0.01 M EDTA, 1 M boric acid). Gels were stained with ethidium bromide and then visualized and photographed under UV light. The size of each PCR product was estimated by using standard molecular weight markers (100-bp ladder; Pharmacia Biotech, Uppsala, Sweden).
Analysis of whole-cell protein profiles by SDS-PAGE.
Isolates were grown on Todd-Hewitt sheep blood agar plates at 37°C for 24 h. Preparation of extracts and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described by Merquior et al. (17). The profiles were initially compared by visual inspection. Densitometric analysis, normalization of the densitometric traces and interpolation of the protein profiles with the Pearson correlation coefficient and the UPGMA (for unweighted pair-group method with arithmetic averages) method were performed by using the Molecular Analyst Fingerprinting Plus software package (version 1.12) of the Image Analysis System (Bio-Rad Laboratories, Hercules, Calif.).
Analysis of DNA amplification products obtained by RAPD-PCR.
Isolates were grown in brain heart infusion broth at 37°C for 18 to 24 h. The DNA was extracted by boiling as previously described (20). Random(ly) amplified polymorphic DNA-PCR (RAPD-PCR) assays were carried out with primer 1254 (5′-CCGCAGCCAA-3′; synthesized by Gibco-BRL, Gaithersburg, Md.) according to the method of Pacheco et al. (20) with some modifications. The PCR mixture consisted of 2.5 mM MgCl2, 1× PCR buffer without MgCl2 (10 mM Tris-HCl, 50 mM KCl [pH 8.3]), 0.625 mM concentrations of each of the four deoxynucleoside triphosphates, 30 pmol of primer, 1 U of Taq DNA polymerase (Boehringer Mannheim), and 3 μl of bacterial lysate containing DNA in a total volume of 30 μl. Amplification was obtained with a GeneAmp PCR System 2400 (Perkin-Elmer, Branchburg, N.J.) under the following conditions: four cycles of 94°C for 5 min, 37°C for 5 min, and 72°C for 5 min, followed by 30 cycles of 94°C for 1 min, 37°C for 1 min, and 72°C for 2 min, with a final extension step at 72°C for 10 min. RAPD-PCR products were analyzed after electrophoresis on 1.8% agarose gels and staining with 0.5 μg of ethidium bromide/ml. A 100-bp DNA ladder (Gibco-BRL) was used as a molecular weight marker. The profiles derived from DNA amplification of bacterial isolates were analyzed and compared as performed for the analysis of whole-cell protein profiles after SDS-PAGE. The Dice coefficient, in conjunction with the application of the Pearson correlation coefficient, was used to calculate the similarities among the electrophoretic profiles in order to evaluate the best coefficient for evaluating the RAPD-PCR results.
Analysis of the chromosomal DNA restriction profiles by PFGE.
Chromosomal DNA was prepared in agarose plugs and treated with the restriction endonuclease SmaI as previously recommended by Teixeira et al. (23). The fragments were separated by pulsed-field gel electrophoresis (PFGE) in 1.2% agarose gels in a CHEF-DRIII system (Bio-Rad), with pulse times increasing from 2 to 30s, over 22 h at 11°C, at a voltage gradient of 6 V/cm. The restriction profiles were analyzed and compared by using the Dice coefficient and clustering by the UPGMA by using the Molecular Analyst Fingerprinting Plus software package, version 1.12, of the Image Analysis System (Bio-Rad).
Discriminatory power of molecular typing techniques.
The discriminatory power of the RAPD-PCR and PFGE techniques for typing S. porcinus strains was evaluated by using the discrimination index as described by Hunter and Gaston (11) as given by the equation: D = 1 − [1/N (N − 1)] Σ nj (nj − 1), where D is the numerical index of discrimination, N is the total number of strains, and nj is the number of strains pertaining to the jth type. The discrimination indexes were obtained considering the comparison of the profiles by visual inspection and by the automated analysis referred to for each methodology.
RESULTS
Information related to clinical sources, sex and age of patients, and year and local of isolation of the S. porcinus isolates recovered from human sources included in the present investigation is indicated in Table 1. Of the 25 human isolates, 16 (64%) were associated with female genitourinary tract infections. The other sources were blood (two isolates), wounds (three isolates), and urine (one isolate). The clinical source of five isolates was not available. Nineteen (76%) of the strains were recovered from female patients, two (8%) were from male patients, and for four (16%) of the isolates the sex of the patient was not recorded.
After cultivation on sheep blood agar, most strains presented a wide zone of complete hemolysis surrounding the colonies. Strains SS-9, SS-607, SS-662, 834-94, 3657-98, and 3658-98 were slower-growing variants exhibiting evident growth on agar blood plates after 24 h of incubation if under an atmosphere containing 5% CO2.
The serological characteristics of the isolates are presented in Tables 1 and 2. Most of the isolates (22 isolates; 88%) obtained from human sources harbored the NG1 (C1) antigen, and 3 (12%) reacted with group P antigen, whereas the isolates from nonhuman sources had diverse group antigens.
All of the 42 strains included in the present study (comprising the 25 human isolates and the 17 reference strains) had the following characteristics. They were gram-positive, catalase-negative cocci; they produced leucine aminopeptidase (LAPase) and l-pyrrolidonylarylamidase (PYRase); they were susceptible to vancomycin; they were positive for arginine and esculin hydrolysis; and they produced acids from maltose, mannitol, ribose, sorbitol, sucrose, and trehalose. Most strains were resistant to bacitracin and positive in the CAMP test. All S. porcinus isolates had negative results for hydrolysis of esculin in the presence of bile; starch hydrolysis; production of pigment; production of gas in Lactobacillus deMan, Rogosa, and Sharpe broth; and motility and production of polysaccharides from both broth and agar containing 5% sucrose. They also failed to form acid from arabinose, inulin, raffinose, and sorbose. Variable results were observed for other tests as shown in Table 3.
TABLE 3.
Phenotypic characteristics of S. porcinus isolated from human and nonhuman sources as determined by conventional physiological testsa
| Physiologic characteristic | % Positive strains:
|
|
|---|---|---|
| From nonhuman sources | From human sources | |
| Growth in 6.5% NaCl broth | 76 | 100 |
| Growth at 10°C | 11 | 37 |
| Growth at 45°C | 17 | 31 |
| Utilization of piruvate | 82 | 52 |
| Hydrolysis of hippurate | 0 | 63 |
| Litmus milk (acidification/clot) | 100/41 | 94/26 |
| Acetoin productionb | 94 | 100 |
| Susceptibility to bacitracin | 0 | 4 |
| Production of acids from: | ||
| Glycerol | 29 | 38 |
| Lactose | 52 | 0 |
| Melibiose | 23 | 0 |
| MGPc | 29 | 28 |
All strains had positive results in the following tests: LAPase and PYRase activity; susceptibility to vancomycin, arginine, and esculin hydrolysis; and production of acids from maltose, mannitol, ribose, sorbitol, sucrose, and trehalose. All strains had negative results for hydrolysis of esculin in the presence of bile (BE reaction), starch hydrolysis, production of pigment, production of gas, motility, and production of acids from arabinose, inulin, raffinose, and sorbose.
Voges-Proskauer test.
MGP, methyl-α-d-glucopyranoside.
The physiologic characteristics obtained with the Rapid ID 32 Strep system are presented in Table 4. S. porcinus isolates were positive for the production of alanine-phenylalanine-proline arylamidase, alcaline phosphatase and β-glucuronidase and for the production of acids from maltose, sucrose, and trehalose. Negative results were observed for production of α-galactosidase, β-manosidase, and urease and for the production of acids from l-arabinose, d-arabitol, melezitose, and cyclodextrin. Only 6 (14%) of the 42 S. porcinus strains were correctly identified using the manufacturer's suggested numerical profile system. Most of the isolates (32 isolates; 76%) were identified as Streptococcus uberis biogroup I, 7% (three isolates) as S. agalactiae and 2% (one isolate) as Streptococcus dysgalactiae. On the other hand, the two strains of S. pyogenes and the five strains of S. agalactiae, used as controls, were correctly identified by the manufacturer's profile system.
TABLE 4.
Phenotypic characteristics of S. porcinus isolated from human and nonhuman sources, as determined by the RAPID ID 32 Strep system, and comparison with expected results and results of related species
| Phenotypic characteristic | % of positive resultsa
|
|||||
|---|---|---|---|---|---|---|
|
S. porcinus
|
S. agalactiae | S. pyogenes | ||||
| Strains from nonhuman sources | Strains from human sources | Total | Expected resultsb | |||
| Arginine dihydrolase | 94 | 100 | 100 | 100 | 100 | 100 |
| α-Galactosidase | 0 | 0 | 0 | 80 | 0 | 0 |
| Alanine-phenylalanine-proline-arylamidase | 100 | 100 | 100 | 100 | 100 | 100 |
| β-Glucosidase | 76 | 96 | 88 | 85 | 0 | 0 |
| β-Glucuronidase | 100 | 100 | 100 | 0 | 50 | 0 |
| Alcaline phosphatase | 100 | 100 | 100 | 100 | 100 | 100 |
| Glycyl-tryptophane arylamidase | 0 | 5 | 3 | 0 | 0 | 0 |
| N-Acetyl-β-glucosaminidase | 0 | 0 | 0 | 1 | 0 | 0 |
| Pyroglutamic acid arylamidase | 12 | 5 | 8 | 1 | 0 | 100 |
| Production of acids from: | ||||||
| Lactose | 76 | 72 | 73 | 30 | 50 | 50 |
| Maltose | 100 | 100 | 100 | 100 | 100 | 100 |
| Mannitol | 82 | 100 | 92 | 99 | 0 | 50 |
| Melebiose | 47 | 0 | 19 | 0 | 0 | 0 |
| Melezitose | 0 | 0 | 0 | 0 | 0 | 0 |
| Methyl-β-d-glucopyranoside | 64 | 28 | 42 | 50 | 100 | 50 |
| Pullulan | 82 | 100 | 92 | 74 | 100 | 0 |
| Raffinose | 5 | 8 | 7 | 0 | 0 | 0 |
| Ribose | 82 | 100 | 92 | 80 | 100 | 50 |
| Sucrose | 100 | 100 | 100 | 100 | 100 | 100 |
| Sorbitol | 76 | 100 | 90 | 74 | 0 | 0 |
| Tagatose | 76 | 0 | 30 | 0 | 0 | 0 |
| Trehalose | 100 | 100 | 100 | 100 | 100 | 100 |
| Acetoin production | 94 | 100 | 97 | 100 | 50 | 0 |
| Hydrolysis of: | ||||||
| Hippurate | 100 | 92 | 95 | 1 | 100 | 0 |
| Glycogen | 0 | 4 | 2 | 0 | 0 | 50 |
All strains presented negative results in tests for detecting the production of α-galactosidase, β-galactosidase, β-mannosidase, N-acetyl-β-glucosaminidase, and urease and the production of acids from cyclodextrin, d-arabitol, and l-arabinose.
Results indicated by the manufacturer for the identification of S. porcinus.
The results of antimicrobial susceptibility tests are shown in Table 5. Most of the 42 S. porcinus isolates were susceptible to all antimicrobials tested, except for 26 (61.9%) strains that were resistant to tetracycline. The majority (20 isolates [76.9%]) of the tetracycline-resistant strains (MIC of >8 μg/ml) were isolated from human sources. Considering each subpopulation according to the origin, resistance to tetracycline was found in 80% (20 of 25) of the isolates recovered from human sources and in 35.2% (6 of 17) of those from nonhuman sources. PCR testing allowed the detection of the tet(M) gene and the tet(O) gene in 23 (88.5%) and 8 (30.8%) of the 26 tetracycline-resistant isolates, respectively. Both tet(M) and tet(O) genes were simultaneously found in 5 (19.2%) strains (Tables 1 and 2). None of the isolates carried the two additional genes tested [tet(K) and tet(L)]. The tet(M) and tet(O) genes were exclusively found in 14 (70%) and 2 (10%) of the 20 tetracycline-resistant isolates obtained from human sources, respectively, whereas both genes were simultaneously found in 4 (20%) of these isolates. Among the six tetracycline-resistant isolates obtained from nonhuman sources, four (66.6%) and one (16.6%) isolates carried the tet(M) gene and the tet(O), respectively, and one (16.6%) carried both genes.
TABLE 5.
MICs of different antimicrobial agents for S. porcinus strains isolated from human and nonhuman sources
| Antimicrobial agent | Source | MIC (μg/ml)a
|
|||
|---|---|---|---|---|---|
| Range | Mode | MIC50 | MIC90 | ||
| Amoxicillin | H | <0.03-0.06 | <0.03 | <0.03 | 0.06 |
| NH | <0.03-0.06 | 0.06 | 0.06 | 0.06 | |
| Cefotaxime | H | <0.06 | <0.06 | <0.06 | <0.06 |
| NH | <0.06 | <0.06 | <0.06 | <0.06 | |
| Cefuroxime | H | <0.12-0.25 | <0.12 | <0.12 | <0.12 |
| NH | <0.12-0.12 | <0.12 | <0.12 | <0.12 | |
| Chloramphenicol | H | <2-4 | 4 | 4 | 4 |
| NH | 4 | 4 | 4 | 4 | |
| Clindamycin | H | <0.6-0.25 | 0.12 | 0.12 | 0.12 |
| NH | 0.12-0.25 | 0.12 | 0.12 | 0.25 | |
| Erythromycin | H | <0.06-0.12 | 0.12 | 0.12 | 0.12 |
| NH | <0.06-0.12 | <0.06 | <0.06 | 0.12 | |
| Levofloxacin | H | <0.5-1 | 1 | 1 | 1 |
| NH | <0.5-1 | <0.5 | <0.5 | 1 | |
| Meropenem | H | <0.06-0.06 | <0.06 | <0.06 | 0.06 |
| NH | <0.06 | <0.06 | <0.06 | <0.06 | |
| Penicillin | H | 0.06 | 0.06 | 0.06 | 0.06 |
| NH | 0.06 | 0.06 | 0.06 | 0.06 | |
| Rifampin | H | <1 | <1 | <1 | <1 |
| NH | <1 | <1 | <1 | <1 | |
| Tetracycline | H | <1->8 | >8 | >8 | >8 |
| NH | <1->8 | <1 | <1 | >8 | |
| Trimethoprim-sulfamethoxazole | H | <0.12/2.38-0.25/4.75 | <0.12/2.38 | <0.12/2.38 | 0.25/4.75 |
| NH | <0.12/2.38-0.25/4.75 | 0.25/4.75 | 0.25/4.75 | 0.25/4.75 | |
| Trovafloxacin | H | <0.25 | <0.25 | <0.25 | <0.25 |
| NH | <0.25 | <0.25 | <0.25 | <0.25 | |
| Vancomycin | H | <0.12-0.5 | 0.5 | 0.5 | 0.5 |
| NH | 0.25-0.5 | 0.25 | 0.25 | 0.5 | |
MIC50, the MIC capable of preventing the growth of 50% of the strains; MIC90, the MIC capable of preventing the growth of 90% of the strains.
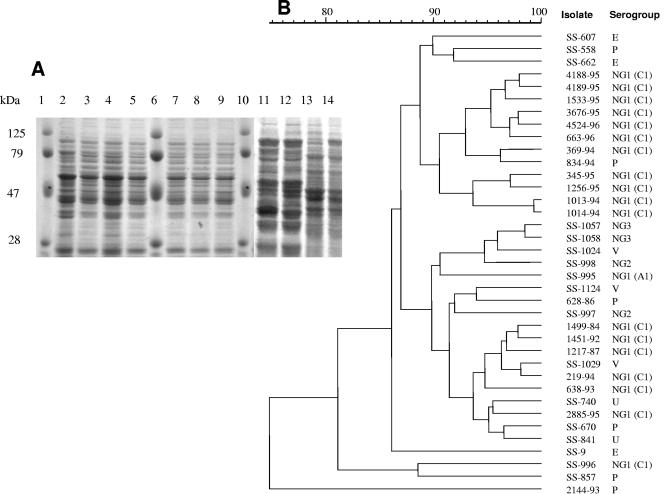
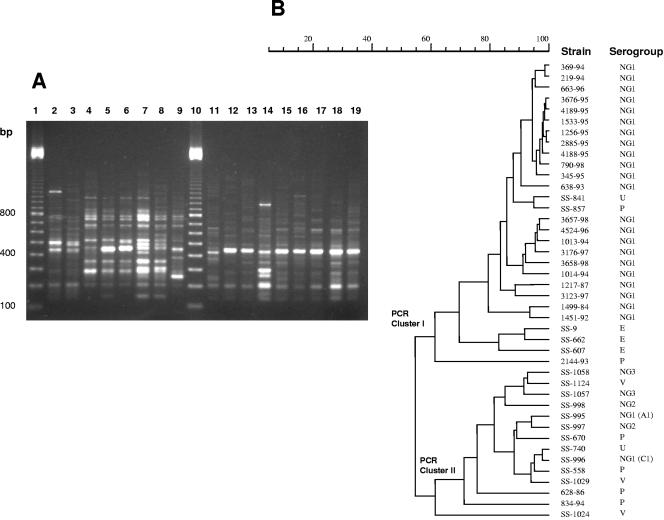
Analysis of whole-cell protein profiles revealed the high similarity among protein profiles of S. porcinus strains, which were different from profiles of the other streptococcal species tested (S. agalactiae and S. pyogenes) (Fig. 1A). Apart from quantitative differences, S. porcinus strains had virtually indistinguishable or highly related protein profiles, with percentages of similarity higher than 75% (Fig. 1B). Major differences were identified in the 30- to 100-kDa region. The use of primer 1254 for RAPD-PCR fingerprinting generated reproducible profiles composed of a number of bands (7 to 15) suitable for the determination of percentages of similarity (Fig. 2). The dendrogram obtained using the Pearson correlation coefficient (Fig. 2) presented a more suitable structure. Considering the data in the dendrogram, the 42 S. porcinus isolates were divided into electrophoretic types with similarities ranging from 50 to 98%. The dendrogram obtained from automated analysis showed two different clusters (I and II) defined by the comparison of similarities among the densitometric curves of the profiles (Fig. 2). All group NG1 (C1) human isolates were included in cluster I, whereas most of the isolates obtained nonhuman sources were included in cluster II. The percentage of similarity between these two major clusters was ca. 50%. Isolates belonging to cluster II (mostly from nonhuman sources) displayed higher diversities than the isolates included in cluster I.
FIG. 1.
(A) SDS-PAGE profiles of whole-cell protein extracts of representative strains of S. porcinus, S. agalactiae, and S. pyogenes. Lanes 1, 6, and 10, molecular size markers (in kilodaltons); lane 2, S. porcinus SS-1029; lane 3, S. porcinus 1533-95; lane 4, S. porcinus 2885-95; lane 5, S. porcinus 3676-95; lane 7, S. porcinus 4188-95; lane 8, S. porcinus 4189-95; lane 9, S. porcinus 663-96; lane 11, S. agalactiae SS-617; lane 12, S. agalactiae SS-1073; lane 13, S. pyogenes SS-103; lane 14, S. pyogenes SS-745. (B) Dendrogram resulting from computer-assisted analysis of protein profiles of S. porcinus strains included in the present study. UPGMA was used for clustering the correlation coefficients (expressed as percentages). The Pearson product moment correlation coefficient was used for calculating the percentage of similarities among the profiles. The scale represents the average percentage of similarity.
FIG. 2.
(A) RAPD-PCR profiles of representative S. porcinus isolates. Lanes 1 and 10, molecular size markers (100-bp ladder); lane 2, SS-9; lane 3, SS-662; lane 4, SS-670; lane 5, SS-841; lane 6, SS-857; lane 7, SS-997; lane 8, SS-998; lane 9, SS-1024, lane 11, 1217-87, lane 12, 1451-92; lane 13, 638-93; lane 14, 834-94; lane 15, 1013-94; lane 16, 345-95; lane 17, 4188-95; lane 18, 3176-97; and lane 19, 790-98. (B) Dendrogram resulting from computer-assisted analysis of the RAPD-PCR profiles of S. porcinus isolates. The Pearson product moment correlation coefficient was used for calculating the percentage of similarities among the profiles.
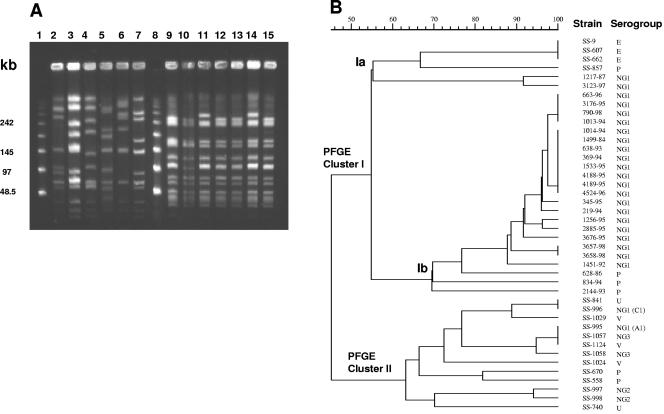
In a quite similar way, the genetic diversity among S. porcinus isolates was observed by using analysis of SmaI-restricted DNA after PFGE. Most of the human isolates had fragmentation profiles more closely related to each other than those of isolates obtained from nonhuman sources (Fig. 3). The majority of isolates harboring group antigen NG1 (C1) presented PFGE banding profiles with percentages of similarity higher than 90% (Fig. 3). The analysis of PFGE profiles generated a dendrogram which defined two separated clusters of strains (I and II) (Fig. 3). Cluster I could be divided in two subgroups of similarity (Ia and Ib). Isolates from nonhuman sources also presented levels of genetic diversity higher than those obtained from human clinical specimens. Interestingly, as seen for RAPD-PCR profiles, S. porcinus isolates of serogroup E clustered with higher similarity in comparison to clustering of PFGE restriction profiles of isolates belonging to other serogroups and separately from the rest of nonhuman strains. The dendrogram structure derived from the analysis of PFGE profiles was slightly different from that derived from analysis of RAPD-PCR profiles.
FIG. 3.
(A) PFGE profiles of chromosomal DNA of S. porcinus isolates after digestion with SmaI. Lanes 1 and 8, molecular size markers (in kilobases; lambda DNA concatemers ranging from 48.5 to 1,018.5 kb); lane 2, SS-1029; lane 3, SS-558; lane 4, SS-740; lane 5, SS-857; lane 6, SS-997; lane 7, SS-1058; lane 9, 1499-84; lane 10, 369-94; lane 11, 1013-94; lane 12, 4188-95; lane 13, 4189-95; lane 14, 663-96; and lane 15, 4524-96. (B) Dendrogram resulting from computer-assisted analysis of the PFGE profiles of S. porcinus isolates. The Dice coefficient was used for calculating the percentages of similarity among the profiles.
A comparison of the discrimination indexes obtained for the RAPD-PCR and PFGE typing systems is shown in Table 6.
TABLE 6.
Comparison of discriminatory power of the RAPD-PCR and PFGE techniques for typing S. porcinus strains
| Techniquea | No. of strains studied | No. of electrophoretic types | No. of strains belonging to the most common type | Discrimination index |
|---|---|---|---|---|
| RAPD-PCR | ||||
| VI | 42 | 8 | 22 | 0.663 |
| AA | 42 | 42 | 2 | 1.000 |
| PFGE | ||||
| VI | 42 | 16 | 19 | 0.787 |
| AA | 42 | 27 | 16 | 0.959 |
RAPD-PCR, polymorphism analysis of DNA by RAPD-PCR; VI, visual inspection; AA, automated analysis; PFGE, polymorphism analysis of DNA by PFGE.
DISCUSSION
The role of S. porcinus as an emerging pathogen affecting human beings or as a previously unrecognized etiological agent of human infections is unclear. The identification of this species in clinical specimens obtained from humans may not have been often observed for several reasons.
S. porcinus isolates may exhibit some characteristics that are similar to those of S. agalactiae or S. pyogenes. S. agalactiae isolates are beta-hemolytic (usually narrow zones), have positive results for the CAMP test, hydrolize arginine and hippurate, produce alkaline phosphatase, are resistant to bacitracin, and produce acids from ribose and trehalose. These microorganisms are known as a common etiological agent of genitourinary tract infections leading to intrauterine or postpartum complications and newborn diseases. Many healthy women asymptomatically carry S. agalactiae in this anatomic site and remain unaffected clinically (22). It is not known whether S. porcinus is carried asymptomatically or not. On the other hand, most S. porcinus strains show positive results for the PYR test, and a few may even be susceptible to bacitracin, resembling S. pyogenes strains, and this may be another explanation for possible misidentification.
The majority of the isolates included in the present study were recovered from clinical specimens related to the genitourinary tract and had characteristics that resemble S. agalactiae as described above. Physiologic characteristics that can help in distinguishing these two species include the wide zone of complete hemolysis produced by S. porcinus strains after cultivation on sheep blood agar, in contrast to the narrow zone produced by S. agalactiae; the production of acetoin observed in the Voges-Proskauer test; the production of PYRase; and the production of acids from mannitol and sorbitol. The inclusion of these tests in a routine basis for the identification of beta-hemolytic streptococci in clinical laboratories would be helpful for a more precise detection and evaluation of the occurrence of S. porcinus infections.
The use of a commercial system (the Rapid ID 32 Strep system) can also lead to misidentification of S. porcinus, as we observed in the present work. Most of the strains were identified as S. uberis. This system seems to be useful for the rapid identification of beta-hemolytic streptococci more commonly associated with human infections (mainly group A and group B streptococci). The limited performance of different systems for the identification of less frequently isolated human or veterinary pathogens has been previously reviewed (1, 28).
An evidence that supports the role of S. porcinus as a newly emerging pathogen is associated with the fact that this microorganism has only been isolated from human beings in the last two decades. Also, the description of this species, as well as the development of studies for a more detailed physiological characterization and definition of the phylogenetic position of S. porcinus, has only been published in a relatively recent period (2, 4, 6, 14). On the other hand, the apparent increase in the frequency of isolation of S. porcinus from human clinical sources may essentially be related to improvements in the identification. As already discussed, this microorganism may have previously been misidentified, since many microbiology laboratories only applied one or a few presumptive identification tests, such as the CAMP test, the PYR test, and evaluation of susceptibility to bacitracin, for the identification of beta-hemolytic streptococci from human sources. Furthermore, some S. porcinus strains cross-reacted with commercial group B serogrouping reagents, as noted earlier (24).
Most of the physiologic characteristics observed for the isolates included in the present study were similar to those described in previous reports (4, 6, 10, 15, 16, 29). However, differences were detected, the most striking being those related to the findings of Collins et al. (4) and Wessman (29), who described the S. porcinus strains as negative for the hippurate test and for production of PYRase. As observed in the present investigation, as well as in other studies (6, 10, 15, 16), S. porcinus strains can be positive for the hippurate test and for production of PYRase. We observed positive PYR test reactions for all S. porcinus strains tested, although some of the reactions were weak. The use of different methodologies for the phenotypic characterization may be a cause of the differences observed.
The only antimicrobial resistance marker found among strains included in the present study was resistance to tetracycline. Resistance was more common among isolates from human sources. Two different genes, tet(M) and tet(O), were found to be associated with tetracycline resistance in S. porcinus, with tet(M) being the predominant among both human and nonhuman isolates. To our knowledge, this is the first report of the presence of these genes in members of this bacterial species. The high prevalence of tetracycline resistance among S. porcinus isolates may be related to the presence of common genetic elements in chromosomal DNA of these microorganisms, such as transposons Tn916 and Tn3701, which code for resistance to tetracycline and are widely distributed among other members of the genus Streptococcus (3).
Analysis of whole-cell protein (WCP) profiles by a standardized method was shown to be a suitable tool for the differentiation of streptococcal species (26, 27). WCP profiles are considered to reflect excellent approximations of genome information, providing indirect analysis of genotypic characteristics (5). Our results indicated that analysis of WCP profiles constitute a relatively simple and a reproducible procedure for the characterization of S. porcinus strains and may be useful for differentiating them from physiologically related beta-hemolytic streptococcal species.
The human isolates included in the present study were obtained from patients living in different locations, and the periods of isolation were also diverse, suggesting the lack of common sources of transmission.
We successfully applied RAPD-PCR and PFGE analysis to investigate the genetic diversity of S. porcinus isolates from different sources. The results of both RAPD-PCR and PFGE analysis revealed a variety of profiles reflecting the substantial genetic diversity among isolates of this species and indicating the potential ability of these techniques for typing these microorganisms. RAPD-PCR analysis, with primer 1254, clearly revealed that most of the S. porcinus isolates from human sources were grouped in the same cluster. When automated analysis was applied by using the Pearson correlation coefficient, two main clusters (I and II) were identified, representing the majority of the isolates from human and nonhuman sources, respectively. The clustering into two groups of similarity was in agreement with the dendrogram structure obtained after analysis of the PFGE profiles, suggesting the lack of clonal relationship among S. porcinus isolates from different hosts. These findings suggest the existence of a few clonal groups of S. porcinus adapted to the human host.
The distinctive PFGE profiles obtained after digestion of chromosomal DNA with SmaI and by computer analysis generated a dendrogram with a more defined distribution of clusters. All of the isolates from human sources were grouped into cluster I and were distinct (48% of similarity) from most of the isolates obtained from nonhuman sources. The PFGE results were in agreement with those obtained by RAPD analysis. A higher similarity among S. porcinus isolates harboring group E antigen was also observed, suggesting a common ancestor for these isolates. Several distinct ancestors may explain the differences in clonality among S. porcinus isolates from nonhuman sources. Both the RAPD-PCR and the PFGE methodologies presented high values for discrimination index (≥0.90); however, PFGE showed higher accuracy for the distinction of clones. Although RAPD-PCR is less time-consuming, restriction endonuclease analysis by using PFGE for typing of S. porcinus provided profiles with a more homogeneous distribution and intensity of bands, leading to more clear-cut and easy-to-interpret data. These observations support the usefulness of these techniques for typing of S. porcinus isolates, although the dendrogram structures resulting from the use of different methods revealed distinct distribution of clusters.
In summary, although the number of isolates is still limited, the present report provides information on the largest collection of S. porcinus recovered from human sources examined to date, and it is the first to present a comparison of the molecular properties of this microorganism. The data indicate the high relationship among isolates from humans in contrast to a larger diversity among isolates probably not related to the zoonotic transmission of strains. Therefore, specific clones may be associated with infection in different hosts. We describe a variety of methods and techniques that can be helpful in the precise identification and characterization of S. porcinus. In addition to the three tests (susceptibility to bacitracin, production of PYRase, and CAMP factor) used in the routine conventional test scheme for the presumptive identification of beta-hemolytic streptococci, we recommend the inclusion of supplementary tests, such as acetoin production and the production of acids from mannitol and sorbitol, in order to differentiate S. porcinus from S. agalactiae and S. pyogenes. S. porcinus strains usually present positive results in all of these three tests, and S. agalactiae strains are negative. S. porcinus strains can be differentiated from S. pyogenes on the basis of usually positive results for the CAMP test and of negative results for susceptibility to bacitracin. The use of more accurate procedures to properly detect and characterize members of this species will clarify their role in both human and animal infections.
Acknowledgments
This study was supported in part by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Financiadora de Estudos e Projetos (FINEP), the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and the Ministério da Ciência e Tecnologia (MCT/PRONEX).
REFERENCES
- 1.Bascomb, S., and M. Manafi. 1998. Use of enzyme tests in characterization and identification of aerobic and facultatively anaerobic gram-positive cocci. Clin. Microbiol. Rev. 11:318-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 3.Bouguénec, C. L., G. de Cespédès, and T. Horaud. 1990. Presence of chromosomal elements resembling the composite structure Tn3710 in streptococci. J. Bacteriol. 172:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., J. A. E. Farrow, V. Katic, and O. Kandler. Taxonomic studies on streptococci of serological groups E, P, U, and V: description of Streptococcus porcinus sp., nov. Syst. Appl. Microbiol. 5:402-413.
- 5.Costas, M. 1990. Numerical analysis of sodium dodecyl-sulphate-polyacrylamide gel electrophoresis protein patterns for the classification, identification and typing of medically important bacteria. Electrophoresis 11:382-391. [DOI] [PubMed] [Google Scholar]
- 6.Facklam, R. R., J. Elliott, N. Pigott, and A. R. Franklin. 1995. Identification of Streptococcus porcinus from human sources. J. Clin. Microbiol. 33:385-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam, R. R., and J. A. Washington II. 1991. Streptococcus and related catalase-negative gram-positive cocci, p. 238-257. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 8.Garvey, J. S., N. E. Cremer, and D. H. Sussdorf. 1977. Gel diffusion, p. 313-327. In Methods in immunology: a laboratory test for instruction and research, 3rd ed. W. A. Benjamin, Inc., Boston, Mass.
- 9.Harrel, W. K., H. Ashworth, L. E. Britt, J. R. George, S. B. Gray, J. H. Green, H. Gross, and J. E. Johnson. 1978. Preparation of streptococci grouping antisera, p. 53-63. In Procedural manual for production of bacterial, fungal, and parasitic reagents. Center for Disease Control, Atlanta, Ga.
- 10.Hommez, J., L. A. Devriese, F. Castryck, and C. Miry. 1991. Beta-hemolytic streptococci from pigs: bacteriological diagnosis. J. Vet. Med. B 38:441-444. [DOI] [PubMed] [Google Scholar]
- 11.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsumi, M., Y. Kataoka, T. Takahashi, N. Kikuchi, and T. Hiramune. 1997. Bacterial isolation from slaughtered pigs associated with endocarditis, especially the isolation of Streptococcus suis. J. Vet. Med. Sci. 59:75-78. [DOI] [PubMed] [Google Scholar]
- 13.Katsumi, M., Y. Kataoka, T. Takahashi, N. Kikuchi, and T. Hiramune. 1998. Biochemical and serological examination of β-hemolytic streptococci isolated from slaughtered pigs. J. Vet. Med. Sci. 60:129-131. [DOI] [PubMed] [Google Scholar]
- 14.Kilian, M. 1998. Streptococcus and Lactobacillus, p. 633-667. In L. Collier, A. Balows, and M. Sussman (ed.), Topley and Wilson's microbiology and microbial infections, 9th ed., vol. 2. Edward Arnold, London, United Kingdom. [Google Scholar]
- 15.Lämmler, C., and K.-H. Bähr. 1996. Characterization of Streptococcus porcinus serogroup P isolated from an aborted fetus of a pig. Med. Sci. Res. 24:177-178. [Google Scholar]
- 16.Lämmler, C., N. Cirak, and J. Smola. 1998. Studies on biochemical and further characteristics of Streptococcus porcinus. J. Vet. Med. B 45:235-243. [DOI] [PubMed] [Google Scholar]
- 17.Merquior, V. L. C., J. M. Peralta, R. R. Facklam, and L. M. Teixeira. 1994. Analysis of electrophoretic whole-cell protein profiles as a tool for characterization of Enterococcus species. Curr. Microbiol. 28:149-153. [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing: ninth information supplement. Approved standard M100-S9. NCCLS, Wayne, Pa.
- 19.Oberhofer, T. R. 1986. Value of the l-pyrrolidonyl-β-naphtylamide hydrolysis test for identification of select gram-positive cocci. Diagn. Microbiol. Infect. Dis. 4:43-47. [DOI] [PubMed] [Google Scholar]
- 20.Pacheco, A. B. E., B. E. C. Guth, K. C. C. Soares, L. Nishimura, D. F. de Almeida, and L. C. S. Ferreira. 1997. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plagemann, O. 1988. Streptococcus porcinus: a cause of abortion in sows. J. Vet. Med. B 35:770-772. [PubMed] [Google Scholar]
- 22.Schuchat, A. 2001. Group B streptococcal disease: from trials and tribulations to triumph and trepidation. Clin. Infect. Dis. 33:751-756. [DOI] [PubMed] [Google Scholar]
- 23.Teixeira, L. M., M. G. S. Carvalho, V. L. C. Merquior, A. G. Steigerwalt, D. J. Brenner, and R. R. Facklam. 1997. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J. Clin. Microbiol. 35:2778-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, T., and R. R. Facklam. 1997. Cross-reactions of reagents from streptococcal grouping kits with Streptococcus porcinus. J. Clin. Microbiol. 35:1885-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 26.Vandamme, P., B. Pot, E. Falsen, K. Kerstersand L. A Devriese. 1996. Taxonomic study of Lancefield streptococcal groups C, G and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int. J. Syst. Bacteriol. 46:774-781. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme, P., U. Torck, E. Falsen, B. Pot, H. Goossens, and K. Kersters. 1998. Whole-cell protein electrophoretic analysis of viridans streptococci: evidence for heterogeneity among Streptococcus mitis biovars. Int. J. Syst. Bacteriol. 48:117-125. [DOI] [PubMed] [Google Scholar]
- 28.Watts, J. L., and R. J. Yancey, Jr. 1994. Identification of veterinary pathogens by use of commercial identification systems and new trends in antimicrobial susceptibility testing of veterinary pathogens. Clin. Microbiol. Rev. 7:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessman, G. E. 1986. Biology of the group E streptococci: a review. Vet. Microbiol. 12:297-328. [DOI] [PubMed] [Google Scholar]