Abstract
Current HIV-1 genotyping assays were developed using subtype B viruses prevalent in Western countries. It is not clear whether these assays are appropriate for use among African patients, who are likely to be infected with non-B subtypes. We evaluated the Bayer TRUGENE HIV-1 genotyping (TG) assay using prospectively collected samples from HIV-1-infected individuals who acquired infection in either sub-Saharan Africa or the West (Europe, North America, and Australia). Plasma samples from 208 individuals with an HIV-1 viral load of >1,000 copies/ml were tested using version 1 primers supplied with the TG assay. If these failed, an alternative primer set version 1.5 was used. Of the 208 individuals, the likely origin of infection was Africa (n = 104), Western (n = 87) and “Others” (i.e., all other geographic locations or origin not certain; n = 17). Among the three groups, the version 1 primers were successful in 85 (82%), 77 (89%), and 13 (76%) individuals, respectively (P = 0.1). Of the remaining 32 samples, 30 were successfully amplified by using the version 1.5 primers. HIV-1 subtypes deduced from the reverse transcriptase sequences correlated with the likely origin of infection: Africa (28A, 3B, 33C, 13D, 6G, 4J, 2K, 5CRF01_AE, and 10CRF02_AG), Western (86B and 1K), and Others (1A and 16B). The success of the version 1 primers correlated with viral load (P < 0.014) and not with HIV-1 subtypes. A protocol based on version 1 primers, followed by 1.5 primers, was successful in sequencing 99% of the samples in this cohort.
Human immunodeficiency virus type 1 (HIV-1) is the major pathogen responsible for the worldwide pandemic of AIDS. There are significant genetic variations between viruses acquired from different geographic regions. Of the three main HIV-1 lineages—M (main), O (outlier), and N (non-M, non-O)-groups O and N are confined to West Africa, whereas group M accounts for the majority of the strains worldwide. However, even within group M, there are further subtypes, as well as numerous chimerical or recombinant forms, some of which are in common circulation (14). HIV-1 subtype B is the most well studied since it is the main subtype in Western countries. As a consequence, most of the research and diagnostic reagents currently in use were developed based on experience with subtype B (17). However, subtype B only accounts for 12% of the total disease burden globally. The non-B subtype itself is a very heterogeneous group, with subtypes C and A and the circulating recombinant forms CRF01_AE and CRF02_AG together accounting for 77% of the world's HIV infections (12).
With the advent of highly active antiretroviral therapy (HAART) and increased access to antiretroviral drugs even in resource poor countries, the development of drug resistance has become a significant problem. Studies evaluating the clinical utility of genotypic and phenotypic antiretroviral drug resistance testing have all pointed to the value and need for resistance testing as standard of care (1, 4, 5, 10, 11, 18). The U.S. Department of Health and Human Services, the International AIDS Society-USA, the European Guidelines group, and the British HIV Association all recommend the use of drug resistance assays for selection of new regimens after virologically confirmed drug failure. It is also recommended that resistance testing should be performed in cases of primary infection to detect transmitted resistance, in drug-naive patients starting therapy for the first time, in infected pregnant women and their babies, and also in the sources of high-risk body fluid exposures in order to guide the use of appropriate postexposure prophylaxis (8, 13, 16).
Sequence-based genotyping is the most widely used HIV-1 resistance assay. Many laboratories use in-house sequencing, but a number of commercial assays are also used widely. As with many other HIV diagnostic tests, resistance assays have been validated primarily in the West using samples from patients infected with HIV-1 subtype B. The TRUGENE genotyping kit (TG; Bayer HealthCare, Berkeley, CA) is a popular commercial genotyping assay used worldwide. It has been evaluated by using coded plasma samples from nine HIV-1 subtype B-infected patients in six independent laboratories and was found to have accuracies of 98.7% at the nucleotide level and 97.6% at the codon level (7). The TG assay was shown to have good concordance compared to other methods and performed well with non-B subtypes (6, 9). However, another study using a panel of selected samples with diverse genotypes found that the original version of the TG assay failed to produce a usable sequence in 12 of 24 non-B subtype samples (2).
In view of the conflicting evidence on the performance of the TG assay with non-B subtype of HIV-1, we undertook an evaluation to compare the outcome of performing routine resistance tests on African patients who were likely to be infected with HIV-1 non-B subtypes and Western patients with likely infection with subtype B virus.
(Presented in part at the 12th Conference on Retroviruses and Opportunistic Infections, 22 to 25 February 2005, Boston, Mass.)
MATERIALS AND METHODS
Study population and specimens.
Samples were tested from HIV-1-infected patients attending the HIV clinics of Guy's and St. Thomas' Hospital Trust in South London between August 2000 and April 2003. The ethnicity of the HIV patient population in South London consists of 45.7% White, 39.1% Black African, 5.7% Black Caribbean, and 9.4% others (3a). A high proportion of the HIV-1-infected Black African patients were recent immigrants from Africa. The standard of care during the study period was to perform a genotypic resistance test on patients taking HAART, with virologic evidence of treatment failure after initial treatment success or when there was a poor response to a newly introduced regimen. Genotyping was also indicated during the study period in cases of primary HIV-1 infection and among infected pregnant women. A policy of baseline resistance testing of new diagnoses or prior to starting of HAART was not initiated until after completion of this evaluation.
During the study period of 33 months, a genotypic resistance test was conducted on 208 plasma samples from 208 patients, all with an HIV-1 viral load of >1,000 copies/ml, as determined by the branched chain DNA assay (Bayer). The origin of infection for each patient was assessed by the requesting clinician based on the patient's country of origin, risk factors, and exposure history. Three main categories were identified as Africa; the West (which included Europe, North America, and Australia); and Others, which included all other geographic locations and those in whom the exact location of infection could not be determined.
TG assay.
All assay procedures were performed according to the manufacturer's instructions. This assay provides automated DNA sequencing of all 297 bp of the protease (PR) gene and the majority of the reverse transcriptase (RT) gene (741 bp) from a 1.3-kb fragment of copy DNA (cDNA) of the HIV-1 pol gene. Briefly, nucleic acid was extracted from plasma samples by using the QIAGEN viral RNA extraction kit (QIAGEN, Hilden, Germany). HIV-1 cDNA was produced by RT-PCR of the extracted RNA using the primer sets supplied by the manufacturer. The original TG kit provided a version 1 primer set. Population-based sequencing of the RT-PCR template was carried out by using bidirectional fluorophore-labeled primers (P2 for PR between codon 7 and 99, RT beginning for the codons 38 to 142 and RT middle for the codons 138 to 247 of RT) and dideoxy terminator cycle sequencing technology (Bayer). Consensus sequences were generated from each patient sample by using the Bayer OpenGene DNA sequencing software.
If the original TG version 1 primers failed to produce a usable sequence, the RT-PCR was repeated with an alternative primer set, version 1.5 primers, supplied by the same manufacturer. The version 1.5 primers (also known as CS1) target a consensus sequence within HIV-1 pol gene and were developed as an improvement to the version 1 primers. Generation of a usable sequence that covered PR codons 7 to 99 and RT codons 38 to 247 was considered a successful outcome. Samples that also failed with the version 1.5 primers were referred to the United Kingdom Health Protection Agency Reference laboratory at Birmingham and/or to the Virco laboratory in Belgium for further investigations.
Patient consensus sequences were compared to those from a database of known antiretroviral drug resistance mutations by using a rule-based system developed by an international expert panel for interpretation of drug resistance (15). The nucleotide sequences were aligned, and phylogenetic analysis was performed by the neighbor-joining tree method with CLUSTAL W software (http://www.clustalw.genome.ad.jp). HIV-1 subtype determination was carried out phylogenetically by comparing the branching patterns of patient sequences against consensus sequences representing a variety of HIV-1 subtypes available in the Los Alamos HIV-1 sequence database (http://hiv-web.lanl.gov.html). The RT sequence was used as the basis for subtype allocation in this analysis.
Statistical analysis.
The statistical package SPSS 12.0.1 for windows (Lead Technologies, Chicago, IL) was used for data analysis. Clinical information used in the analysis included age, sex, HIV-1 viral load, CD4 count, the clinical reason for the genotypic resistance test, HIV-1 subtypes, geographic origin of infection, and the success or failure of the original version 1 and 1.5 primers in producing a usable sequence. A chi-square test was used for analysis of categorical variables, and a Student t test was used for continuous variables. Logarithmically transformed HIV-viral loads were compared between samples successfully and unsuccessfully amplified using version 1 primers and plotted in the boxplot format.
RESULTS
Of the 208 patients, 104 had a likely source of infection from Africa; 81 were likely to be infected within Europe, and together with three patients infected in North America and three from Australia, these formed the Western group. There were seven patients who were likely to have been infected by a source from the Caribbean, six who were likely to have been infected by a source from South America, and two who were likely to have been infected by a source from Asia. Two further patients had risk factors linked to more than one geographical location and their likely source of infection could not be determined. Those that were not in the African or Western group were analyzed together (Others, n = 17).
There were significantly more female than male patients in the African group compared to Western or Others (Table 1). The African patients were more likely to have a lower CD4 count, probably due to more advanced disease presentation in this group. These ethnic differences in presentation of HIV-1 infection were previously recognized in a patient cohort from South London (3). The mean age of the groups and the HIV-1 viral load of the samples tested were similar.
TABLE 1.
Comparison of the clinical information, HIV-1 subtypes, and TG assay success rate between Western, African, and Others groups of patients
| Parameter | Value for group
|
|||
|---|---|---|---|---|
| Western | African | Others | Pa | |
| General | ||||
| No. of samples | 87 | 104 | 17 | |
| Mean age in yr (range) | 39 (24-60) | 37 (8-66) | 36 (18-60) | 0.071 |
| Sex ratio (M:F) | 83:4 | 41:63 | 14:3 | <0.001 |
| Mean log viral load | 4.14 | 4.04 | 4.03 | 0.355 |
| Mean CD4 | 346 | 206 | 333 | <0.001 |
| Indications for genotyping test (no. of samples) | 0.133 | |||
| Treatment failure | 69 | 87 | 14 | |
| Primary infection | 10 | 1 | 1 | |
| Poor response to new regimen | 4 | 7 | 1 | |
| Pregnancy | 0 | 6 | 1 | |
| Restart after treatment | 1 | 3 | 0 | |
| Others | 3 | 0 | 0 | |
| HIV-1 subtype (no. of samples) | <0.001 | |||
| A | 0 | 28 | 1 | |
| B | 86 | 3 | 16 | |
| C | 0 | 33 | 0 | |
| D | 0 | 13 | 0 | |
| G | 0 | 6 | 0 | |
| J | 0 | 4 | 0 | |
| K | 1 | 2 | 0 | |
| CRF01_AE | 0 | 5 | 0 | |
| CRF02_AG | 0 | 10 | 0 | |
| Outcome of genotyping test (no. of samples [%]) | 0.1 | |||
| Version 1 primer success | 77 (89) | 85 (82) | 13 (76) | |
| Version 1.5 primer success | 9/10 (90) | 19/19 (100) | 3/4 (75) | |
| Version 1.5 primer failure | 1/10 (10) | 0 | 1/4 (25) | |
As determined by chi-square tests for categorical variables between the Western, African, and Others groups and independent sample t tests for continuous variables between the Western and African groups.
The main indication for a resistance test was treatment failure (170 of 208 [82%]). This was followed by primary infection (n = 12; 6%) and poor response to a new regimen (n = 12; 6%). Seven patients had resistance tests requested as a result of pregnancy, and six of these were African in origin. Seven patients had a resistance test requested on indications that were not standard of care at the time of the study: four had a resistance test requested during treatment interruption in preparation for restarting HAART, and three others had a baseline resistance test requested on an earlier sample since the clinician was considering a treatment change despite successful current therapy. There was no difference in the indications for a resistance test between the three groups.
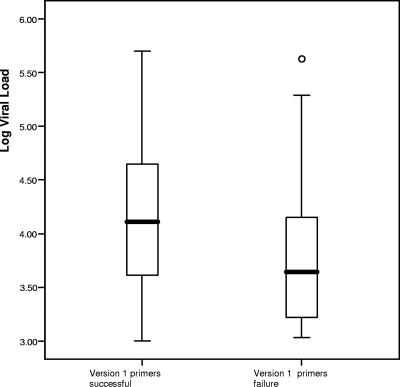
The TG version 1 primers were successful in producing a full sequence in 175 of the 208 samples (84%). There was no statistically significant difference in success rate between the three groups (Table 1). However, the mean HIV-1 viral load of the successful samples was significantly higher than the samples that failed (mean log10 viral load of 4.14 versus 3.79, P = 0.014) (Fig. 1). The success rate of samples with an HIV-1 viral load of ≥5,000 copies/ml was 89.3% (125 of 140). This compared to a success rate of 73.5% (50 of 68) when the viral load was <5,000 copies/ml (P = 0.004). The 33 samples that failed to produce usable sequences with the original version 1 primers were further tested with the alternative version 1.5 primers, and 31 of 33 (94%) were successful in producing a full sequence. The two samples that failed with version 1.5 primers, one each from the Western and Others groups, were referred to two other laboratories that used a different amplification strategy. A full sequence was obtained for the Western sample (subtype B), whereas a partial sequence was obtained for the Others group sample (subtype A). As with the version 1 primers, there was also no statistically significant difference in the success rate between the three groups with version 1.5 primers.
FIG. 1.
Boxplot comparing log10 HIV-1 viral load of samples with successful or failure version 1 primers (mean log10 viral load of 4.14 versus 3.79 [two-tailed independent sample t test P = 0.014]). One sample in the failure group, marked as a circle in the plot, was identified by SPSS as an outlier.
HIV-1 subtypes were allocated on the basis of the RT sequences obtained from these samples. As expected with the Western group, 86 of 87 (99%) were subtype B, and the majority of the Others group (16 of 17; 94%) were also subtype B. The major subtypes in the African group were non-B: C (n = 33; 32%), A (n = 28; 27%), D (n = 13; 13%), and CRF01_AG (n = 10; 10%) (P < 0.001).
The clinical information was reanalyzed using the subtype information (Table 2). The results largely mirrored that of the analysis between the three patient groups in that there were significantly more females among the non-B subtypes, with significantly lower CD4 counts than the subtype B group. Treatment failure as an indication for a resistance test was comparable between B and non-B subtypes, whereas pregnancy as an indication for testing was exclusively found among the non-B infected patients. Primary infection as an indication for a resistance test was more prevalent among subtype B-infected patients (P = 0.006). There was no significant difference in HIV-1 viral load between B and non-B subtypes. The success rates of version 1 primers with B and non-B subtypes were 87.6% (92 of 105) and 80.6% (83 of 103), respectively (P = 0.165), and for version 1.5 primers they were 92.3% (12 of 13) and 95% (19 of 20), respectively (P = 1.0). There was therefore no significant difference in the success or failure with either primer set in relation to HIV-1 subtypes (Table 3).
TABLE 2.
Comparison of the clinical information, origin of infection, and TG assay success rate between subtype B- and non-B-infected patients
| Parameter | Value for group infected by subtype:
|
||
|---|---|---|---|
| B | Non-B | Pa | |
| General | |||
| No. of samples | 105 | 103 | |
| Mean age in yr (range) | 39 (24-60) | 37 (8-66) | 0.051 |
| Sex ratio (M:F) | 98:7 | 40:63 | <0.001 |
| Mean log viral load | 4.12 | 4.05 | 0.48 |
| Mean CD4 | 334 | 215 | <0.001 |
| Indications for genotyping test (no. of samples) | 0.006 | ||
| Treatment failure | 85 | 85 | |
| Primary infection | 11 | 1 | |
| Poor response to new regimen | 5 | 7 | |
| Pregnancy | 0 | 7 | |
| Restart after treatment interruption | 1 | 3 | |
| Others | 3 | 0 | |
| Origin of HIV-1 infection (no. of samples) | <0.001 | ||
| Western | 86 | 1 | |
| African | 3 | 101 | |
| Others | 16 | 1 | |
| Outcome of genotyping test (no. of samples [%]) | 0.363 | ||
| Version 1 primer success | 92 (88) | 83 (81) | |
| Version 1.5 primer success | 12/13 (92) | 19/20 (95) | |
| Version 1.5 primer failure | 1/13 (8) | 1/20 (5) | |
As determined by chi-square tests for categorical variables between the B and non-B subtypes and independent sample t tests for continuous variables between B and non-B subtypes.
TABLE 3.
Proportion of each HIV-1 subtype that failed with TG version 1 and 1.5 primers
| HIV-1 subtype | No. failed/total no. (%)
|
|
|---|---|---|
| Version 1 primer failure | Version 1.5 primer failure | |
| A | 7/29 (24) | 1/7 (14) |
| B | 13/105 (12) | 1/13 (8) |
| C | 4/33 (12) | 0/4 (0) |
| D | 4/13 (31) | 0/4 (0) |
| G | 1/6 (17) | 0/1 (0) |
| J | 2/4 (50) | 0/2 (0) |
| K | 1/3 (33) | 0/1 (0) |
| CRF01_AE | 1/5 (20) | 0/1 (0) |
| CRF02_AG | 0/10 (0) | |
DISCUSSION
Several studies have tried to establish the performance of the TG genotyping assay with non-B subtypes HIV-1 (2, 6, 9). These studies generally involved very small sample numbers (range, 22 to 35). Two of these three studies also used a highly selected panel of samples with known HIV-1 subtypes for the evaluation (2, 9). The present study is unique in that a large number of prospectively collected clinical samples (n = 208) in which a resistance test was clinically indicated were being evaluated. South London is also unique in that there are approximately equal numbers of HIV-1 patients who acquired their infection from the West and sub-Saharan Africa, thus allowing an evaluation of the TG assay with a number of diverse subtypes in a nonselective clinical setting. Our approach involved analysis of the performance of the assay between different patient groups and confirmed the validity of the results by reanalysis of the data relative to the HIV-1 subtypes. A good correlation was demonstrated between patient groups and the expected HIV-1 subtype.
The initial report on the use of the TG assay with non-B subtypes was promising (6). However, the TG assay with the original version 1 primers performed very poorly in a subsequent study by Beddows et al. (2) with only 16 of 30 (53%) of the selected panel of samples generating usable sequences. The study by Jagodzinski et al. (9), with a panel of HIV-1 isolates, also highlighted the deficiency of the version 1 primers in that 3 of 31 isolates (9.7%) failed and all were non-B subtypes (one C and two G). An improvement was observed with the use of the alternative version 1.5 primers. These primers were successful with all HIV-1 subtypes using the same panel of isolates, despite the need to test at a higher dilution (5,000 copies/ml instead of >20,000 copies/ml) due to a limited quantity of the original viral stock.
The overall success rate of the original version 1 primers in our hands was 84%. The use of the alternative version 1.5 primers significantly improved the outcome in that only two samples persistently failed genotyping. We did not retest previously successful samples with the version 1.5 primers, but Jagodzinski et al. (9) have demonstrated that the alternative primer set was able to produce sequences for all of the selected genotypes at a much lower viral load. Our results suggest that a higher HIV-1 viral load is associated with a greater chance of success with the original version 1 primers. Although a variable effect of viral load with some samples was observed and the 0.35-log difference (Fig. 1) seems to be within the acceptable limit of assay variability, further analyses confirmed a higher likelihood of success in samples with viral loads of ≥5,000 compared to <5,000 copies/ml (89.3% versus 73.5%; P = 0.004). It is likely that the alternative version 1.5 primers have a higher performance level than version 1 primers due to more efficient annealing to most HIV-1 subtype sequences. A higher viral load in the sample allowed some degree of compensation and may therefore increase the success rate of the version 1 primers, regardless of the subtype.
Since the completion of our study, the manufacturer of the TG assay has replaced the version 1 primers in the TG kits with the version 1.5 primers. Our experience of this new-format TG assay is that the initial success rate is much higher. Among the last 100 clinical samples that we have tested, only seven initially failed to sequence: two were due to the use of samples with low viral loads (<1,000 copies/ml), four were successfully sequenced when the assay was repeated, and only one sample failed repeatedly (data not shown). This observation is consistent with our evaluation data indicating that the version 1.5 primers are more efficient.
In conclusion, in this large-scale prospective evaluation of the TG assay with clinical samples, we found that the assay was equally effective for testing samples from patients infected in Africa or Western countries. There was no significant difference in assay performance with B and non-B subtypes. The replacement of version 1 primers by 1.5 primers in the current test is expected to improve the efficiency of the TG genotyping assay.
REFERENCES
- 1.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. L. Abrams, B. J. Brizz, J. P. A. Ioannidis, T. C. Merigan, et al. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 2.Beddows, S, S. Galpin, S. H. Kazmi, A. Ashraf, A. Johargy, A. J. Frater, N. White, R. Braganza, J. Clarke, M. McClure, and J. Weber. 2003. Performance of two commercially available sequence-based HIV-1 genotyping systems for the detection of drug resistance against HIV type 1 group M subtypes. J. Med. Virol. 70:337-342. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, A. E., S. Murad, S. O'Shea, A. de Ruiter, C. Watson, and P. J. Easterbrook. 2005. Ethnic differences in stage of presentation of adults newly diagnosed with HIV-1 infection in south London. HIV Med. 6:59-65. [DOI] [PubMed] [Google Scholar]
- 3a.CDSC. 2003. Survey of prevalent diagnosed HIV infections. CDSC, London, United Kingdom.
- 4.Deeks, S. G., N. S. Hellmann, R. M. Grant, N. T. Parkin, C. J. Petropoulos, M. Becker, W. Symonds, M. Chesney, and P. A. Volberding. 1999. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J. Infect. Dis. 176:1375-1381. [DOI] [PubMed] [Google Scholar]
- 5.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. B. Boucher, J. M. Schapiro, and P. Delamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 6.Erali, M., S. Page, L. G. Reimer, and D. R. Hillyard. 2001. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J. Clin. Microbiol. 39:2157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant, R. M., D. R. Kuritzkes, V. A. Johnson, J. W. Mellors, J. L. Sullivan, R. Swanstrom, R. T. D'Aquila, M. Van Gorder, M. Holodniy, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 41:1586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 9.Jagodzinski, L. L., J. D. Cooley, M. Weber, and N. L. Michael. 2003. Performance characteristics of human immunodeficiency virus type 1 (HIV-1) genotyping systems in sequence-based analysis of subtypes other than HIV-1 subtype B. J. Clin. Microbiol. 41:98-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantor, R., and D. Katzenstein. 2004. Drug resistance in non-subtype B HIV-1. J. Clin. Virol. 29:152-159. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzi, P., M. Opravil, B. Hirschell, J. P. Chave, H. J. Furrer, H. Sax, T. V. Perneger, L. Perrin, L. Kaiser, and S. Yerly. 1999. Impact of drug resistance mutations on virologic response to salvage therapy. AIDS 13:F71-F21. [DOI] [PubMed] [Google Scholar]
- 12.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2000. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Pozniak, A., B. Gazzard, J. Anderson, A. Babiker, D. Churchill, S. Collins, M. Fisher, M. Johnson, S. Khoo, C. Leen, C. Loveday, G. Moyle, M. Nelson, B. Peter, A. Phillips, D. Pillay, E. Wilkins, I. Williams, M. Youle, et al. 2003. British HIV Association (BHIVA) guidelines for the treatment of HIV-infected adults with antiretroviral therapy. HIV Med. 4(Suppl. 1):1-41. [PubMed] [Google Scholar]
- 14.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 15.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The EuroGuidelines Group for HIV Resistance. 2001. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS 15:309-320. [DOI] [PubMed] [Google Scholar]
- 17.Walter, E. A., B. Gilliam, J. A. Delman, K. Spooner, J. T. Morris. N. Aronson, S. A. Wegner, N. L. Michael, and L. J. Jagodzinski. 2000. Clinical implications of identifying non-B subtypes of human immunodeficiency virus type 1 infection. Clin. Infect. Dis. 31:798-802. [DOI] [PubMed] [Google Scholar]
- 18.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy has failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]