Abstract
IS6110 fingerprinting of Mycobacterium tuberculosis is the standard identification method in studies on transmission of tuberculosis. However, intensive epidemiological investigation may fail to confirm transmission links between patients clustered by IS6110-restriction fragment length polymorphism (RFLP) typing. We applied typing based on variable numbers of tandem repeats (VNTRs) of mycobacterial interspersed repetitive units (MIRUs) to isolates from 125 patients in 42 IS6110 clusters, for which thorough epidemiological data were available, to investigate the potential of this method in distinguishing epidemiologically linked from nonlinked patients. Of seven IS6110 clusters without epidemiological links, five were split by MIRU-VNTR typing, while nearly all IS6110 clusters with proven or likely links displayed conserved MIRU-VNTR types. These results provide molecular evidence that not all clusters determined on the basis of multibanded IS6110 RFLP patterns necessarily reflect transmission of tuberculosis. They support the use of MIRU-VNTR typing as a more reliable and faster method for transmission analysis.
IS6110 restriction fragment length polymorphism (RFLP) typing of Mycobacterium tuberculosis has been used extensively in studies on tuberculosis transmission and is one of the most widely applied and standardized molecular typing methods (1, 6, 10, 31, 34, 35). M. tuberculosis isolates from epidemiologically linked patients generally show identical IS6110 RFLP patterns, thus comprising transmission clusters. Consequently, the finding that a substantial proportion of tuberculosis cases in industrialized countries is clustered by DNA fingerprinting is interpreted as a reflection of a high rate of recent transmission (2, 5, 8, 14, 22, 27, 28, 32, 39). However, IS6110-based RFLP fingerprints are not always reliable indicators of epidemiological linkage between tuberculosis patients (7, 33). Even the most meticulous analysis of all available data on possible contacts between clustered patients does not reveal epidemiological links in all cases (33). Furthermore, because in many settings tuberculosis often results from casual contacts, the majority of the links can be identified only after combining the genotyping of the M. tuberculosis isolates with intensive epidemiologic investigation. In addition, opportunities for early and thus more-efficient prevention and intervention are limited by the fact IS6110 RFLP typing is labor intensive and requires weeks for culturing M. tuberculosis.
Typing methods based on mycobacterial interspersed repetitive unit (MIRU)-VNTR analysis offer a potential solution to the drawbacks faced using IS6110 RFLP typing. MIRU-VNTR analysis determines the number of tandem repeats at multiple independent loci (12, 29). This PCR-based method is highly reproducible and much faster than IS6110-RFLP typing and displays a discriminatory power close to that of IS6110-RFLP, especially in low-incidence areas (9, 15, 18, 23, 25, 30). Previous studies have demonstrated the ability of MIRU-VNTR typing to split certain IS6110 clusters, suggesting that the use of IS6110 alone may overestimate the existence of transmission clusters (4, 9, 13, 15, 19, 21, 23). Therefore, MIRU-VNTR typing has been proposed as an efficient first-line typing method, to be followed by IS6110-RFLP subtyping of the resulting MIRU-VNTR clusters (9, 25). However, all but one of these observations were made on the basis of M. tuberculosis isolates harboring few copies of IS6110, for which IS6110 RFLP typing is known to have limited discriminatory power. Furthermore, epidemiological information on the clustered cases was limited.
To further evaluate the usefulness of MIRU-VNTR typing for conducting population-based studies of recent transmission, we applied secondary MIRU-VNTR typing and spoligotyping (17) to isolates from patients in IS6110 RFLP clusters selected from a previous population-based study, for whom extensive and well-structured epidemiological data were available (33). Our aim was to determine in a representative manner the potential of MIRU-VNTR typing to distinguish epidemiologically linked patients from unlinked patients in IS6110 RFLP clusters.
MATERIALS AND METHODS
Study population.
In a prospective, population-based study conducted in the province of North Holland, The Netherlands (population, 2,500,000), we previously investigated tuberculosis cases for which DNA fingerprint clustering was an indication of epidemiological linkage and therefore of recent transmission (33). Furthermore, we determined which of these patients could have been detected earlier by contact tracing. The study included all tuberculosis patients residing in the study area and reported to the Municipal Health Services, as is mandatory in The Netherlands. Of 664 patients included from 1 July 1998 to 1 July 2000, 483 (73%) had a positive M. tuberculosis culture. Patients who were part of an IS6110 cluster with one or more other patients from North Holland and diagnosed within the 2-year period preceding or following such patients' diagnosis (n = 155) were assigned to transmission groups (TG) according to the likelihood of epidemiological linkage between patients. The assignments were based on the results of extensive interviews of the patients before and after the results of RFLP typing became available, combined with sociodemographic and clinical data.
For the present study, the isolates obtained from 114 of the 155 patients in IS6110 clusters were subtyped using MIRU-VNTR. Isolates to be subtyped were chosen based on the availability of DNA at the mycobacteria laboratory at the Diagnostic Laboratory for Infectious Diseases and Perinatal Screening of the National Institute of Public Health and the Environment; the 114 patients did not differ significantly from the other 41 patients with respect to age, sex, or Dutch versus non-Dutch ethnicity and nationality (t test and χ2 test; P > 0.05). In addition, isolates from 11 patients from outside the above-described study period were included when a cluster was represented by only a single isolate within the study period. In the resulting sample of 125, assignments into the various transmission groups differed slightly between this study and the previous one because of the inclusion of these 11 isolates. The number of isolates subtyped by MIRU-VNTR per TG and the description of the epidemiological link belonging to each TG are depicted in Table 1.
TABLE 1.
Characteristics of the TG classified in this study
| TG | No. of patients | Description |
|---|---|---|
| 1 | 44 | Patients with a clear epidemiological link, confirmed by IS6110-PGRS typing |
| 2 | 13 | Patients with links as clear as those in TG1, confirmed by IS6110-PGRS typing, who should have been, but were not, detected by contact tracing |
| 3 | 54 | Patients with an epidemiological link which was initially unclear but became likely after IS6110-PGRS typing and the second interview |
| 4 | 14 | Patients for whom meticulous analysis of all available data ruled out an epidemiological link but whose IS6110-PGRS typing results suggested that they belonged to a certain cluster |
IS6110/PGRS RFLP and spoligotyping.
All isolates were subjected to IS6110 RFLP typing, as described previously (34). When strains harbored fewer than five IS6110 copies, subtyping using the polymorphic GC-rich tandem repeat (PGRS) was performed as described earlier (36). Isolates were considered to belong to a cluster when no differences were found in IS6110 or PGRS-banding patterns. Spoligotyping was performed according to a previously described method (17). Computer-assisted analysis of IS6110-PGRS RFLP patterns and spoligotyping patterns was done using Bionumerics software, version 3.5 for Windows (Applied Maths, Kortrijk, Belgium), as described previously (16, 37).
VNTR analysis.
MIRU-VNTR typing relies on PCR amplification of different MIRU-VNTR regions by use of primers specific for the flanking regions of these MIRUs and on the determination of the sizes of the amplicons, which reflect the numbers of the amplified MIRU copies (12, 20, 25, 29). The 125 M. tuberculosis isolates were genotyped by multiplex PCR amplification as described previously (30). The MIRU loci used in this study correspond to a subset of 6 out of 12 previously defined MIRU loci containing VNTRs (29) and 6 additional loci containing VNTRs of other interspersed sequences (12, 20, 24). The VNTRs used in this study are shown in Table 2. They were selected from a wider set of loci on the basis of their variability in unrelated isolates, stability in clonally related isolates, and potential for robust PCR amplification and reliable size analysis (P. Supply, unpublished data). In this report, they are collectively designated MIRU-VNTRs. Briefly, the target genetic sequences were amplified using fluorescently labeled primers and 40 PCR cycles (26, 30). The samples were subjected to electrophoresis using a 96-well ABI 377 automated sequencer. Sizing of the PCR fragments and assignment of the various VNTR alleles in the 12 loci were done using the GeneScan and customized Genotyper software packages (PE Applied Biosystems). The genotypes are expressed as a numerical code representing the number of MIRU-VNTRs in each of the 12 loci.
TABLE 2.
Conditions for multiplex PCRs of 12 MIRU-VNTR loci
| Multiplex | Locusa | VNTR length (bp) | MgCl2 concn (mM) | PCR primer pairs (5′ to 3′, with labeling indicated)b | Reference |
|---|---|---|---|---|---|
| Mix A | VNTR 0580 | 77 | 3.0 | GCGCGAGAGCCCGAACTGC (FAM) | 30 |
| GCGCAGCAGAAACGTCAGC | |||||
| VNTR 2996 | 51 | 3.0 | TAGGTCTACCGTCGAAATCTGTGAC | 30 | |
| CATAGGCGACCAGGCGAATAG (VIC) | |||||
| VNTR 0802 | 54 | 3.0 | GGGTTGCTGGATGACAACGTGT (NED) | 30 | |
| GGGTGATCTCGGCGAAATCAGATA | |||||
| Mix B | VNTR 0960 | 53 | 2.0 | GTTCTTGACCAACTGCAGTCGTCC | 30 |
| GCCACCTTGGTGATCAGCTACCT (FAM) | |||||
| VNTR 1644 | 53 | 2.0 | TCGGTGATCGGGTCCAGTCCAAGTA | 30 | |
| CCCGTCGTGCAGCCCTGGTAC (VIC) | |||||
| VNTR 3192 | 53 | 2.0 | ACTGATTGGCTTCATACGGCTTTA | 30 | |
| GTGCCGACGTGGTCTTGAT (NED) | |||||
| Mix F | VNTR 0424 | 51 | 1.5 | CTTGGCCGGCATCAAGCGCATTATT | 38 |
| GGCAGCAGAGCCCGGGATTCTTC (FAM) | |||||
| VNTR 0577 | 58 | 1.5 | CGAGAGTGGCAGTGGCGGTTATCT (VIC) | 38 | |
| AATGACTTGAACGCGCAAATTGTGA | |||||
| Mix G | VNTR 2401 | 58 | 3.0 | CTTGAAGCCCCGGTCTCATCTGT (FAM) | 38 |
| ACTTGAACCCCCACGCCCATTAGTA | |||||
| VNTR 3690 | 58 | 3.0 | CGGTGGAGGCGATGAACGTCTTC (VIC) | 38 | |
| TAGAGCGGCACGGGGGAAAGCTTAG | |||||
| VNTR 4156 | 59 | 3.0 | TGACCACGGATTGCTCTAGT | 38 | |
| GCCGGCGTCCATGTT (NED) | |||||
| Individual | VNTR 1982 | 78 | 1.5 | CCGGAATCTGCAATGGCGGCAAATTAAAAG | 26 |
| TGATCTGACTCTGCCGCCGCTGCAAATA (FAM) |
Locus designation according to the position (in kilobase pairs) on the M. tuberculosis H37Rv chromosome. VNTR 00580, 2996, 0802, 0960, 1644, and 3192 correspond to MIRU locus 4 (alias ETRD), 26, 40, 10, 16, and 31 (alias ETRE), respectively (12, 29). VNTR 577, 1982, and 4156 correspond to ETRC, QUB-18, and QUB 4156, respectively (12, 20, 24).
FAM, 6-carboxyfluorescein.
RESULTS
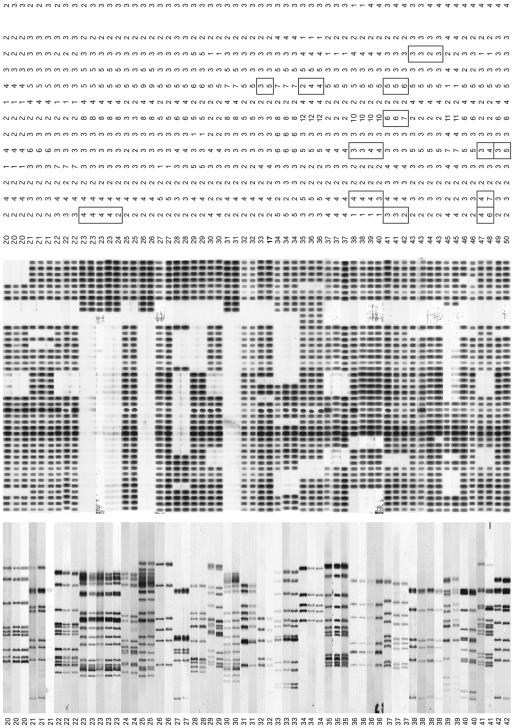
The results of secondary MIRU-VNTR typing of the M. tuberculosis isolates from 125 patients, previously assigned to 42 IS6110 RFLP clusters, are shown in Fig. 1 and Table 3.
FIG. 1.
IS6110-PGRS RFLP and MIRU-VNTR genotypes of the M. tuberculosis isolates from 125 patients in 42 IS6110-PGRS clusters, as assigned to four transmission groups (for explanation of transmission groups, see Table 1). Designations of MIRU-VNTR loci are given according to the position (in kilobase pairs) on the M. tuberculosis H37Rv chromosome. Alias designations are in parentheses. Spoligo, spoligotyping.
TABLE 3.
Comparison of discriminatory power of MIRU-VNTR typing versus IS6110-PGRS RFLP typing in relation to epidemiological data
| Epidemiological link between clustered patients (transmission group) | IS6110 clusters involveda | No. of patients | No. of IS6110-PGRS clusters | No. (%) of IS6110-PGRS clusters split by MIRU-VNTR typing |
|---|---|---|---|---|
| Evident link (TG1 and TG2) | 1-23, 36 | 57 | 24 | 1 (4.2) |
| Likely link (TG3) | 15-35, 37, 38 | 54 | 23 | 3 (13) |
| No link established (TG4) | 36-42 | 14 | 7 | 5 (71.4) |
See Fig. 1 for designated cluster numbers.
Isolates from a total of 125 patients were examined.
Among the 24 IS6110-PGRS clusters comprising 57 patients with a proven epidemiological link (TG1 and TG2), only one (cluster 8), including two patients, was subdivided by MIRU-VNTR typing. Of the 23 IS6110-PGRS clusters comprising 54 patients with a likely epidemiological link (TG3), three (clusters 23, 32, and 34) were subdivided by MIRU-VNTR typing. In clusters 23 and 34, one isolate was distinguished from four and two other isolates, respectively, while cluster 32 included two isolates subdivided by MIRU-VNTR typing. In all cases where IS6110-PGRS clusters of TG1, TG2, or TG3 were split by MIRU-VNTR typing, the differences were minor and always concerned only one MIRU locus.
In contrast, of the seven IS6110 clusters without established epidemiological links (TG4), five (71.4%) were split by MIRU-VNTR typing. This difference in the proportion of subdivided clusters is highly significant (TG4 versus TG1, TG2, and TG3, P = 0.00097 [Fishers' exact test]). MIRU-VNTR typing identified a total of 12 genotypes among isolates from 14 patients in these groups.
As an average, two MIRU-VNTR loci changed in each of the five TG4 clusters subdivided by MIRU-VNTR typing. This number was thus higher than the number of changes observed for the TG1, TG2, and TG3 clusters that were split by MIRU-VNTR typing (Fig. 1).
In total, 9 (21%) of the 42 IS6110-PGRS RFLP clusters were subdivided by MIRU-VNTR typing. The numbers of IS6110 RFLP bands in these isolates ranged from 5 to 15.
Conversely, two examples (between clusters 9 and 17 and between cluster 16 and one isolate of cluster 32) were found in which isolates with distinct IS6110-PGRS RFLP patterns (59.4 and 64.1% similarity, respectively) exhibited identical VNTR patterns (Fig. 1). These isolates also exhibited distinct spoligotyping patterns. For these isolates the discriminative power of IS6110 RFLP typing and spoligotyping was thus higher than that of the MIRU-VNTR typing.
In contrast, all isolates within the same IS6110-PGRS cluster exhibited identical spoligotyping patterns, except for isolates in cluster 38. One of the two TG4 isolates in cluster 38 contained one fewer direct repeat spacer than the other isolates of that IS6110 cluster. This isolate had a MIRU-VNTR type identical to that of the two other isolates within the same IS6110 cluster.
Comparison of the spoligotypes and IS6110 fingerprints detected in this study with those in the international spoligotype and IS6110-RFLP databases revealed that the corresponding isolates belong to a wide variety of genotype families (11, 26). These families were distributed to identical degrees among the different “transmission groups.” All IS6110 clusters, except clusters 1, 15, 32, and 34, displayed IS6110 profiles consisting of six or more bands.
DISCUSSION
Molecular typing using IS6110 has generally been used as the standard method in studies of the transmission of tuberculosis, on the assumption that IS6110 RFLP-based clustering of cases is the result of recent transmission. However, even meticulous analysis of all epidemiological data fails to confirm transmission links among a number of patients within IS6110-PGRS RFLP clusters. For 14% of patients grouped into IS6110-PGRS clusters in our previous study, an epidemiological link was virtually ruled out after two consecutive series of extensive interviews of the patients (33).
Failures to detect recent transmission chains indicated by IS6110-PGRS clusters are classically attributed to the limited sensitivity of conventional contact investigation, but our findings suggest that some reflect limitations in the resolution of IS6110-PGRS RFLP. In the present study, MIRU-VNTR typing was found to be the most accurate first-line method to exclude epidemiological links.
In our study, 71.4% (5/7) of the IS6110-PGRS clusters comprising patients for whom an epidemiological link was not detected (TG4) and whose isolates showed RFLP fingerprints with high copy numbers of IS6110 were subdivided by MIRU-VNTR typing. For these patients, meticulous analysis of all data, including the results of a second interview, indicated that they were unlikely to have met during the infectious period of the index patient. Examples include two immigrants from the same country, one with extrapulmonary tuberculosis diagnosed in 1998 after living in The Netherlands for 10 years and the other with infectious pulmonary tuberculosis that was diagnosed at immigration in 2000. Although a common but unidentified source for the two TG4 strains with matching MIRU-VNTRs cannot be ruled out completely, the thorough analysis of our epidemiological data makes this event less likely. The resolution of the other TG4 clusters by MIRU-VNTR typing fully corroborates the absence of a direct epidemiological link indicated by intensive contact investigation.
Conversely, in contrast to TG4, almost all patients in IS6110-PGRS clusters with an evident (TG1 and TG2) or likely (TG3) epidemiological link remained clustered after MIRU-VNTR typing. In addition, in the few IS6110-PGRS clusters in TG1, TG2, and TG3 that were split by MIRU-VNTR typing, the MIRU-VNTR types differed only by a single MIRU locus, compared to differences in up to three loci for the TG4 clusters split by MIRU-VNTR. TG1 and TG2 included relatives or close friends of an infectious index patient who were identified by contact tracing (TG1) or who should have been—but were not—identified by that intervention (TG2). The latter group includes persons in close contact with an index patient but not mentioned by him or her, persons not reached for the contact investigation by the Municipal Health Service, and persons not complying with that investigation. Only one IS6110-PGRS TG1 cluster, including two patients, was subdivided by MIRU-VNTR typing out of the 24 clusters comprising a total of 57 patients in TG1 and TG2. This latter observation most likely reflects a rare and stochastic MIRU-VNTR mutation event, possibly DNA replication or homologous recombination dependent, in clonal populations originating from recent transmission (30). This hypothesis is consistent with the low proportion of changes in MIRU-VNTR profiles observed among serial isolates from chronically infected patients (25).
In TG3, including 54 patients in 23 IS6110-PGRS clusters, an epidemiological link which was initially unclear became likely after IS6110 typing and a second interview. This group includes patients living in the same apartment complex or regularly using the same tram service as an index patient, as well as homeless persons who used housing facilities with an index patient before he or she was diagnosed and thereafter became untraceable. Only 3 of these 54 patients were not matched by identical MIRU-VNTR patterns within their three respective clusters. Notably, while one of these three clusters included isolates with 17 IS6110 copies (cluster 23), the two others (clusters 32 and 34) included isolates with only 5 IS6110 copies. Whereas the MIRU-VNTR difference in cluster 23 also probably reflects a rare MIRU-VNTR variation between clonally transmitted isolates (see above), another explanation may be possible for the differences observed in clusters 32 and 34. As noted earlier, a total of five IS6110 copies is considered the upper limit in defining the low-copy-number isolates. For such isolates, IS6110 RFLP is thought to be of limited specificity for detection of recent transmission even when complemented by spoligotyping and PGRS typing. Therefore, the possibility is not totally excluded that the patients whose low-copy-number isolates were distinguished by MIRU-VNTR typing were actually infected by as-yet-unidentified contacts, different from the contacts of the other patients of clusters 32 or 34.
A comparison of the overall discriminatory powers of MIRU-VNTR and IS6110-RFLP typing was beyond the scope of this study. It was designed to test the potential of MIRU-VNTR to split epiunlinked IS6110-PGRS clusters but not the potential of IS6110-PGRS to split epiunlinked MIRU-VNTR clusters. It is conceivable that if MIRU-VNTR typing had been used as initial typing method, a part of the MIRU-VNTR clusters would have been subdivided by IS6110-PGRS RFLP typing. In this study, two epidemiologically unlinked clusters with different IS6110-PGRS RFLP patterns had identical MIRU-VNTR patterns, consistent with the slightly lower resolution power of MIRU-VNTR typing compared to that of IS6110-RFLP typing (9, 25). Nevertheless, it is of considerable practical advantage to use MIRU-VNTR as the first-line method for transmission analysis, especially when combined with spoligotyping, as these two clusters had distinct spoligotypes, as has already been observed in previous studies (9).
In contrast to IS6110-PGRS-based RFLP typing, PCR-based MIRU-VNTR typing or spoligotyping can be applied with crude DNA extracts from heat-killed mycobacterial cultures as soon as they become positive, avoiding culture delays of several weeks. Depending upon the initial bacterial load, the total time needed to obtain the genotypes when starting from the clinical specimen can be reduced to a few days (3). Epidemiological investigations based on genotyping data can thus begin sooner, providing opportunities for the more rapid detection of secondary cases, latent tuberculosis infections, and sites of transmission (33). By combining MIRU-VNTR typing and spoligotyping, results can more easily be read and exchanged between laboratories than is the case with the complex IS6110-PGRS patterns. Therefore, the first-line use of MIRU-VNTR typing instead of IS6110-PGRS RFLP typing should provide a more rapid and more reliable insight into the dynamics of tuberculosis transmission both within and among communities or countries.
It can be concluded from this study that for a number of cases, estimated to be 14% in The Netherlands, the use of IS6110-PGRS RFLP typing will result in false clustering of tuberculosis patients, even if the isolates have high copy numbers of IS6110. The population-based study presented here included patients from both the general population and from tuberculosis risk groups in rural and urban areas representative of The Netherlands. Since the country's sociodemographic and disease incidence characteristics are similar to those of most developed countries, we predict that our observations can be widely generalized.
Taking into consideration the technical advantages and the accuracy gained for reliable exclusion of epidemiological links, a combination of MIRU-VNTR typing and spoligotyping instead of RFLP-based methods would thus be recommendable in many settings for first-line screening of potential tuberculosis case clusters.
Acknowledgments
This study was supported by The Netherlands Organization for Health Research and Development (ZonMw), Den Haag, The Netherlands, and by the Institut National de la Santé et de la Recherche Médicale (INSERM), Institut Pasteur de Lille, Lille, France.
Philip Supply is a researcher of the Centre National de la Recherche Scientifique (CNRS). We thank Philippe Boutin for providing laboratory facilities. Miriam Dessens, Annemarie van den Brand, Marianne van Prehn, and Tridia van der Laan are acknowledged for their contribution to the practical work. We also thank Lucy D. Philips for editorial review.
REFERENCES
- 1.Agerton, T., S. Valway, B. Gore, C. Pozsik, B. Plikaytis, C. Woodley, and I. M. Onorato. 1997. Transmission of a highly drug-resistant strain of Mycobacterium tuberculosis. Community outbreak and nosocomial transmission via a contaminated bronchoscope. JAMA 278:1073-1077. [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiological methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, R. E., D. M. Gascoyne-Binzi, S. H. Gillespie, A. Dickens, S. Qamer, and P. M. Hawkey. 2001. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:2453-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149-1156. [DOI] [PubMed] [Google Scholar]
- 6.Bock, N. N., J. P. Mallory, N. Mobley, B. DeVoe, and B. Brooks Taylor. 1998. Outbreak of tuberculosis associated with a floating card game in the rural south: lessons for tuberculosis contact investigations. Clin. Infect. Dis. 27:1221-1226. [DOI] [PubMed] [Google Scholar]
- 7.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 8.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodriguez, I. Garcia, P. Cabrera, C. Lafoz, S. Samper, H. Takiff, O. Alfonso, J. M. Pavon, M. J. Torres, D. van Soolingen, D. E. Enarson, and C. Martin. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164:1165-1170. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley, C. L., P. M. Small, G. F. Schecter, G. K. Schoolnik, R. A. McAdam, W. R. Jacobs, Jr., and P. C. Hopewell. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus: an analysis using restriction fragment length polymorphisms. N. Engl. J. Med. 326:231-235. [DOI] [PubMed] [Google Scholar]
- 11.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kalembo, H. M. Li, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 13.Gascoyne-Binzi, D. M., R. E. Barlow, R. Frothingham, G. Robinson, T. A. Collyns, R. Gelletlie, and P. M. Hawkey. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J. Clin. Microbiol. 39:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng, E., B. Kreiswirth, C. Driver, J. Li, J. Burzynski, P. DellaLatta, A. LaPaz, and N. W. Schluger. 2002. Changes in the transmission of tuberculosis in New York City from 1990 to 1999. N. Engl. J. Med. 346:1453-1458. [DOI] [PubMed] [Google Scholar]
- 15.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohammed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans, P. W. M., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. W. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, D. Frommel, M. Zribi, and J. D. A. van Embden. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia and the Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 17.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. R. Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. B. M. C. Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lok, K. H., W. H. Benjamin, M. E. Kimerling, V. Pruitt, D. Mulcahy, N. Robinson, N. B. Keenan, and N. E. Dunlap. 2002. Molecular typing of Mycobacterium tuberculosis strains with a common two-band IS6110 pattern. Emerg. Infect. Dis. 8:1303-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savine, E., W. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shamputa, I. C., L. Rigouts, L. A. Eyongeta, N. A. El Aila, A. van Deun, A. H. Salim, E. Willery, C. Locht, P. Supply, and F. Portaels. 2004. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J. Clin. Microbiol. 42:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 28.Solsona, J., J. A. Caylà, E. Verdú, M. P. Estrada, S. Garcia, D. Roca, B. Miquel, P. Coll, F. March, and the Cooperative Group for Contact Study of Tuberculosis Patients in Ciutat Vella. Molecular and conventional epidemiology of tuberculosis in an inner city district. Int. J. Tuber. Lung Dis. 5:724-731. [PubMed]
- 29.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 30.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valway, S. E., M. P. C. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. Scott Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 32.Van Deutekom, H., J. J. J. Gerritsen, D. van Soolingen, E. J. C. van Ameijden, J. D. A van Embden, and R. A. Coutinho. 1997. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin. Infect. Dis. 25:1071-1077. [DOI] [PubMed] [Google Scholar]
- 33.Van Deutekom, H., S. P. Hoijng, P. E. W. de Haas, M. W. Langendam, A. Horsman, D. van Soolingen, and R. A. Coutinho. 2004. Clustered tuberculosis cases: do they represent recent transmission and can they be detected earlier? Am. J. Respir. Crit. Care Med. 169:806-810. [DOI] [PubMed] [Google Scholar]
- 34.Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. W. M. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 36.Van Soolingen, D., P. E. W. de Haas, P. W. M. Hermans, P. M. A. Groenen, and J. D. A. van Embden. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 31:1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nyamadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, G. D. van der Spuy, R. Johnson, V. N. Chihota, C. Locht, P. Supply, and P. D. van Helden. 2004. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J. Clin. Microbiol. 42:5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weis, S. E., J. M. Pogoda, Z. Yang, M. D. Cave, C. Wallace, M. Kelley, and P. F. Barnes. 2002. Transmission dynamics of tuberculosis in Tarrant county, Texas. Am. J. Respir. Crit. Care Med. 166:36-42. [DOI] [PubMed] [Google Scholar]