Abstract
The synthesis and transportation proteins of the Vi capsular polysaccharide of Salmonella enterica serovar Typhi (serovar Typhi) are encoded by the viaB operon, which resides on a 134-kb pathogenicity island known as SPI-7. In recent years, Vi-negative strains of serovar Typhi have been reported in regions where typhoid fever is endemic. However, because Vi negativity can arise during in vitro passage, the clinical significance of Vi-negative serovar Typhi is not clear. To investigate the loss of Vi expression at the genetic level, 60 stored strains of serovar Typhi from the Faisalabad region of Pakistan were analyzed by PCR for the presence of SPI-7 and two genes essential for Vi production: tviA and tviB. Nine of the sixty strains analyzed (15%) tested negative for both tviA and tviB; only two of these strains lacked SPI-7. In order to investigate whether this phenomenon occurred in vivo, blood samples from patients with the clinical symptoms of typhoid fever were also investigated. Of 48 blood samples tested, 42 tested positive by fliC PCR for serovar Typhi; 4 of these were negative for tviA and tviB. Three of these samples tested positive for SPI-7. These results demonstrate that viaB-negative, SPI-7-positive serovar Typhi is naturally occurring and can be detected by PCR in the peripheral blood of typhoid patients in this region. The method described here can be used to monitor the incidence of Vi-negative serovar Typhi in regions where the Vi vaccine is used.
Salmonella enterica subsp. enterica serovar Typhi is primarily but not exclusively the etiologic agent of a systemic infection in humans known as enteric (typhoid) fever. Unlike many other Salmonella serovars, serovar Typhi is restricted to human populations. Despite improvements in sanitation and healthcare in many developing countries, typhoid fever remains an important public health problem in areas of undeveloped and developing countries. The World Health Organization estimates that the current annual global burden of typhoid is approximately 22 million new cases, 5% of which are fatal (7, 19).
The pathogenesis of the members of the genus Salmonella is attributed, in part, to the acquisition of horizontally transferred DNA, including plasmids, prophage, and gene islands (3, 4, 24, 25, 31). Arguably the most important form of horizontally transferred DNA with respect to the pathogenesis of the salmonellae is the possession of large gene islands that carry genes that are transcribed in a coordinated manor and directly impinge on the pathogenic potential of the bacterium. These gene islands (termed pathogenicity islands) are missing from closely related nonpathogenic strains, are often but not always flanked by small direct repeats, and are frequently associated with tRNA genes (10, 11).
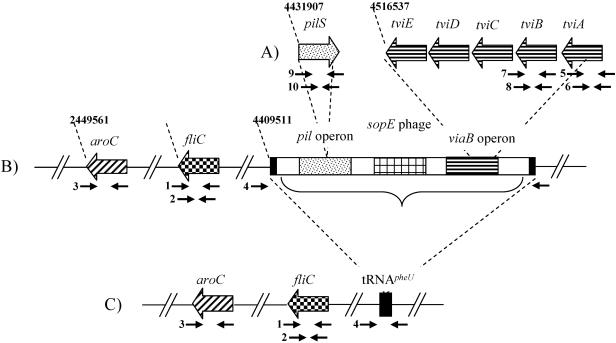
The serovar Typhi CT18 genome sequence identified five previously described Salmonella pathogenicity islands (SPIs) and also predicted five more gene islands that had coding sequences implicated in pathogenicity (31). One such island, known as SPI-7, is inserted in between two partially duplicated copies of the tRNApheU gene located at positions 4409511 and 4543074, respectively, in the serovar Typhi CT18 genome sequence (Fig. 1) (33). SPI-7 encodes genes responsible for several pathogenic traits, including a type IV pilus, implicated in aiding attachment to eukaryotic cells (45), and the sopE prophage (27, 28, 41), which harbors a gene encoding an effector protein secreted via a type III secretion system within its tail fiber genes. In addition, SPI-7 carries the viaB operon, which encodes the genes responsible for the synthesis and transportation for the virulence (Vi) capsule (15, 42).
FIG. 1.
Map of regions investigated in this study, including SPI-7 and the viaB locus. The gene names and regions are highlighted, as are the approximate location of the primer pairs (see Table 1) and the location of the regions on the chromosome (according to the serovar Typhi CT18 genome sequence). (A) Map corresponding to the pilS gene and a section of the viaB locus (tviE to tviA) within SPI-7. The schematic locations of primer pairs 9 and 10 (pilS), 7 and 8 (tviB), and 5 and 6 (tviA) are shown. (B) Location of the aroC and fliC genes and SPI-7 and the primer pairs specific for each region: 3 (aroC), 1 and 2 (fliC), and 4 (SPI-7 flanking). Regions of interest within SPI-7 are depicted and include the flanking tRNA sequences (black boxes), the pil operon (dotted box), the sopE phage (checked box), and the viaB operon (striped box). (C) Same arrangement as in panel B, but with the absence of SPI-7. Thus, this region would be predicted to generate an amplicon with primer pair 4.
The viaB operon is only found in organisms that can produce the Vi polysaccharide and has no corresponding homologues in Escherichia coli. The organisms that have been documented as producing Vi are S. enterica serovars Typhi, Dublin, and Paratyphi C and Citrobacter freundii (14). Interestingly, the Salmonella serovars that can produce Vi also possess an SPI-7 element. The viaB region in serovar Typhi consists of 10 genes. tviA, tviB, tviC, tviD, and tviE are involved in synthesis of the capsule. Export of the capsule is controlled by the proteins encoded by vexA, vexB, vexC, vexD, and vexE (42). In addition, rcsB and rcsC (viaA) and the ompR-envZ two-component regulatory system regulate the production of the Vi polysaccharide (32).
Traditionally, the production of Vi by serovar Typhi is a distinguishing feature of the bacterium, and agglutination using Vi antisera is a routine procedure for the identification of serovar Typhi in research and diagnostic laboratories (19, 22). The precise biological role of the Vi polysaccharide remains unclear. It is, however, believed to prevent phagocytosis and complement-mediated killing when the bacteria are outside eukaryotic cells but inside the host (36), although there are data that suggest that the rates of internalization of encapsulated and unencapsulated serovar Typhi into macrophages are equivalent (16). These experimental data imply that Vi may be important in the survival of the bacterium inside the macrophage but not in cellular invasion of the macrophage or the intestinal wall. Volunteer studies have indicated that Vi-positive strains of serovar Typhi are more virulent in humans than Vi-negative strains, although Vi production is not essential for the infection process in humans (18).
Despite the role of the Vi antigen as a distinguishing feature of serovar Typhi, serovar Typhi isolates lacking the Vi capsular polysaccharide antigen during slide agglutination with Vi typing antisera is not uncommon. Vi-negative isolates have been reported in several countries, including India and Malaysia (1, 20, 26). In fact, in 2000, serovar Typhi isolates that were Vi negative by molecular probes were responsible for an epidemic of multidrug-resistant typhoid fever in Kolkata, India (37). It is however, possible that Vi agglutination-negative serovar Typhi reported from clinical microbiology laboratories may be Vi positive but demonstrate a downregulation of Vi or loss of viaB on culturing. These findings have been substantiated by the description of additional serovar Typhi isolates lacking SPI-7 (29). SPI-7 may therefore be able to excise from the chromosome and act in similar fashion to a conjugative transposon (33). However, all of these studies were performed on serovar Typhi isolates that had been cultured and in many cases stored. The loss of SPI-7 and Vi negativity could therefore arise by selection during isolation or storage. The significance of Vi-negative serovar Typhi in vivo is thus currently ambiguous. Recently, in Karachi, Pakistan, only 1 of more than 2,000 stored clinical isolates of serovar Typhi was found to be Vi negative by immunofluorescence detection of Vi and PCR detection of the SPI-7 locus (43). There was complete correlation between Vi expression and the presence of SPI-7.
We have investigated here the frequency of Vi-negative serovar Typhi among clinical isolates from typhoid patients living within the Faisalabad region of Pakistan. In order to establish the possible presence of Vi-negative serovar Typhi in this region of Pakistan, stored serovar Typhi cultures were screened by PCR for the presence of viaB and SPI-7. Also, to investigate the viaB region within serovar Typhi strains without the need for culture or storage, PCR was performed directly on total DNA extracted from the blood of patients with suspected typhoid fever (23, 35). Typhoid fever continues to be a major health problem in Pakistan, and these data provide insight into the occurrence of naturally occurring Vi-negative strains in the blood of typhoid patients and in turn may inform future vaccine development (2).
MATERIALS AND METHODS
Bacterial strains and clinical samples.
This study was carried out with 60 isolates of serovar Typhi collected from hospitals in the Faisalabad region (population of approximately 10 million) of Pakistan between March 2002 and September 2002. Strains were isolated from unvaccinated patients clinically diagnosed with typhoid fever, i.e., fever for ≤3 days with enlarged spleen, headache, malaise, abdominal discomfort, and/or agitation. Initially, the strains were cultured from blood samples and identified by conventional biochemical and serologic methods (8). After primary isolation, serovar Typhi strains were plated on MacConkey agar, subcultured in Trypticase soy broth overnight, and tested for Vi antigen (antisera; Bio-Stat); aliquots were preserved in 20% glycerol and stored at −20°C for further use. When required, an aliquot of the stored serovar Typhi isolates was revived in Trypticase soy broth for 24 h at 37°C, and total genomic DNA was extracted from the Trypticase soy broth by the conventional phenol-chloroform method (38).
This study also included the assessment of blood samples taken in February 2005 from 48 unvaccinated patients suspected of having typhoid fever. Patients were of both sexes and a broad age range; these patients had 2 ml of blood collected in tubes containing anticoagulant (20 mM potassium EDTA) The blood was stored at 4°C and prepared for PCR within 48 h of collection.
DNA extraction from blood samples.
DNA from blood samples was extracted by procedure described by Haque et al. (12). Briefly, 1 ml of blood containing 20 mM potassium EDTA as anticoagulant was centrifuged at 10,000 rpm (Sorvall Legend RT) for 5 min. Plasma was separated for serology. The pellet was resuspended in 1 ml of lysis buffer (0.2% Triton X-100 in Tris-HCl [pH 8.0]). The mixture was gently aspirated several times to encourage efficient hemolysis. The tube was centrifuged at 12,000 rpm (Sorvall Legend RT) for 6 min, the supernatant was discarded, and the procedure was repeated. The pellet was washed with distilled water. The supernatant was removed, and the pellet was subsequently resuspended in 20 to 30 μl of distilled water. The tubes were sealed and then sterilized in boiling water for 20 min.
PCR conditions and primers.
The oligonucleotides utilized in this study were supplied by Sigma (Dorset, United Kingdom) and are presented in Table 1. The PCR and thermal cycling conditions for DNA from stored bacterial cultures were as follows. A total of 100 μl of a PCR mix contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 150 pmol of each primer, 95 nmol of each deoxynucleoside triphosphate, 1 μl of Taq polymerase (Fermentas), 20 μl of template, and distilled water up to 100 μl. The reaction mixture was subjected to 30 cycles of 94°C for 30 s, 50 to 60°C for 30 s (see Table 1 for primer annealing temperatures), and 72°C for 1 to 2 min (see Table 1 for primer extension times), followed by 5 min at 72°C (MasterCycler; Eppendorf, Hamburg, Germany). Samples were separated immediately by gel electrophoresis on 2% agarose gels at 100 V for 60 min and then photographed using Eagle Eye (Stratagene).
TABLE 1.
Oligonucleotides used in this studya
| Target gene | Primer pairb | Annealing temp (°C) | Elongation time (min) | Predicted amplification size (bp) | Primer | Sequence (5′-3′) | Source or reference |
|---|---|---|---|---|---|---|---|
| fliC | 1 | 53 | 1 | 495 | ST1 | TATGCCGCTACATATGATGAG | Song et al. (40) |
| ST2 | TTAACGCAGTAAAGAGAG | ||||||
| fliC nested | 2 | 50 | 1 | 363 | ST3 | ACTGCTAAAACCACTACT | Song et al. (40) |
| ST4 | TGGAGACTTCGGTCGCGTAG | ||||||
| aroC | 3 | 57 | 2 | 639 | aroCsfor | GGCACCAGTATTGGCCTGCT | Kidgell et al. (21) |
| aroCsrev | CATATGCGCCACAATGTGTTG | ||||||
| tRNApheU | 4 | 57 | 2 | 1,275 | DE0032-F | GCTCAGTCGGTAGAGCAGGGGATT | Pickard et al. (33) |
| DE0083-R | TCATCTTCAGGACGGCAGGTAGAATG | ||||||
| tviA | 5 | 55 | 2 | 599 | V1 | GTTATTTCAGCATAAGGAG | Hashimoto et al. (13) |
| V2 | ACTTGTCCGTGTTTTACTC | ||||||
| tviA nested | 6 | 55 | 1 | 307 | V3 | GTGAACCTAAATCGCTACAG | Hashimoto et al. (13) |
| V4 | CTTCCATACCACTTTCCG | ||||||
| tviB | 7 | 57 | 2 | 846 | tviB-F | CGAGTGAAACCGTTGGTACA | Wain et al. (43) |
| tviB-R | CAATGATCGCATCGTAGTGG | ||||||
| tviB nested | 8 | 60 | 2 | 774 | tviB-in-F | GAATCGGGGAGATATTGTGG | This study |
| tviB-in-R | TGCCATACTCTCGTCTTACC | ||||||
| pilS | 9 | 58 | 1 | 502 | pilS-F | GTATCAACATTAAATCCATGC | This study |
| pilS-R | CGTTACTTTCGCATCGGTGTG | ||||||
| pilS nested | 10 | 57 | 1 | 335 | pilS-F-inner | ATCATTGGGGTGATAGCC | This study |
| pilS-R-inner | GCAGATTGCGGAACTTTG |
For amplification where total DNA from whole blood acted as a template, the PCR and cycling conditions were as follows. A total of 100 μl of PCR mix contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 200 pmol of each primer, 125 nmol of each deoxynucleoside triphosphate, 1.5 μl of Taq polymerase (Fermentas), 20 μl of template (purified total DNA from whole blood), and distilled water (up to 100 μl). The reaction mixture was subjected to 30 cycles of 94°C for 1 min, 50 to 60°C for 30 s (see Table 1 for primer annealing temperatures), and 72°C for 1 to 2 min (see Table 1 for primer extension times), followed by 7 min at 72°C (Mastercycler). PCR products were handled as described above.
RESULTS
Detection of serovar Typhi from stored isolates by PCR.
Blood samples from patients suspected of having typhoid fever are routinely cultured in order to confirm the clinical diagnosis. The identification of serovar Typhi is confirmed by standard laboratory procedures including biochemical testing and agglutination using relevant antisera (09 and Vi). In Faisalabad a significant proportion (ca. 15%) are agglutination negative for Vi, suggesting that these serovar Typhi isolates cannot produce the Vi antigen. In order to explore this phenomenon, 60 stored serovar Typhi samples, isolated between March 2002 and September 2002 in the Faisalabad region of Pakistan, were recultured and investigated.
Initially, it was essential to confirm that the cultured bacteria were serovar Typhi; this was accomplished via PCR targeted to the aroC and fliC genes. The aroC gene and the fliC gene were amplified with the primer pairs aroCsfor-aroCsrev (primer pair 3) and ST1-ST2 (primer pair 1), respectively (Table 1 and Fig. 1). The aroC primers are specific for Salmonella, have been utilized in previous studies (21, 43), and are predicted to generate an amplicon of 639 bp with Salmonella DNA. The fliC primers amplify a serovar Typhi-specific region in the flagellin gene and have been previously described as being highly sensitive for the detection of serovar Typhi within clinical specimens (9, 17, 39, 40). The fliC primers (primer pair 1) are predicted to produce an amplicon of 495 bp with serovar Typhi DNA (Table 1). All 60 cultured bacteria gave amplifications of the predicted size with both the aroC and the fliC primers, thus confirming that all 60 strains were serovar Typhi (Fig. 2).
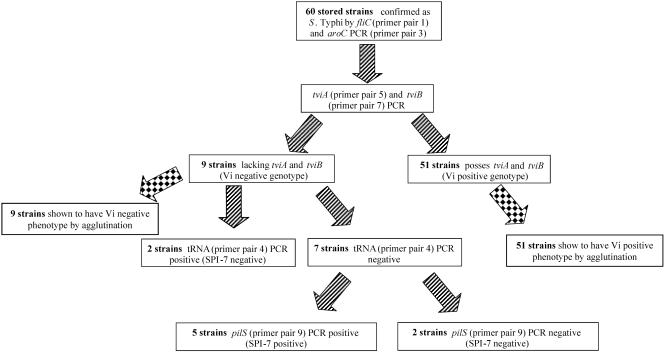
FIG. 2.
Sequential investigation of the Vi status of stored serovar Typhi strains from Faisalabad, Pakistan. A flow diagram shows the results of investigating the ability of 60 stored isolates from Faisalabad, Pakistan, to express Vi. The 60 strains were investigated at all regions displayed in Fig. 1, which included fliC (primer pair 1), aroC (primer pair 3), tviA (primer pair 5), tviB (primer pair 7), tRNA pheU (primer pair 4), and pilS (primer pair 9).
PCR detection of viaB operon.
A recent study of Vi-negative serovar Typhi in Karachi, Pakistan, demonstrated that Vi-negative serovar Typhi could be detected by a multiplex PCR method. In order to utilize the methodology described by Wain et al. (43) on clinical samples with small amounts of DNA, the PCRs were not multiplexed, and each set of primers was used individually. Detection of the viaB operon was performed on the 60 stored serovar Typhi isolates by PCR amplification of two genes essential for Vi production; tviA and tviB (Fig. 1). Primer pair 5 is specific to the DNA sequence within the tviA gene and is predicted to generate an amplicon of 599 bp if tviA is present. The primers specific for the tviB gene were tviB-F and tviB-R (primer pair 7) and were predicted to generate an amplicon of 846 bp if tviB is present (Table 1). The 60 strains confirmed as serovar Typhi by aroC and fliC PCR amplification were subjected to PCR with the tviA and tviB specific primers. Of the 60 strains confirmed as serovar Typhi, 9 (ca. 15%) failed to give any amplification with the tviA primers. Also, the same nine strains failed to give any visible amplification for tviB (Fig. 2). Since tviA and tviB are essential for Vi production, these nine strains were thought to be incapable of Vi expression. Agglutination with Vi antisera was in accordance with these results. In conclusion, nine strains were found to lack the tviA and tviB genes and were therefore genotypically Vi negative. These same nine strains were found to be Vi negative by agglutination and were therefore both genotypically and phenotypically Vi negative. The remaining 51 strains were found to possess both tviA and tviB genes and were found to be phenotypically Vi positive by Vi agglutination.
Assessing the presence or absence of SPI-7.
The viaB locus is located on SPI-7, which is inserted between two partially duplicated tRNApheU genes (as discussed above); however, SPI-7 may be unstable and can potentially excise from the chromosome (5, 29). This mechanism was hypothesized to be responsible for the inability of the nine samples above to generate a visible amplification for tviA and tviB. These nine samples were subjected to PCR to confirm the presence or the absence of SPI-7. Primers DE0032-F and DE0083-R (primer pair 4) have been previously utilized to demonstrate the lack of an insertion at the tRNApheU locus (33, 43). These primers are predicted to generate a PCR amplicon of 1,275 bp (Table 1) if the island is absent. SPI-7 is 134 kb in length; therefore, the presence of the island is outside the constraints of the PCR. In addition, the presence of the island was confirmed by PCR to a separate loci within SPI-7, a gene within the type IV pilus cluster that encodes the major pilin subunit, pilS (44, 45). The pilS primers (pilS-F and pilS-R [primer pair 9]) were predicted to generate an amplicon of 502 bp if the gene (and therefore the island) was present.
Two of the nine confirmed serovar Typhi strains gave amplification across the tRNA gene that indicated the absence of SPI-7 (Fig. 1 and 2). In addition, these two isolates failed to give any amplification with the pilS primers, thus confirming the absence of SPI-7. The remaining seven strains failed to give any amplification with primers DE0032-F and DE0083-R, suggesting the presence of SPI-7. This was subsequently confirmed by the amplification of the pilS gene in five strains; this result suggested that SPI-7 was present but that there had been a deletion within SPI-7 that included the viaB locus. Two strains, however, failed to give any amplification with the tRNA primers or the pilS primers, suggesting the absence of SPI-7 with rearrangement around the tRNApheU, which resulted in the failure of the DE0032-F and DE0083-R to anneal, or the loss of two regions (pil and tviB) within SPI-7. Of these nine samples, two were confirmed to be lacking SPI-7, five appeared to have part of SPI-7 but not tviB, and two did not produce amplification across the tRNA gene or at two sites within SPI-7.
These results indicate that serovar Typhi which is unable to produce Vi can be detected in stored cultures from this region in Pakistan. It is also apparent that there appear to be three distinct mechanisms. One such mechanism has been extensively described and involves the excision and consequential loss of SPI-7. The second mechanism suggests that SPI-7 has again been lost, but a rearrangement at the tRNA locus prevents the production of a visible PCR amplicon across the tRNA junction. The final mechanism appears to be novel, whereby the presence of SPI-7 is suggested, but the viaB operon cannot be detected, which implies a deletion that removes part or all of the viaB operon. Further characterization of cultured isolates was not considered useful, and so DNA was extracted directly from patient's blood. The detection of serovar Typhi isolates that are lacking the genes responsible for Vi production can be detected on cultured strains via a simple PCR assay. However, it is not clear whether the loss of the genes responsible for Vi production is due to the lack of a positive selective pressure by culturing and/or storage or whether these strains are circulating in populations where typhoid is endemic. In order to investigate this, a PCR assay was performed directly on DNA extractions from blood samples of patients suspected of contracting typhoid. This procedure was selected in order to remove the influence of culturing and storage.
Sensitivity of PCR (fliC and tviA genes) for application on blood samples.
The detection of serovar Typhi circulating in peripheral blood has been demonstrated by several studies to be a highly sensitive and reproducible method of diagnosis (9, 17, 40). Indeed, a nested PCR detecting the fliC (H-d1) has been implied to be the “gold standard” in typhoid detection, giving a higher sensitivity than that of blood culture and the Widal test (35). Due to the nature of the PCR, this method is specific for serovar Typhi and will not generate an amplification with other invasive Salmonella strains, such as serovar Paratyphi A or Sendai (17).
In order to develop a methodology that could be utilized for the detection of Vi-negative serovar Typhi in blood samples, nested PCR for the fliC gene was compared with nested PCR of the tviA gene as described in Hashimoto et al. (13). DNA was extracted from a culture of a reference serovar Typhi strain by phenol-chloroform method, and serial dilutions were made in distilled water. Each dilution was used as a template for PCR. Both conventional PCR and nested PCR were performed on the serial dilutions for the fliC gene by using primer pair 1 (ST1-ST2 [primary]) and primer pair 2 (ST3-ST4 [nested]) (Table 1) and also for the tviA gene using primer pair 5 (V1-V2 [primary]) and primer pair 6 (V3-V4 [nested]) (Table 1 and Fig. 1). The predicted sizes of the amplicons for serovar Typhi were 495 and 363 bp (fliC [conventional/nested]) and 599 and 307 bp (tviA [conventional/nested]), respectively. The calculations were made according to the recommendations of Song et al. (40).
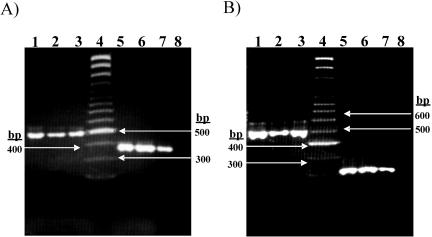
The results of the experiments assessing the sensitivity of the fliC primers and the tviA nested primer pairs are shown in Fig. 3. The amplicons from the conventional PCR and the nested PCR from both the fliC primers the tviA primers were of the expected size. The results presented in Fig. 3 demonstrate that conventional PCR for fliC can detect serovar Typhi DNA in 105 CFU/ml, whereas the nested fliC PCR is significantly more sensitive and can detect serovar Typhi DNA in a concentration as low as 5 CFU/ml. The nested tviA PCR was equal in sensitivity to the nested fliC PCR. These results are in agreement with the results of previous studies on the sensitivity of nested PCR for the detection of serovar Typhi in clinical samples (13, 35, 40).
FIG. 3.
Investigating the sensitivity of the fliC and tviA nested PCR. (A) PCR amplification of the fliC gene with primer pairs 1 and 2 (Table 1 and Fig. 1). Lanes 1, 2, and 3 show PCR amplification of the fliC gene with primer pair 1 with 107, 106, and 105 CFU/ml, respectively. The size of the amplicon was 495 bp. Lanes 5, 6, and 7 show PCR amplification with primer pair 2 with product from primer pair 1 as the template (nested) with 102, 10, and 5 CFU/ml, respectively. The size of the amplicon was 363 bp. Lane 4 is Generuler SM 0323 (Fermentas), and lane 8 is a negative control (no template). (B) PCR amplification of the tviA gene with primer pairs 5 and 6 (Table 1 and Fig. 1). Lanes 1, 2, and 3 show PCR amplification of the tviA gene with primer pair 5 with 107, 106, and 105 CFU/ml, respectively. The size of amplicon was 599 bp. Lanes 5, 6, and 7 show PCR amplification of tviA with primer pair 6 with products from primer pair 5 as a template (nested) with 102, 10, and 5 CFU/ml, respectively. The size of amplicon was 307 bp. Lane 4 is Generuler SM 0323 (Fermentas), and lane 8 is a negative control (no template).
Detection of Vi-negative serovar Typhi in blood samples from typhoid patients.
The nested PCR for the tviA and tviB genes was used to test whether serovar Typhi lacking genes essential for Vi could be detected in the peripheral blood of typhoid patients. Blood samples were collected from 48 patients with clinically diagnosed typhoid fever admitted to various hospitals in the region during February 2005. Blood samples were taken from these patients, and total DNA was extracted.
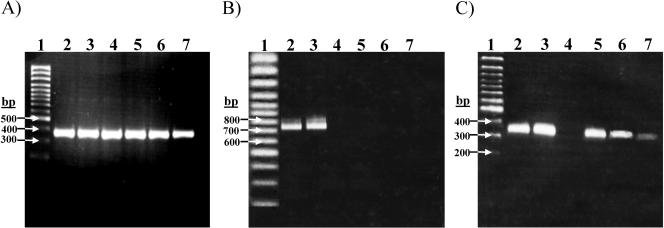
Of the 48 blood samples, 42 tested positive for serovar Typhi DNA by means of the nested fliC (primer pairs 1 and 2, Table 1) PCR. The 42 samples that were confirmed of containing serovar Typhi were then investigated further in order to detect the presence or absence of the viaB locus. Nested PCR for the tviA and tviB genes was performed. Four samples (9%) tested negative by nested PCR for both tviA and tviB (Fig. 4.). In addition, three of these samples tested positive for the pilS gene by nested PCR (Fig. 4), thus indicating the presence of SPI-7 but the absence of the viaB locus. The remaining sample did not produce any amplification for tviA, tviB, or pilS. None of these four samples produced any visible amplification across the tRNA junction with primer pair 4. These PCR assays were repeated and performed simultaneously with the control fliC primers to confirm the result; the repeated assay was in complete accordance with the initial experiment.
FIG. 4.
Detection of Vi-negative serovar Typhi in the peripheral blood of typhoid patients by nested PCR. (A) The detection of serovar Typhi in the blood of patients suspected of having typhoid fever by nested fliC PCR on total DNA extracted from the blood of patients. Primer pair 1 (Table 1 and Fig. 1) was used for the primary reaction, and primer pair 2 was used for the secondary reaction; the product from primary reaction was used as a template in the secondary reaction. Lanes 2 to 7 show nested fliC PCR from six typhoid patients blood samples with amplification size 363 bp. Lane 1 is Generuler SM 0323 (Fermentas). (B) Nested tviB PCR with primer pairs 7 (primary) and 8 (nested); the lanes and samples correspond to those in panel A. (C) Nested pilS PCR with primer pairs 9 (primary) and 10 (nested). Lanes and samples correspond to those in panel A.
DISCUSSION
The characterization of serovar Typhi for expression of Vi capsular polysaccharide is necessary to define the role of Vi in the pathogenesis and epidemiology of typhoid fever. Serovar Typhi lacking Vi capsular polysaccharide antigen has been known and reported worldwide for several decades. However, most of the reports of the Vi-negative isolates are based on the serological tests with Vi typing antisera and, since Vi expression is particularly sensitive to the osmolarity of the selected growth media, this would be a phenotypic rather than a genotypic event (32). Recently, molecular evidence of the loss of Vi antigen has suggested that Vi-negative strains can be derived by the excision of SPI-7 (30) or by a spontaneous base change in the viaB operon (5, 29, 43). It has been postulated that after long-term storage or repeated culturing on laboratory media Vi-negative strains would predominate. This spontaneous loss of Vi expression upon culture and/or storage implies a strong positive selective pressure that serves to maintain the Vi capsule in the natural niche of serovar Typhi. This theory raises doubt as to the existence of Vi-negative strains of serovar Typhi in the field. Typhoid is a common disease in Pakistan and, using stored cultures and blood samples from typhoid patients, a two-pronged strategy was adopted to investigate whether serovar Typhi that is unable to express Vi could be detected in this region without culture and storage.
Initially, we sought to determine whether Vi-negative serovar Typhi could be detected in stored serovar Typhi strains from the Faisalabad region of Pakistan. The stored strains (more than 1 year old) were recultured, and the identity was confirmed by PCR targeting of the aroC and fliC genes (Fig. 1). Of 60 strains examined, 9 were negative for both the tviA and tviB genes (Fig. 2) and were therefore designated as genotypically Vi negative. tviA and tviB were both selected to ensure that Vi negativity was not due to spontaneous mutation; if both genes were missing, we assumed that the genes had been lost. Our results showed the presence of Vi-negative serovar Typhi isolates that had lost a significant amount, if not all, of the viaB operon.
Further investigations were carried out to assess the nature of the deletion and to detect the presence or absence of SPI-7. Using the methodology of Wain et al. (43), primer pair 4 (Fig. 1) confirmed the presence or absence of SPI-7. If missing, an amplicon would be produced across the tRNA junction (1,275 bp) and, if present, the distance between the primers is such that an amplicon would not be generated. The presence of SPI-7 was also confirmed by using an additional second primer pair from another region within SPI-7. Two serovar Typhi strains were found to have lost SPI-7. In addition, two stored strains were PCR negative for tviA, tviB, and pilS but did not produce any PCR product across the tRNApheU junction. These strains may have also undergone rearrangement that has deleted SPI-7 and a portion of the adjacent region. One of the SPI-7-negative serovar Typhi strains investigated by Nair et al. (29) appeared to have also lost an adjacent section at 5′ end of the island which encompassed the phoN gene. Moreover, detailed microarray investigations have predicted that some Salmonella isolates that are without SPI-7 are also missing genes adjacent to the 5′ side of tRNApheU (6, 34). The results shown here can be explained by these data; SPI-7 is absent but, due to a further deletion event including phoN, a PCR amplification across the tRNA junction cannot be generated.
The remaining five strains appeared to retain the island but have somehow deleted all or part of the viaB locus. These results are in contrast to a recent study whereby only 1 in 2,000 stored strains from the Aga Kahn University in Karachi, Pakistan, tested negative for tviB, having lost the whole of SPI-7 (43). A rearrangement or deletion within or adjacent to the viaB locus may be a novel mechanism of Vi negativity, although the exact method remains unclear and requires further investigation. It is known that insertion sequence elements within the viaB locus can diminish and also completely prevent Vi production; this has been shown to be the case in Citrobacter freundii (30) and recently in the serovar Typhi sequence strain CT18 (Stephen Baker, unpublished data). However, this mechanism could not be confirmed in isolates in the present study.
PCR amplification on the total DNA from blood samples revealed that 4 samples of the 42 that were confirmed to contain serovar Typhi did not contain the tviA or tviB genes. Three of these samples tested positive for the pilS gene, demonstrating the presence of SPI-7. The data presented here suggest that serovar Typhi that is incapable of Vi production (genotypically Vi negative) can be detected in the peripheral blood of typhoid patients by nested PCR in this region of Pakistan. In addition, it would appear that the mechanism responsible for this genotype in these cases is based upon the deletion of the viaB operon and not excision of the whole of SPI-7. Furthermore, this implies that the strains found to be lacking the viaA and viaB genes after culture and storage may have been missing these genes prior to isolation, and the loss of these genes may be independent of isolation and culture. It is logical to think that under the correct conditions, environmental loss of one operon (viaB) of 15 kb is much more likely than the loss of the whole island (SPI-7) of 134 kb. Our results appear to support this assumption; moreover, serovar Typhi strains circulating within this geographic region of Pakistan may be more prevalent due to the loss of Vi expression by this mechanism. However, the exact nature and size of the deletion could not be established and is currently under investigation.
The data presented here may pose further questions on the role of Vi in serovar Typhi infection. It has been established previously that Vi is not essential for the development of typhoid fever (18). This appears to be the case for other human-restricted invasive Salmonella serovars, such as serovars Paratyphi A and Sendai, which both induce a disease clinically indistinguishable from typhoid independent of Vi production. Indeed, is now known that in this region of Pakistan serovar Typhi with the inability to produce Vi can be detected without culturing; therefore, these serovar Typhi strains may be circulating in this typhoid-endemic region. It is currently unknown whether these circulating Vi-negative serovar Typhi strains are more or less virulent than their Vi-positive counterparts. The trigger behind the development and the spread of these serovar Typhi Vi-negative organisms also remains unclear. These findings suggest the molecular surveillance of Vi production by serovar Typhi in regions where Vi-based vaccines are used would be sensible.
Our findings show that Vi-negative serovar Typhi strains are not only artifacts of storage but can exist naturally. However, although deletion of SPI-7 was shown to play a role in some cases, the majority of the samples tested positive for SPI-7, and a localized deletion within or around the viaB locus appears to be more common. It is also noteworthy that typhoid vaccination in this region of Pakistan is not routine, and none of the patients involved in the present study was vaccinated against typhoid. We do not know the transmission potential of these Vi-negative strains, but the implications of these data require further investigation, in particular, the accurate estimation of circulating Vi-negative serovar Typhi strains in Vi-vaccinated, recently vaccinated, and unvaccinated populations. The methodology presented here will be useful in this effort.
Acknowledgments
This work was funded by the by the National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan, and the Higher Education Commission of Pakistan. S.B., J.W., and G.D. are supported by the Wellcome Trust.
REFERENCES
- 1.Arya, S. C. 2000. Salmonella typhi Vi antigen-negative isolates in India and prophylactic typhoid immunization. Natl. Med. J. India 13:220. [PubMed] [Google Scholar]
- 2.Arya, S. C., and K. B. Sharma. 1995. Urgent need for effective vaccine against Salmonella paratyphi A, B, and C. Vaccine 13:1727-1728. [DOI] [PubMed] [Google Scholar]
- 3.Baumler, A. J. 1997. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 5:318-322. [DOI] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bueno, S. M., C. A. Santiviago, A. A. Murillo, J. A. Fuentes, A. N. Trombert, P. I. Rodas, P. Youderian, and G. C. Mora. 2004. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J. Bacteriol. 186:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. W. H. O. 82:346-353. [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, W. H. 1986. Edwrads and Ewing's identification of the Enterobacteriaceae, 4th ed. Elsevier Science Publishing Co., Inc., New York, N.Y.
- 9.Frankel, G., S. M. Newton, G. K. Schoolnik, and B. A. Stocker. 1989. Unique sequences in region VI of the flagellin gene of Salmonella typhi. Mol. Microbiol. 3:1379-1383. [DOI] [PubMed] [Google Scholar]
- 10.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function, and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 12.Haque, A., N. Ahmed, A. Peerzada, A. Raza, S. Bashir, and G. Abbas. 2001. Utility of PCR in diagnosis of problematic cases of typhoid. Jpn. J. Infect. Dis. 54:237-239. [PubMed] [Google Scholar]
- 13.Hashimoto, Y., Y. Itho, Y. Fujinaga, A. Q. Khan, F. Sultana, M. Miyake, K. Hirose, H. Yamamoto, and T. Ezaki. 1995. Development of nested PCR based on the ViaB sequence to detect Salmonella typhi. J. Clin. Microbiol. 33:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto, Y., and A. Q. Khan. 1997. Comparison of ViaB regions of Vi-positive organisms. FEMS Microbiol. Lett. 157:55-57. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, Y., N. Li, H. Yokoyama, and T. Ezaki. 1993. Complete nucleotide sequence and molecular characterization of ViaB region encoding Vi antigen in Salmonella typhi. J. Bacteriol. 175:4456-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose, K., T. Ezaki, M. Miyake, T. Li, A. Q. Khan, Y. Kawamura, H. Yokoyama, and T. Takami. 1997. Survival of Vi-capsulated and Vi-deleted Salmonella typhi strains in cultured macrophage expressing different levels of CD14 antigen. FEMS Microbiol. Lett. 147:259-265. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, K., K. Itoh, H. Nakajima, T. Kurazono, M. Yamaguchi, K. Moriya, T. Ezaki, Y. Kawamura, K. Tamura, and H. Watanabe. 2002. Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J. Clin. Microbiol. 40:633-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9:810-816. [DOI] [PubMed] [Google Scholar]
- 19.Ivanoff, B. C. C. L. 2003. The diagnosis, prevention, and treatment of typhoid fever. World Health Organization, Geneva, Switzerland.
- 20.Jegathesan, M. 1983. Phage types of Salmonella typhi isolated in Malaysia over the 10-year period 1970-1979. J. Hyg. 90:91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 22.Le Minor, L. 1992. The genus Salmonella, 2nd ed., vol. III. Springer-Verlag, New York, N.Y.
- 23.Massi, M. N., T. Shirakawa, A. Gotoh, A. Bishnu, M. Hatta, and M. Kawabata. 2003. Rapid diagnosis of typhoid fever by PCR assay using one pair of primers from flagellin gene of Salmonella typhi. J. Infect. Chemother. 9:233-237. [DOI] [PubMed] [Google Scholar]
- 24.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 25.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 26.Mehta, G., and S. C. Arya. 2002. Capsular Vi polysaccharide antigen in Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 40:1127-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 29.Nair, S., S. Alokam, S. Kothapalli, S. Porwollik, E. Proctor, C. Choy, M. McClelland, S. L. Liu, and K. E. Sanderson. 2004. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J. Bacteriol. 186:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou, J. T., C. J. Huang, H. S. Houng, and L. S. Baron. 1992. Role of IS1 in the conversion of virulence (Vi) antigen expression in Enterobacteriaceae. Mol. Gen. Genet. 234:228-232. [DOI] [PubMed] [Google Scholar]
- 31.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug-resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 32.Pickard, D., J. Li, M. Roberts, D. Maskell, D. Hone, M. Levine, G. Dougan, and S. Chatfield. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash, P., O. P. Mishra, A. K. Singh, A. K. Gulati, and G. Nath. 2005. Evaluation of nested PCR in diagnosis of typhoid fever. J. Clin. Microbiol. 43:431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbins, J. D., and J. B. Robbins. 1984. Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella typhi. J. Infect. Dis. 150:436-449. [DOI] [PubMed] [Google Scholar]
- 37.Saha, M. R., T. Ramamurthy, P. Dutta, and U. Mitra. 2000. Emergence of Salmonella typhi Vi antigen-negative strains in an epidemic of multidrug-resistant typhoid fever cases in Calcutta, India. Natl. Med. J. India 13:164. [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Song, J. H., H. Cho, M. Y. Park, Y. S. Kim, H. B. Moon, Y. K. Kim, and C. H. Pai. 1994. Detection of the H1-j strain of Salmonella typhi among Korean isolates by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 50:608-611. [DOI] [PubMed] [Google Scholar]
- 40.Song, J. H., H. Cho, M. Y. Park, D. S. Na, H. B. Moon, and C. H. Pai. 1993. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J. Clin. Microbiol. 31:1439-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson, N., S. Baker, D. Pickard, M. Fookes, M. Anjum, N. Hamlin, J. Wain, D. House, Z. Bhutta, K. Chan, S. Falkow, J. Parkhill, M. Woodward, A. Ivens, and G. Dougan. 2004. The role of prophage-like elements in the diversity of Salmonella enterica serovars. J. Mol. Biol. 339:279-300. [DOI] [PubMed] [Google Scholar]
- 42.Virlogeux, I., H. Waxin, C. Ecobichon, and M. Y. Popoff. 1995. Role of the viaB locus in synthesis, transport, and expression of Salmonella typhi Vi antigen. Microbiology 141(Pt. 12):3039-3047. [DOI] [PubMed] [Google Scholar]
- 43.Wain, J., D. House, A. Zafar, S. Baker, S. Nair, C. Kidgell, Z. Bhutta, G. Dougan, and R. Hasan. 2005. Vi antigen expression in Salmonella enterica serovar Typhi clinical isolates from Pakistan. J. Clin. Microbiol. 43:1158-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, X. L., C. Morris, and J. Hackett. 1997. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major “pathogenicity island” of Salmonella typhi. Gene 202:139-146. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]