Abstract
The INNO-LiPA Rif.TB assay for the identification of Mycobacterium tuberculosis complex strains and the detection of rifampin (RIF) resistance has been evaluated with 360 smear-positive respiratory specimens from an area of high incidence of multidrug-resistant tuberculosis (MDR-TB). The sensitivity when compared to conventional identification/culture methods was 82.2%, and the specificity was 66.7%; the sensitivity and specificity were 100.0% and 96.9%, respectively, for the detection of RIF resistance. This assay has the potential to provide rapid information that is essential for the effective management of MDR-TB.
Portugal has a frequency of new cases of tuberculosis (TB) of 45.0 per 100,000 inhabitants (37), of which 21% are multidrug-resistant TB (MDR-TB) cases, and of these, >5% are resistant to four or five first-line anti-TB drugs (1, 23, 35). Despite control measures that have reduced the incidence of MDR-TB (6), in the major cities Lisbon and Oporto new cases exceed 50.0 per 100,000 inhabitants (19) and MDR-TB occurs in excess of 15% of cases (6, 36). The clinical laboratory has a major role in the control of TB (4, 20), inasmuch as effective management of TB patients from areas that have high rates of MDR-TB is dependent on the rapid identification of Mycobacterium tuberculosis complex strains and their antibiotic susceptibility profiles (3, 35, 37). The role of the laboratory is even more critical for the management of AIDS patients who also have MDR-TB (3).
In Portugal, as is the case worldwide, the vast majority of M. tuberculosis complex strains with resistance to rifampin (RIF) are also resistant to isoniazid (INH), and although monoresistance to INH is common (25), monoresistance to RIF is rare (3, 6, 17, 24, 29). Thus, RIF resistance can be used for the identification of MDR-TB infections (8, 30). This makes it possible to treat MDR-TB patients aggressively (with four or five drugs) while sparing non-MDR-TB patients from areas with high MDR-TB frequencies from said therapy (3, 8, 21); a marked reduction in the frequency of noncompliance would consequently be expected (3, 8, 17, 21).
Ninety-five percent of M. tuberculosis strains with resistance to RIF contain distinct mutations located within an 81-bp (27- codon) region of the beta subunit of the RNA polymerase (rpoB) gene (29, 31). Several methods can rapidly detect these specific mutations and thereby identify RIF-resistant M. tuberculosis complex strains (14, 22, 31, 32). One of these methods is the line probe assay INNO-LiPA Rif.TB (Innogenetics, Zwijndrecht, Belgium), a commercially available kit not yet approved by the U.S. Food and Drug Administration which identifies M. tuberculosis complex strains and RIF resistance within a very short period of culture time (16, 26, 28). We have evaluated this assay for the identification of M. tuberculosis complex strains and the detection of mutations in the rpoB gene linked to RIF resistance directly from acid-fast smear-positive respiratory specimens obtained from patients who presented with tuberculosis (clinical symptoms and radiological evidence). The assays were performed in parallel with conventional isolation, identification, and susceptibility testing procedures routinely used in our mycobacteriology clinical laboratory as part of the “TB Fast Track Program,” modeled after that of the New York State Department of Health (7, 8, 27). This program is under the supervision of the TB Task Force of Greater Lisbon, a cooperative joint venture involving the major hospitals of the Greater Lisbon area (33). From September 2002 to September 2003, a total of 360 acid-fast positive respiratory specimens consisting of sputa (n = 318), bronchoalveolar lavage fluids (n = 23), and bronchial secretions (n = 19) from patients presenting with presumptive active TB were received in our laboratory; each specimen was accompanied by a physician-completed questionnaire that included pertinent patient demographics, clinical history, and MDR-TB risk factors. The patients, all from the Greater Lisbon area, ranged in age from 14 to 89 years (average, 42 years) and were mainly male (73.8%). The three major MDR-TB risk factors reported were, in order of importance, prior anti-TB treatment, contact with other MDR-TB patients, and origin from an area with a known high incidence of MDR-TB. The human immunodeficiency virus status was determined for only 150 patients (41.7%), and of these, 82 patients were coinfected with human immunodeficiency virus. Anti-TB treatment had already been initiated for 189 patients (52.5%) at the time of specimen collection.
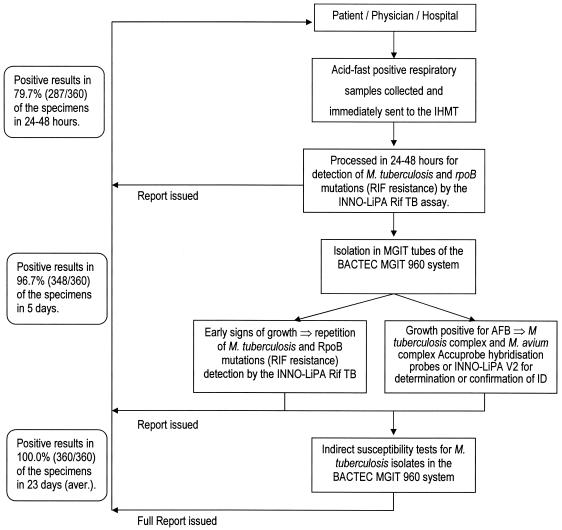
The TB Fast Track work algorithm, restricted to the work week of Monday to Friday, is summarized in Fig. 1. Briefly, all specimens received were processed by the conventional mycobacteriological NaOH-NALC method (15), and aliquots were collected for acid-fast staining (Ziehl-Neelsen stain), for inoculation of MGIT tubes employed by the BACTEC MGIT 960 system (Becton-Dickinson Diagnostic Instrument Systems, Towson, Md.) according to the manufacturer's instructions, and for extraction of total DNA with a QIAamp DNA mini kit (QIAGEN, GmbH, Hilden, Germany). The identification of M. tuberculosis complex and Mycobacterium avium complex strains present in full-grown cultures in the BACTEC MGIT 960 culture system was made with the aid of the Accuprobe system (Gen-Probe Inc., San Diego, California). A positive identification of M. tuberculosis was followed by susceptibility assays for RIF, INH, ethambutol, streptomycin, and pyrazinamide afforded by the BACTEC MGIT 960 system. The identification of M. tuberculosis and detection of rpoB gene mutations were made directly from acid-fast-bacillus (AFB)-positive respiratory specimens with the aid of the INNO-LiPA Rif.TB assay according to the following protocol, adapted for direct detection (5, 34). Five microliters of total DNA extract was used in a 50-μl PCR mixture containing the outer LiPA primers (Table 1), 1 microliter of the first-round product was transferred to a 50-μl second-round reaction mixture containing the inner primers biotinylated at the 5′ end (Table 1), and 10 microliters of the labeled PCR product was denatured and hybridized to membrane-bound capture-specific oligonucleotide probes, followed by a color detection step. One probe is specific for M. tuberculosis complex strains, five partially overlapping probes hybridize exclusively to the rpoB gene wild-type sequence, and four probes hybridize with amplicons carrying the following mutations: R2:D516V, R4a:H526Y, R4b:H526D, and R5:S531L (26). The absence of hybridization of one or more of the wild-type probes indicates a mutation that will be identified by hybridization with one of the mutation probes. The assay was used with appropriate controls, as described in previous evaluations (13, 30), namely, inclusion of a negative control containing no mycobacterial DNA (distilled sterile water) and a positive control containing DNA from an M. tuberculosis H37Rv (ATCC 27294) MGIT culture. An initial screening of the kit with nontuberculosis mycobacterial DNA samples (30) was also carried out. To control for potential inhibition of PCR amplification, a second separate aliquot of the extracted DNA (5 μl) served as the source for a parallel INNO-LiPA Rif.TB nested PCR supplemented with 5 μl of M. tuberculosis H37Rv genomic DNA. Negative amplification results in the presence of the spiked DNA were considered to be due to inhibition of amplification. For those specimens where no amplification was obtained by the direct application of the INNO-LiPA Rif.TB assay to the AFB-positive specimens, the amplification procedure was repeated as soon as a positive growth index was detected in the BACTEC MGIT 960 system tubes. If no amplification occurred, then the AFB present in the culture were subjected to the nucleic acid amplification (NAA) INNO-LiPA MYCOBACTERIA v2 assay (Innogenetics), which identifies 16 nontuberculosis mycobacterial strains (20). The reference BACTEC 460 TB system (Becton-Dickinson) was used when samples presented a RIF susceptibility status that differed from that provided by the assay (12). Preliminary reports were sent to the physician at the completion of each phase, and a full report was issued after the completion of the conventional isolation, identification, and susceptibility tests (Fig. 1). Each specimen identified as having a mutation in the rpoB gene was subjected to sequence analysis of both DNA strands of a 350-bp fragment of the rpoB gene (24).
FIG. 1.
TB Fast Track work algorithm. IHMT, Instituto de Higiene e Medicina Tropical; aver., average.
TABLE 1.
Primers and conditions used for this studya
| Primer | Primer sequence (5′-3′) | References |
|---|---|---|
| LIPA-Outerprimer1 | GAGAATTCGGTCGGCGAGCTGATCC | 5, 34 |
| LIPA-Outerprimer2 | CGAAGCTTGACCCGCGCGTACACC | 5, 34 |
| LIPA-Innerprimer1 | GGTCGGCATGTCGCGGATGG | 5, 34 |
| LIPA-Innerprimer2 | GCACGTCGCGGACCTCCAGC | 5, 34 |
| RPOB-1 | GGGAGGGGATGACCACCCA | 13, 24 |
| RPOB-2 | GCGGTACGGCGTTTCGATGAAC | 13, 24 |
The INNO-LiPA Rif.TB thermal cycling conditions were as follows. The first- and second-round PCRs consisted of 40 cycles of denaturation (95°C for 30 s), annealing (62°C for 30 s), and extension (72°C for 30 s). The rpoB amplification thermal cycling conditions consisted of 30 cycles of denaturation (94°C for 60 s), annealing (60°C for 60 s), and extension (72°C for 60 s).
Only the primary results, with no adjustment of false-negative and false-positive results, were used for calculating sensitivity, specificity, and positive and negative predictive values for the INNO-LiPA Rif.TB assay. The isolation of M. tuberculosis complex strains in culture and identification by the Accuprobe M. tuberculosis complex DNA probe served as the “gold standard” for comparison to the results obtained with the INNO-LiPA Rif.TB assay. The susceptibility to RIF obtained with the BACTEC MGIT 960 system served as the “gold standard” for comparison to that obtained with the INNO-LiPA Rif.TB assay.
A total of 287 (79.7%) of the 360 acid-fast positive respiratory specimens assayed gave positive amplification results, of which 281 were identified as true-positive results by the culture isolation and M. tuberculosis Accuprobe identification procedures (Table 2). Retroactive reviews of the medical records of the six patients whose specimens yielded false-positive results revealed that all six patients were confirmed as positive for TB by clinical criteria and that at least three patients presented evidence of recent or current anti-TB therapy. Although these specimens could have been considered true-positive results for the final diagnosis of tuberculosis, for the above-defined assay performance evaluation compared to the “gold standard” afforded by Accuprobe technology, they were considered false-positive results. Of the 73 specimens that were identified by the INNO-LiPA Rif.TB assay as negative for the presence of M. tuberculosis, 12 were shown to be true-negative results by culture isolation/M. tuberculosis Accuprobe identification. Of these 12 specimens, 2 were identified as containing M. avium complex isolates, 1 was identified as containing Mycobacterium chelonae, and the remaining 9 were negative for any mycobacteria. These last nine specimens came from patients whose radiological work-ups did not support a diagnosis of active TB. For the remaining 61 false-negative specimens, the nested PCR was repeated with the use of the DNA spike. Nineteen of these false-negative specimens yielded acid-fast smears with very small numbers of bacilli, and no DNA was detectable after extraction. For the remaining 42 specimens, extracted DNAs were visualized by agarose gel electrophoresis, and the repetition of the nested PCR revealed inhibition of amplification for 32 of the 42 specimens. At this time, we are unable to explain the negative results for amplification of the remaining 10 specimens. Nevertheless, the 61 specimens were considered false-negative specimens for the identification of the M. tuberculosis complex by the INNO-LiPA Rif.TB assay (Table 2). The assay yielded a sensitivity of 82.2% and a specificity of 66.7% for the direct identification of M. tuberculosis complex strains. The positive and negative predictive values for the identification of the M. tuberculosis complex were 97.9% and 16.4%, respectively (Table 2). This performance is compatible with that of other commercially available NAA assays certified for direct application to smear-positive respiratory samples (8, 9, 18).
TABLE 2.
Correlation of the INNO-LiPA Rif.TB method for direct identification of M. tuberculosis complex in acid-fast-bacillus-positive respiratory specimens with conventional isolation in culture (BACTEC 960 system) followed by Accuprobe M. tuberculosis complex identification
| INNO-Lipa Rif.TB test result for M. tuberculosisb | No. of specimens with correlation to conventional test resulta
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 281 | 6 | 287 |
| Negative | 61 | 12 | 73 |
| Total | 342 | 18 | 360 |
Identification by Accuprobe M. tuberculosis complex probes and conventional biochemical tests with isolated cultures.
Sensitivity of INNO-LiPA Rif.TB direct identification compared to M. tuberculosis complex identification after isolation in culture, 82.2%; specificity, 66.7%; positive predictive value, 97.9%; negative predictive value, 16.4%.
All M. tuberculosis complex isolates (n = 342) were subjected to susceptibility assays with the five first-line anti-TB drugs, and the overall resistance patterns are summarized in Table 3. Among the drug-resistant isolates, 8.8% were multidrug resistant (INH plus RIF), and 5.6% were resistant to all five drugs (Table 3). The correlation of the RIF resistance/susceptibility results obtained by standard indirect susceptibility testing with those obtained with the INNO-LiPA Rif.TB kit is summarized in Table 4. Thirty-one M. tuberculosis complex isolates shown to be resistant to RIF by the standard susceptibility test were detected by the INNO-LiPA Rif.TB assay as carrying the following mutations: S531L (29 isolates), H526Y, and D516V (1 isolate each). Sequencing of the rpoB gene region confirmed the presence of the detected point mutations. The INNO-LiPA Rif.TB assay failed to detect any mutation in one sample that was repeatedly identified as RIF resistant by both the BACTEC MGIT 960 and BACTEC 460TB systems. No point mutation in the rpoB gene was detected by sequencing, and thus this phenotype must be due to another, as yet undescribed cause (10, 29, 31). The predominant mutation detected in the rpoB gene is S531L, the one most frequently detected in European and Asian countries (2, 11, 13, 16, 26).
TABLE 3.
Resistance patterns of the 342 M. tuberculosis complex isolates obtained in this study
| Resistance patterna | No. of isolates |
|---|---|
| HRSEZ | 19 |
| HRSE | 1 |
| HRSZ | 2 |
| HREZ | 5 |
| HRS | 2 |
| HR | 1 |
| SE | 1 |
| HS | 10 |
| H | 6 |
| R | 2 |
| S | 33 |
| Susceptible to all five drugs | 260 |
| Total | 342 |
H, isoniazid; R, rifampin; S, streptomycin; E, ethambutol; and Z, pyrazinamide. Resistance was determined with the indirect drug susceptibility assay of the BACTEC MGIT 960 system.
TABLE 4.
Correlation of INNO-LiPA Rif.TB and standard susceptibility tests in the BACTEC 960 system
| INNO-Lipa Rif.TB test result for RIF resistance of M. tuberculosis complexb | No. of isolates with correlation to standard test resulta
|
||
|---|---|---|---|
| Resistant | Susceptible | Total | |
| Resistant | 31 | 0 | 31 |
| RIF Susceptible | 1 | 310 | 311 |
| Total | 32 | 310 | 342 |
Susceptibility was determined by the BACTEC MGIT 960 system's indirect susceptibility assay and confirmed by the BACTEC 460TB system's indirect susceptibility assay when necessary.
Sensitivity of INNO-LiPA Rif.TB M. tuberculosis complex direct detection of rpoB gene mutations that confer RIF resistance in acid-fast-bacillus-positive respiratory specimens compared to RIF resistance determined by standard indirect susceptibility assays, 96.9%; specificity, 100.0%; positive predictive value, 100.0%; negative predictive value, 99.7%.
All 310 RIF-susceptible isolates were correctly identified by the INNO-LiPA Rif.TB assay as being negative for mutations in the rpoB gene. The specificity and sensitivity of the INNO-LiPA Rif.TB assay for the detection of the RIF resistance profile were 100.0% and 96.9%, respectively. The positive and negative predictive values obtained were 100.0% and 99.7%, respectively (Table 4), in agreement with those obtained by others (13, 30). The INNO-LiPA Rif.TB PCR-based hybridization assay, adjusted for the use of a nested PCR to increase its sensitivity, rapidly identifies M. tuberculosis and its resistance to RIF. The assay is simple, convenient, cost-effective, and highly reliable when run in parallel with a conventional TB laboratory diagnostic algorithm. It directly identified 79.7% of the samples within 24 to 48 h of the arrival of the specimens. This percentage was raised to 96.7% with the application of the same amplification and detection protocol to growing cultures of specimens with initial negative direct amplification results. The major limitations found were (i) false-negative results due to inhibition of amplification, (ii) the absence of an internal amplification control, and (iii) the fact that this assay, as well as other NAA assays, identifies DNA from dead mycobacteria from patients undergoing therapy. Regarding the RIF resistance profile, the assay yielded results that were 96.9% in agreement with those obtained much later by the use of a culture-based susceptibility method. The ability to rapidly identify an MDR-TB-type infection affords the effective management of MDR-TB, reduces the frequency of noncompliance since patients that do not have MDR-TB can be treated less aggressively, and therefore contributes to the control of TB.
Acknowledgments
M.V. and C.L. contributed equally to this study.
We are grateful for the participation of the physicians and laboratory personnel from hospitals involved in the TB Task Force Program for Greater Lisbon, especially to our colleagues Gabriela Abreu (Hospital do Barreiro), Isabel Oliveira (Hospital Cascais), Judite Batista (Hospital Egas Moniz), Clara Portugal (Hospital Amadora-Sintra), Ana Rodrigues (Hospital de Almada), Cāndida Canha (Hospital Pulido Valente), Maria José Salgado and Carvalho de Sousa (Hospital Santa Maria), Jesuina Duarte (Hospital de Setúbal), and Filomena Martins (Hospital São Francisco Xavier). We give special thanks to Francisco Antunes, Jaime Pina, José Moniz-Pereira, Fernando Ventura, and Emília Valadas for their support, encouragement, and participation in the local and nationwide meetings held by the TB Task Force of Greater Lisbon.
This study was supported by grant SDH.IC.I.01.17-TB from the Task Force for Greater Lisbon and by the TB-Fast-Track Program from the Calouste Gulbenkian Foundation. We thank the Quilaban Corporation for generous technical support.
REFERENCES
- 1.Antunes, M. L., J. Aleixo-Dias, A. F. Antunes, M. F. Pereira, F. Raymundo, and M. F. Rodrigues. 2000. Anti-tuberculosis drug resistance in Portugal. Int. J. Tuberc. Lung Dis. 4:223-231. [PubMed] [Google Scholar]
- 2.Bartfai, Z., A. Somoskovi, C. Kodmon, N. Szabo, E. Puskas, L. Kosztolanyi, E. Farago, J. Mester, L. M. Parsons, and M. Salfinger. 2001. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 39:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1993. Initial therapy for tuberculosis in the era of multidrug resistance: recommendations of the Advisory Council for the Elimination of Tuberculosis. Morb. Mortal. Wkly. Rep. 42:1-8. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2002. TB elimination cooperative agreement. Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.De Beenhouwer, H., Z. Lhiang, G. Jannes, W. Mijs, L. Machtelinckx, R. Rossau, H. Traore, and F. Portaels. 1995. Rapid detection of rifampin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber. Lung Dis. 76:425-430. [DOI] [PubMed] [Google Scholar]
- 6.Direcção Geral de Saúde. 2004. Programa Nacional de Luta contra a Tuberculose (PNT). Ponto da situação epidemiológica e de desempenho ano 2003, p. 1-12. DGS, Lisboa, Portugal.
- 7.Frieden, T. R., T. Sterling, A. Pablos-Mendez, J. O. Kilburn, G. M. Cauthen, and S. W. Dooley. 1993. The emergence of drug-resistant tuberculosis in New York City. N. Engl. J. Med. 328:521-526. [DOI] [PubMed] [Google Scholar]
- 8.Hale, Y. M., E. P. Desmond, K. C. Jost, Jr., and M. Salfinger. 2000. Access to newer laboratory procedures: a call for action. Int. J. Tuberc. Lung Dis. 4:171-175. [PubMed] [Google Scholar]
- 9.Hale, Y. M., G. E. Pfyffer, and M. Salfinger. 2001. Laboratory diagnosis of mycobacterial infections: new tools and lessons learned. Clin. Infect. Dis. 33:834-846. [DOI] [PubMed] [Google Scholar]
- 10.Heep, M., B. Brandstatter, U. Rieger, N. Lehn, E. Richter, S. Rusch-Gerdes, and S. Niemann. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano, K., C. Abe, and M. Takahashi. 1999. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J. Clin. Microbiol. 37:2663-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inderlied, C. B., and K. A. Nash. 1996. Antimycobacterial agents: in vitro susceptibility testing, spectra of activity, mechanisms of action and resistance, and assays for activity in biologic fluids, p. 127-175. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams and Wilkins, Baltimore, Md.
- 13.Johansen, I. S., B. Lundgren, A. Sosnovskaja, and V. O. Thomsen. 2003. Direct detection of multidrug-resistant Mycobacterium tuberculosis in clinical specimens in low- and high-incidence countries by line probe assay. J. Clin. Microbiol. 41:4454-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapur, V., L. L. Li, S. Iordanescu, M. R. Hamrick, A. Wanger, B. N. Kreiswirth, and J. M. Musser. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Ga.
- 16.Matsiota-Bernard, P., G. Vrioni, and E. Marinis. 1998. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J. Clin. Microbiol. 36:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchison, D. A. 1998. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int. J. Tuberc. Lung Dis. 2:10-15. [PubMed] [Google Scholar]
- 18.Noordhoek, G. T., J. D. A. Van Embden, and A. H. J. Kolk. 1996. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J. Clin. Microbiol. 34:2522-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ordway, D. J., L. Costa, M. Martins, H. Silveira, L. Amaral, M. J. Arroz, F. Ventura, and H. M. Dockrell. 2004. Increased interleukin-4 production by CD8 and gammadelta T cells in health-care workers is associated with the subsequent development of active tuberculosis. J. Infect. Dis. 190:756-766. [DOI] [PubMed] [Google Scholar]
- 20.Padilla, E., V. González, J. M. Manterola, A. Pérez, M. D. Quesada, S. Gordillo, C. Vilaplana, M. A. Pallarés, S. Molinos, M. D. Sánchez, and V. Ausina. 2004. Comparative evaluation of the new version of the INNO-LiPA Mycobacteria and GenoType Mycobacterium assays for identification of Mycobacterium species from MB/BacT liquid cultures artificially inoculated with mycobacterial strains. J. Clin. Microbiol. 42:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons, L. M., A. Somoskovi, R. Urbanczik, and M. Salfinger. 2004. Laboratory diagnostic aspects of drug resistant tuberculosis. Front. Biosci. 9:2086-2105. [DOI] [PubMed] [Google Scholar]
- 22.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 23.Portugal, I., M. J. Covas, L. Brum, M. Viveiros, P. Ferrinho, J. Moniz-Pereira, and H. David. 1999. Outbreak of multiple drug-resistant tuberculosis in Lisbon: detection by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3:207-213. [PubMed] [Google Scholar]
- 24.Portugal, I., S. Maia, and J. Moniz-Pereira. 1999. Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J. Clin. Microbiol. 37:3022-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridzon, R., C. G. Whitney, M. T. McKenna, J. P. Taylor, S. H. Ashkar, A. T. Nitta, S. M. Harvey, S. Valway, C. Woodley, R. Cooksey, and I. M. Onorato. 1998. Risk factors for rifampin mono-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 157:1881-1884. [DOI] [PubMed] [Google Scholar]
- 26.Rossau, R., H. Traore, H. De Beenhouwer, W. Mijs, G. Jannes, P. De Rijk, and F. Portaels. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salfinger, M. 1977. Diagnosis of tuberculosis and other diseases caused by mycobacteria. Infection 25:60-62. [DOI] [PubMed] [Google Scholar]
- 28.Sharma, M., S. Sethi, B. Mishra, C. Sengupta, and S. K. Sharma. 2003. Rapid detection of mutations in rpoB gene of rifampin resistant Mycobacterium tuberculosis strains by line probe assay. Ind. J. Med. Res. 117:76-80. [PubMed] [Google Scholar]
- 29.Somoskovi, A., L. M. Parsons, and M. Salfinger. 2001. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somoskovi, A., Q. Song, J. Mester, C. Tanner, Y. M. Hale, L. M. Parsons, and M. Salfinger. 2003. Use of molecular methods to identify the Mycobacterium tuberculosis complex (MTBC) and other mycobacterial species and to detect rifampin resistance in MTBC isolates following growth detection with the BACTEC MGIT 960 system. J. Clin. Microbiol. 41:2822-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 32.Victor, T. C., A. M. Jordaan, A. van Rie, G. D. van der Spuy, M. Richardson, P. D. van Helden, and R. Warren. 1999. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuber. Lung Dis. 79:343-348. [DOI] [PubMed] [Google Scholar]
- 33.Viveiros, M., and the TB Task Force for Greater Lisbon. 2003. The Fast-Track TB Programme—preliminary results. Rev. Port. Dis. Infect. 2:28. [Google Scholar]
- 34.Watterson, S. A., S. M. Wilson, M. D. Yates, and F. A. Drobniewski. 1998. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 36:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 1998. The W.H.O./IUTALD Global Project on Anti-Tuberculosis Drug Resistance Surveillance 1994-1997, p. 1-227. World Health Organization, Geneva, Switzerland.
- 36.World Health Organization. 2003. Surveillance of tuberculosis in Europe. EuroTB (InVS/KNCV). Draft report on tuberculosis cases notified in 2002, p. 1-18. World Health Organization, Geneva, Switzerland.
- 37.World Health Organization. 2005. Global tuberculosis control—surveillance, planning, financing. W.H.O. report 2005, p. 1-247. World Health Organization, Geneva, Switzerland.