Abstract
We compared real-time LightCycler and TaqMan assays and the GP5+/6+ PCR/enzyme immunoassay (EIA) to assess the human papillomavirus type 16 (HPV16) load in cervical scrape specimens. Both real-time PCR assays determined the HPV16 load in scrape specimens similarly. The level of agreement between these assays and the GP5+/6+ PCR/EIA was low (P = 0.004), suggesting that the latter method is not suited for quantifying HPV16 DNA.
It is now well established that a persistent infection with a high-risk human papillomavirus (hr-HPV) type is a necessary cause of cervical cancer (1, 7, 14). In addition, several studies have shown that an increased human papillomavirus (HPV) DNA load within a cervical smear specimen is associated with an increased risk of cervical intraepithelial neoplasia grade 3 (CIN3) and cervical carcinomas (3, 6, 11, 13). Therefore, viral load assessments as follow-ups of consensus hr-HPV tests may have implications for the identification of women among those with hr-HPV infections that have the highest risk of CIN3 lesions or cervical carcinomas (10). However, data on the possible clinical value of viral load are not consistent, which in part may reflect the use of different quantification assays (4, 8, 9).
Here, we compared a newly developed real-time PCR assay based on fluorescence resonance energy transfer (LightCycler; see Table 1) with a previously described real-time PCR assay based on fluorigenic 5′ nuclease chemistry (TaqMan) (13) and the semiquantitative consensus HPV GP5+/6+ PCR enzyme immunoassay (EIA) (12) for their abilities to assess the HPV16 load in cervical scrapings as a follow-up of GP5+/6+ PCR typing. To that end, a total of 109 cervical scrapings were randomly selected from a population-based screening study among scrape specimens that were HPV16 positive by GP5+/6+ PCR genotyping (12). These comprised 54 samples classified as normal and 55 classified as borderline or mild dyskaryosis (BMD). BMD equals atypical squamous cells (ASC) of undetermined significance/ASC-high-grade squamous intraepithelial lesion/ASC-low-grade squamous intraepithelial lesion, according to the Bethesda 2001 classification (2). The scrape specimens comprised 80 cases with single infections and 29 cases with multiple infections including HPV16. Thirteen randomly chosen GP5+/6+ PCR-negative cervical scrapings were also subjected to real-time PCR assays.
TABLE 1.
Primers, probes, and PCR specifications for the quantification of HPV16 and β-globin targets
| Assay (length of PCR amplicon [bp]) | Sequence (5′ → 3′)a | Position in genomeb | Cycling conditionsc |
|---|---|---|---|
| LightCycler HPV16 (158) | fw: GAGGAGGAGGATGAAATAGATGGT | 658-681 | den: 95°C/3 s |
| re: GCCCATTAACAGGTCTTCCAA | 816-796 | ann: 56°C/15 s | |
| dp: ACAAAAGGTTACAATATTGTAATGGGCTCT | 735-706 | elon: 72°C/9 s | |
| ap: CCGGTTCTGCTTGTCCAGCTGG | 703-682 | ||
| LightCycler β-globin (196) | fw: GAGCCATCTATTGCTTACATTTGC | 62122-62145 | den: 95°C/3 s |
| re: TTGGTCTCCTTAAACCTGTCTTGT | 62318-62295 | ann: 55°C/15 s | |
| dp: CCAGGGCCTCACCACCAACTTC | 62275-62254 | elon: 72°C/10 s | |
| ap: CCACGTTCACCTTGCCCCACAG | 62251-62230 | ||
| TaqMan HPV16 (139) | fw: CAGATACACAGCGGCTGGTTT | 5914-5934 | 13 |
| re: TGCATTTGCTGCATAAGCACTA | 6053-6032 | ||
| Probe: TGACCACGACCTACCTCAACACCTACACAGG | 5936-5966 | ||
| TaqMan β-globin (152) | fw: GAGCCATCTATTGCTTACATTTGC | 403-426 | 13 |
| re: TTGGTCTCCTTAAACCTGTCTTGT | 555-532 | ||
| Probe: TCTACCCTTGGACCCAGAGGTTCTTTGAGT | 471-501 | ||
| GP5+/6+ PCR (141) | fw: TTTGTTACTGTGGTAGATACTAC | 6624-6646 | 12 |
| re: CTTTTTATTGACATTTAGTATAAG | 6741-6764 | ||
| Probe: GTCATTATGTGCTGCCATATCTACTTCAGA | 6662-6691 |
fw, forward primer; re, reverse primer; dp, donor probe (3′ end labeled with fluorescein); ap, acceptor probe (3′ end phosphorylated, HPV16 5′ end labeled with LightCycler-Red-640 and β-globin 5′ end labeled with LightCycler-Red-705). TaqMan probes are at the 5′ end 6-carboxyfluorescein and at the 3′ end 6-carboxytetramethylrhodamine labeled; the EIA probe was 5′ end digoxigenin labeled.
Real-time PCR was performed on 100 ng target DNA using reagents the same as those described by Stevens et al. (10), except that 3.5 mM MgCl2 was used for HPV16. Details of primers, probes, and cycling determinants are given in Table 1. For HPV16 quantification, standard curves of 10-fold dilutions of pHPV16 plasmid DNA (ranging from 10 to 105 fg) spiked in 100 ng human placental DNA were used, whereas those of the β-globin gene contained only human placental DNA (ranging from 3 pg to 30 ng). Samples were tested in duplicate, and the values obtained were averaged. The real-time PCR assays determined the HPV16 load normalized per cell and per scrape specimen, whereas the GP5+/6+ PCR/EIA assayed the load per scrape specimen expressed as EIA optical density (OD) values after a 1-hour substrate incubation (5).
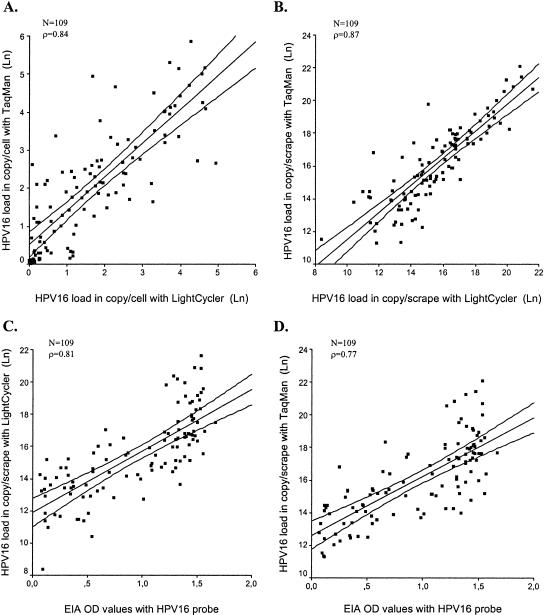
The median HPV16 loads per cell of the 109 cervical samples determined by the LightCycler and TaqMan assays were 4.0 copies/cell (range, 0.0 to 137.4) and 6.7 copies/cell (range, 0.0 to 346.2), respectively. The HPV16 copies/cell values measured by TaqMan were on average 1.6 times higher (95% confidence interval [CI], 1.3 to 2.1) than those measured by LightCycler. The correlation between both assays for determining HPV16 copies/cell was strong (Spearman ρ = 0.84; Fig. 1A) and their agreements were moderate (kappa value calculated after categorization of the variables in tertiles, 0.59). Correlation and agreement were not significantly influenced by the level of dyskaryosis (normal cytology or BMD) or the number of HPV types present in a scrape specimen. None of the 13 GP5+/6+ PCR-negative cervical scrape specimens revealed any HPV16 signal in both LightCycler and TaqMan assays, whereas all revealed β-globin gene PCR signals with both assays within the same range as that of the GP5+/6+ PCR-positive scrapings.
FIG. 1.
Correlation of HPV16 DNA loads as determined by different assays. (A) Correlation of TaqMan (y axis) and LightCycler (x axis) assays stratified for cellular input. (B) Correlation of TaqMan (y axis) and LightCycler (x axis) assays in assessing HPV16 load per scrape specimen. (C) GP5+/6+ PCR/EIA OD values (x axis) plotted against the LightCycler HPV16 load/scrape specimen (y axis). (D) GP5+/6+ PCR/EIA OD values (x axis) plotted against the TaqMan HPV16 load/scrape specimen (y axis). All values determined by real-time PCR are ln normalized in the plots. The inner line in each plot represents the regression line, whereas the outer lines indicate the 99% confidence interval of the regression line. Spearman correlation coefficients (ρ) are indicated in each plot.
Subsequently, the semiquantitative GP5+/6+ PCR/EIA was used to determine the HPV16 load per cervical scrape specimen. The median EIA OD value of the GP5+/6+ PCR after 1-hour substrate incubation was 1.17 (range, 0.07 to 1.67). The median numbers of calculated HPV16 copies/scrape specimen were 5.1 × 106 (range, 3.3 × 104 to 2.5 ×109) and 8.1 × 106 (range, 8.0 × 104 to 4.0 × 109) by LightCycler and TaqMan, respectively. The TaqMan assay determined the number of HPV16 copies/scrape specimen to be on average 1.7 times (95% CI, 1.3 to 2.2) higher than the LightCycler assay did. Conversely, the median amount of cells per scrape specimen as calculated by both techniques did not differ (mean of β-globin ratios, 1.00; 95% CI, 1.00 to 1.01). Correlation and agreement were strong (Spearman ρ = 0.87) and good (kappa value = 0.64), respectively, between the two real-time PCR assays in determining the HPV16 load/scrape specimen (Fig. 1B). These values decreased when comparing either of the two real-time PCR assays with the semiquantitative GP5+/6+ PCR/EIA (Fig. 1C and D). Correlation coefficients were 0.81 and 0.77 and kappa values were 0.53 and 0.50 for GP5+/6+ PCR/EIA values versus LightCycler and TaqMan values, respectively. The number of HPV types present in a scrape specimen or the degree of dyskaryosis did not significantly influence the correlations. By means of a regression model for standardized values, we tested for differential effects of TaqMan and GP5+/6+ PCR/EIA on LightCycler values and found a significantly higher coherence between the results for the different real-time PCR assays than between the results for GP5+/6+ PCR/EIA and the LightCycler assay (P = 0.004).
The fact that TaqMan scored the HPV16 load overall higher by a factor 1.6 to 1.7 than the LightCycler assay did could be due to many factors. However, variables such as HPV16 primer composition and the use of nonlinearized plasmids in the standard curve are most likely to be the underlying cause. Nevertheless, this difference was consistent throughout the whole detection range, making data obtained with these two methods still comparable when using a correction factor. Moreover, data collected so far support the concept that there exist differences in viral load values of several magnitudes between clinically relevant and irrelevant HPV infections (10, 13). Therefore, it is unlikely that a factor difference of 1.6 to 1.7 in HPV16 load measured by the two real-time assays would be of clinical relevance.
It should be realized that this comparative analysis was performed on a random, cross-sectional series of smear specimens with normal cytology or BMD. As a high viral load may be an important determinant of CIN3 lesions or cervical carcinomas (reviewed in reference 9), the choice of these samples could potentially have led to studying a range of viral copy numbers that fall beyond the level that would represent high-grade cervical lesions. However, preliminary HPV16, -18, -31, and -33 viral load analysis by LightCycler on cervical smear specimens of women having underlying high-grade CIN lesions revealed ranges of 9.2 × 104 to 5.3 × 108, 7.6 × 104 to 5.1 × 108, 2.4 × 104 to 9.4 × 108, and 2.0 × 106 to 8.5 × 108, respectively (C. J. Hogenwoning et al., unpublished results). These ranges correspond to the range of HPV16 copies/scrape specimen found by the LightCycler assay (i.e., 3.3 × 104 to 2.5 × 109) analyzed in this comparative study on smear specimens from women with as-yet-unknown clinical outcomes.
In summary, we demonstrated that the TaqMan and the LightCycler real-time PCR technologies produce similar results for the quantification of HPV16 DNA in cervical scrape specimens and that both are therefore useful in the detection of viral load in clinically relevant HPV infections. However, the GP5+/6+ PCR/EIA assay was not adequate for a precise assessment of the HPV load. Our data can be of value for future comparisons of studies involving viralload assessment in relation to the risk of CIN3 lesions or cervical carcinomas. In our setting, one such study is currently in progress.
REFERENCES
- 1.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulk, S., F. J. van Kemenade, L. Rozendaal, and C. J. Meijer. 2004. The Dutch CISOE-A framework for cytology reporting increases efficacy of screening upon standardisation since 1996. J. Clin. Pathol. 57:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuzick, J., G. Terry, L. Ho, T. Hollingworth, and M. Anderson. 1992. Human papillomavirus type 16 in cervical smears as predictor of high-grade cervical intraepithelial neoplasia [corrected]. Lancet 339:959-960. [DOI] [PubMed] [Google Scholar]
- 4.Gravitt, P. E., R. D. Burk, A. Lorincz, R. Herrero, A. Hildesheim, M. E. Sherman, M. C. Bratti, A. C. Rodriguez, K. J. Helzlsouer, and M. Schiffman. 2003. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol. Biomark. Prev. 12:477-484. [PubMed] [Google Scholar]
- 5.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Josefsson, A. M., P. K. Magnusson, N. Ylitalo, P. Sorensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. O. Adami, and U. B. Gyllensten. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355:2189-2193. [DOI] [PubMed] [Google Scholar]
- 7.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 8.Pretet, J. L., V. Dalstein, S. Monnier-Benoit, S. Delpeut, and C. Mougin. 2004. High risk HPV load estimated by Hybrid Capture II correlates with HPV16 load measured by real-time PCR in cervical smears of HPV16-infected women. J. Clin. Virol. 31:140-147. [DOI] [PubMed] [Google Scholar]
- 9.Snijders, P. J., A. J. van den Brule, and C. J. Meijer. 2003. The clinical relevance of human papillomavirus testing: relationship between analytical and clinical sensitivity. J. Pathol. 201:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Stevens, S. J., S. A. Verkuijlen, A. J. Brule, and J. M. Middeldorp. 2002. Comparison of quantitative competitive PCR with LightCycler-based PCR for measuring Epstein-Barr virus DNA load in clinical specimens. J. Clin. Microbiol. 40:3986-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swan, D. C., R. A. Tucker, G. Tortolero-Luna, M. F. Mitchell, L. Wideroff, E. R. Unger, R. A. Nisenbaum, W. C. Reeves, and J. P. Icenogle. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 37:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Duin, M., P. J. Snijders, H. F. Schrijnemakers, F. J. Voorhorst, L. Rozendaal, M. A. Nobbenhuis, A. J. van den Brule, R. H. Verheijen, T. J. Helmerhorst, and C. J. Meijer. 2002. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int. J. Cancer 98:590-595. [DOI] [PubMed] [Google Scholar]
- 14.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]