Abstract
Drug resistance, particularly vancomycin and methicillin resistance, in Staphylococcus aureus continues to emerge as a significant public health threat in both the hospital and community settings. In addition to the limited treatment options, S. aureus strains acquire and express numerous virulence factors that continue to increase its ability to cause a wide spectrum of human disease. As a result, empirical treatment decisions are confounded and there is a heightened need for a diagnostic test (or assay) to rapidly identify antibiotic resistance and specific virulence determinants and indicate the appropriate treatment. To that end we developed a platform using multiplex molecular beacon probes with real-time PCR for the rapid detection of drug resistance-determining genes and virulence factors in S. aureus. In this study, we demonstrate the specificity and sensitivity of our platform for detection of the genes conferring methicillin (mecA) and vancomycin (vanA) resistance as well as a gene encoding the virulence factor Panton-Valentine leucocidin (lukF) in S. aureus isolates.
Staphylococcus aureus constitutes a major public health threat, as it is the leading nosocomially acquired pathogen in the United States (2, 3). This gram-positive species is armed with a wide array of virulence determinants that produce a diverse spectrum of clinical presentations, including the common boil, food poisoning, toxic shock syndrome, osteomyelitis, necrotizing pneumonia, and endocarditis (14). Methicillin-resistant S. aureus (MRSA) strains, which are commonly multidrug resistant and may only be susceptible to vancomycin, present both a treatment and infection control challenge in the hospital setting (2). This problem is further confounded by the recent spread of community-acquired MRSA (CA-MRSA) and the identification of vancomycin-resistant MRSA (VMRSA) strains, which have been documented on three separate occasions in the United States (1, 4, 15, 19, 23). In each case of VMRSA, vancomycin resistance developed as the result of transfer of a plasmid carrying the vanA operon to S. aureus (1, 4, 23).
Clinical microbiology laboratories still depend on phenotypic assays for both identification and antibiotic resistance testing of organisms; currently, it requires 24 to 48 h to definitively identify an MRSA culture by conventional methods. MRSA detection assays have been developed using the nuc gene target to distinguish S. aureus from other species, in conjunction with the mecA gene target to define methicillin resistance (5, 7, 18), and the first commercial test has been approved by the U.S. Food and Drug Administration for MRSA detection, using a molecular beacon that targets the orfX gene, which is juxtaposed to the Staphylococcus chromosomal cassette (SCCmec) element (8).
In this report, we present and validate a strategy for the creation of a multiplex diagnostic platform that uses interchangeable molecular beacons to answer a diverse set of biological questions in an objective binary format. The ultimate goal will be to develop this strategy into an assay that tests primary specimens in a routine clinical microbiology laboratory. Molecular beacons are short oligonucleotide probes possessing a hairpin structure; these probes fluoresce upon excitation, when annealed to their target (24). MRSA and the recently described VMRSA strains were correctly identified among blinded samples, using a four-color multiplex molecular beacon assay designed to both detect these organisms and distinguish them from methicillin-susceptible S. aureus (MSSA) and methicillin-resistant coagulase-negative staphylococci (MR-CNS).
MATERIALS AND METHODS
Bacterial strains.
Bacterial isolates used in this study were from archived collections at Mount Sinai Hospital, Toronto, Ontario, Canada, the Public Health Research Institute Tuberculosis Center, Newark, N.J., and the Wadsworth Center, Albany, N.Y. Many selected MRSA strains from the Public Health Research Institute Tuberculosis Center were previously genotyped on the basis of their spa types (11), and those from Toronto had been selected based on being of distinct species, pulsed-field gel electrophoresis type, and/or MIC antibiogram, and therefore the isolates used in this study included both geographic and genetic diversity (see Table 2). Species and strains were selected such that there were multiple positive and negative controls for the assays. Strains were probed for each particular target in a blinded format, and the identity of each strain was unknown until after analysis. All DNA samples tested were isolated from pure bacterial cultures; primary specimens and mixed cultures were not evaluated in this study.
TABLE 2.
Validation of target specificity
| Target of molecular beacon | Bacterial straina
|
Total (252 strains) | |||||
|---|---|---|---|---|---|---|---|
| MRSA | MSSA | MR-CNS | MS-CNS | VRE | Otherb | ||
| BAC16S | 17/17 | 3/3 | 3/3 | 3/3 | 3/3 | 5/5 | 34 |
| SG16S | 109/109 | 36/36 | 10/10 | 20/20 | 0/5 | 0/29 | 209 |
| mecA | 109/109 | 0/36 | 10/10 | 0/20 | 0/5 | 0/29 | 209 |
| spa | 109/109 | 36/36 | 0/10 | 0/20 | 0/5 | 0/29 | 209 |
| vanA | 0/93 | 0/29 | 0/8 | 0/20 | 20/29c | 0/48 | 228 |
| lukF | 32/61d | 4/31d | 0/2 | 0/2 | 0/2 | 0/2 | 100 |
Number of strains positive for target/number of strains tested. Common nosocomially acquired pathogens were tested to validate the specificity of each probe for its corresponding gene. In total, 252 strains were used for validation of the assay, of which 175 were staphylococci. Strains are genetically and geographically diverse, representing isolates from New York, New Jersey, California, Montana, and Texas as well as Canada and Nigeria. All strains were classified correctly.
Group Other consists of vancomycin-susceptible enterococci (VSE), Klebsiella spp., group A Streptococcus pyogenes, Enterococcus gallinarum, Lactobacillus spp., Listeria monocytogenes, and Micrococcus spp.
The 9 out of 29 VRE strains negative for vanA were shown to be positive for vanB using a different molecular beacon probe.
Strains positive and negative for lukF were confirmed using conventional PCR with gel electrophoresis.
Molecular beacon probes and oligonucleotide primers.
Molecular beacon probes and oligonucleotide primers were designed using the Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, CA) and mfold, the Zuker DNA folding program (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/form1.cgi). Both molecular beacons and primers were validated using the guidelines set in the molecular-beacons website (www.molecular-beacons.org). Single-probe reactions were conducted with each probe to determine its specificity and sensitivity for its target. Selected probes were then combined and tested in a multiplex format. Molecular beacon probes were obtained from Biosearch Technologies (Novato, CA), and oligonucleotide probes were obtained from Integrated DNA Technologies (Coralville, IA). Molecular beacon and oligonucleotide primer sequences are listed in Table 1.
TABLE 1.
Molecular beacon and primer sequences
| Oligonucleotidea | Sequence (5′-3′)b |
|---|---|
| mecA, F | TGGTATGTGGAAGTTAGATTGG |
| mecA, R | ATATGCTGTTCCTGTATTGGC |
| mecA, MB | (FAM)-CGCGATTTCAATATGTATGCTTTGGTCTTTCTGATCGCG-(DABCYL) |
| spa, F | CATTACTTATATCTGGTGGCG |
| spa, R | GTTAGGCATATTTAAGACTTG |
| spa, MB | (CY5)-CGCGAGTTAGGCATATTTAAGACTTGTCGCG-(BHQ-2) |
| SG16S, F | TGGAGCATGTGGTTTAATTCGA |
| SG16S, R | TGCGGGACTTAACCCAACA |
| SG16S, MB | (HEX)-CGCTGACTTACCAAATCTTGACATCCTTCAGCG-(DABCYL) |
| BAC16S, F | TGGAGCATGTGGTTTAATTCGA |
| BAC16S, R | TGCGGGACTTAACCCAACA |
| BAC16S, MB | (ROX)-CGCTGGCGAGCTGACGACARCCATGCACCAGCG-(DABCYL) |
| lukF, F | GCCAGTGTTATCCAGAGG |
| lukF, R | CTATCCAGTTGAAGTTGATCC |
| lukF, MB | (FAM)-CGCGAAGAATTTATTGGTGTCCTATCTCGATCGCG-(DABCYL) |
| vanA, F | AATCGGCAAGACAATATGAC |
| vanA, R | ACCTCGCCAACAACTAAC |
| vanA, MB | (ROX)-GGACGTGTGAGGTCGGTTGTGCGGTATACGTCC-(DABCYL) |
Molecular beacon (MB) and forward (F) and reverse (R) primer sequences for each of the targets probed for in the blinded panel of bacterial species.
Underlined sequences represent stem structure of molecular beacons. DABCYL, 4-(4′-dimethylaminophenylazo)benzoic acid; FAM, fluorescein; HEX, hexachlorofluorescein; ROX, rhodamine; BHQ-2, Black Hole Quencher 2.
RT-PCR.
Real-time PCR (RT-PCR) was performed using an Mx4000 multiplex QPCR system (Stratagene, La Jolla, CA). Oligonucleotide primer pairs and molecular beacon sequences for six unique targets are listed in Table 1. The targets include the 16S rRNA gene sequences conserved in all bacteria (BAC16S), the 16S rRNA gene sequence common to the Staphylococcus genus (SG16S), the protein A gene (spa) specific to Staphylococcus aureus, the mecA and vanA genes that confer resistance to methicillin and vancomycin, respectively, and the lukF gene that encodes a component of the Panton-Valentine leucocidin (PVL). For the reactions using BAC16S, SG16S, spa, and mecA as targets, each 50-μl reaction contained 25 μl iQ Supermix (Bio-Rad, Hercules, CA), 0.5 μM of each primer (Table 1), BAC16S forward and BAC16S reverse (SG16S and BAC16S use the same primers), mecA forward and mecA reverse, spa forward and spa reverse, 0.025 μM of BAC16S molecular beacon probe, 0.4 μM of SG16S molecular beacon probe, 0.1 μM of mecA molecular beacon probe, 0.05 μM of spa molecular beacon probe, and 1 μl of purified genomic DNA (used at approximately 15 to 100 ng/μl); the remainder of the 50 μl was nuclease-free water. Genomic DNA was purified as described previously (12). The thermocycling program used was 1 cycle of 95°C for 10 min followed by 45 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s. The entire RT-PCR takes approximately 3 h from setup to analysis. When the vanA or lukF molecular beacon probe is used in place of BAC16S, the primers for each are used at 0.5 μM and the BAC16S primers are still used for the SG16S molecular beacon. The vanA and lukF molecular beacon probes are used at 0.05 μM and 0.2 μM, respectively.
RESULTS
Genetic targets.
A total of six genetic targets are described in this study. Each was chosen on the basis of being a positive predictor of a clinically significant biological trait associated with Staphylococcus aureus. The optimal oligonucleotide primers and the molecular beacon hybridization probes to each target region were designed using Beacon Designer 2.0 software. Each candidate molecular beacon was synthesized with a fluorescein fluorophore, and its sensitivity and specificity were evaluated against a panel of test DNA samples from diverse genera and species in an RT-PCR assay. This process was necessary for the optimization of conditions for each new probe designed. In this assay, the molecular beacons acted as yes-or-no (qualitative) hybridization probes that provided basic information about taxonomic identification, drug resistance, and virulence.
A conserved region (BAC16S) in the 16S ribosomal gene, present in all bacteria, was selected as a positive PCR internal control target. A second region (SG16S) of the same gene was selected that is specific for the genus Staphylococcus (22). A single PCR amplicon contains both the SG16S- and BAC16S-specific target sequences, but no interference was observed when the two molecular beacons were used together. A molecular beacon probe for sequences in the spa gene was designed to distinguish S. aureus strains (11) from other staphylococcal species. As positive predictors of methicillin and vancomycin resistance, molecular beacon probes for sequences in mecA and vanA, respectively, were synthesized and analyzed against panels of susceptible and resistant bacterial isolates. A molecular beacon probe was also developed for the lukF gene that carries one of the two genes involved in Panton-Valentine leucocidin (PVL) activity, a recently described virulence determinant associated with CA-MRSA cases (10, 15).
Multiplexing of molecular beacons.
The evaluation of the molecular beacons' sensitivities and specificities for their targets in a multiplex assay required the resynthesis of the molecular beacons with distinguishable fluorophores. In this study, we utilized a spectrofluorometric thermal cycler that was able to detect up to four fluorophores in a single reaction. The fluorophores used in these assays (Table 1) were chosen on the basis of correspondence to the filter set equipped on the Stratagene Mx4000 spectrofluorometric thermal cycler used in this study.
A three-color multiplex assay was tested to identify MRSA isolates and distinguish them from MSSA, methicillin-susceptible coagulase-negative Staphylococcus (MS-CNS), MR-CNS, and non-Staphylococcus spp. bacteria. A total of 209 bacterial DNA samples were isolated from MRSA, MSSA, MR-CNS, MS-CNS, vancomycin-resistant Enterococcus faecalis and E. faecium (VRE), and Klebsiella pneumoniae, and these were tested in a single-tube multiplex RT-PCR assay that contained three pairs of oligonucleotide primers and three unique molecular beacons to the SG16S rRNA, spa, and mecA genes, which were, respectively, labeled with hexachlorofluorescein (detected at 556-nm wavelength), Cy5 (detected at 670 nm), and fluorescein (detected at 515 nm) fluorophores. The multiplex assay correctly identified each unknown DNA sample 100% in each test (Table 2).
Four-color multiplex assay.
We have designed the current diagnostic platform to be expandable so as to provide the flexibility to address “emerging” biological questions. As examples, we evaluated three additional molecular beacons to extend the “core” platform of SG16S, spa, and mecA gene targets in an effort to provide more detailed isolate information for clinical use.
The BAC16S molecular beacon in a four-color multiplex assay provides a positive control, since it confirms the presence of bacterial DNA in the sample (16); it also serves as an internal control, since all SG16S-positive samples should also be BAC16S positive. As shown in Table 2, all bacterial DNA samples were positively detected with the BAC16S molecular beacon probe.
The addition of a molecular beacon specific to vanA establishes a robust four-color multiplex assay to detect VMRSA isolates. We synthesized a vanA molecular beacon labeled with rhodamine, a fluorophore compatible with the filter set on the Mx4000, and we tested its specificity and sensitivity in multiplex assays with 228 blinded DNA samples isolated from MRSA, MSSA, MR-CNS, MS-CNS, VRE, vancomycin-susceptible Enterococcus (VSE), and K. pneumoniae strains. In a single-tube multiplex RT-PCR experiment, four resolvable fluorophores linked to SG16S, spa, mecA, and vanA beacons were each evaluated in a 2.5-h assay (Fig. 1A to E).
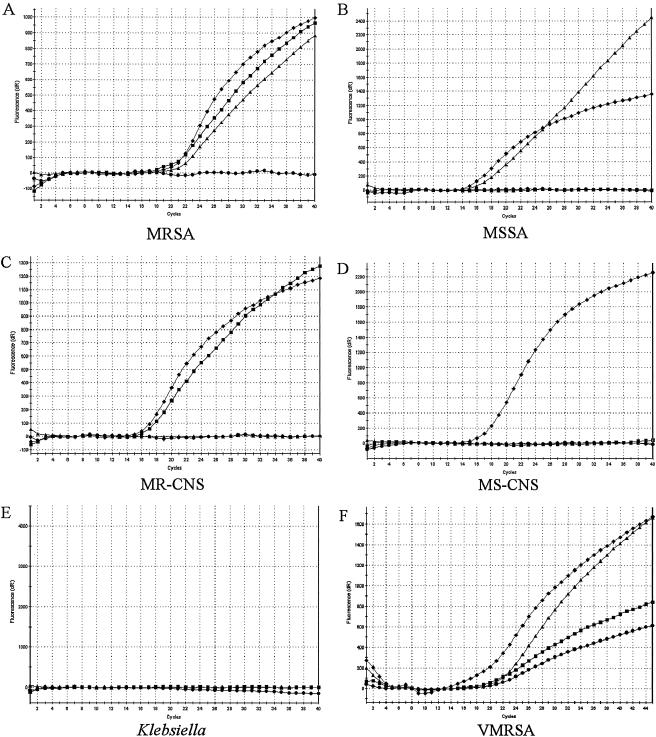
FIG. 1.
Species-level identification and drug resistance determination. Real-time PCR fluorescent signatures are associated with several bacterial species and their resistance determinants. Each sample was probed with four different molecular beacon probes in one reaction and analyzed using the Stratagene Mx4000 software. Symbols: squares, mecA; triangles, spa; diamonds, SG16S; circles, vanA. dR, first derivative of the fluorescence changes with respect to temperature.
As shown in Table 2, the assay correctly identified each test DNA sample. The specificity of each molecular beacon was observed in each assay: SG16S, mecA, and spa did not hybridize to VRE or VSE DNA and vanA did not hybridize to vancomycin-susceptible staphylococci or to the VSE DNA. Among the 29 VRE isolates, 20 were identified with the vanA molecular beacon. The other nine VRE that were negative for vanA were positive for vanB when a different molecular beacon probe was used (data not shown).
During the testing of the four-color multiplex assay, a VMRSA isolate was identified in a nursing home in New York state (1). The New York State Department of Health Wadsworth Center confirmed the identification of this strain and provided DNA samples in a blinded format for use in the evaluation of our multiplex assay. As shown in Fig. 1F, the VMRSA isolate gave positive signals for the presence of SG16S, spa, mecA, and vanA. The laboratory also provided 10 blinded DNA samples that included two tubes of the VMRSA, and MSSA, MRSA, MR-CNS, and several nonstaphylococcal species served as negative controls. Each sample was identified correctly (Fig. 1 and data not shown).
Molecular beacon probes for S. aureus virulence genes.
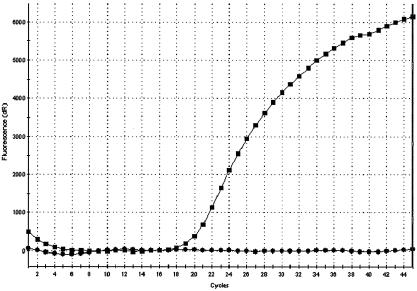
The PVL virulence determinant is encoded by two contiguous genes, lukF and lukS, on a staphylococcal phage (10). Initial analysis using both lukF and lukS molecular beacons showed that all PVL-positive strains had both genes present. As a result, we chose lukF alone for the detection of PVL in S. aureus. DNA from a total of 92 S. aureus isolates was tested with the lukF molecular beacon, and a positive hybridization was identified in 32 of the 61 MRSA strains and 4 of the 31 MSSA strains assayed (Table 2). These results were confirmed by conventional PCR visualized with gel electrophoresis (data not shown). A comparison between PVL-positive and PVL-negative MRSA strains probed with lukF is shown in Fig. 2. The lukF probe for PVL could not be used in the multiplex format, because its primers were incompatible with those for SG16S. The lukF probe is already in use as a tool for detection and expression of PVL in clinical isolates (20).
FIG. 2.
Detection of S. aureus virulence factor PVL in MRSA isolates. Real-time PCR signatures for a PVL-positive MRSA and a PVL-negative MRSA when probed for lukF are shown. PVL-positive MRSA (squares) and PVL-negative MRSA (circles) were probed for lukF in two separate reactions and compared using the Stratagene Mx4000 software. dR, first derivative of the fluorescence changes with respect to temperature.
Sensitivity of the individual molecular beacons.
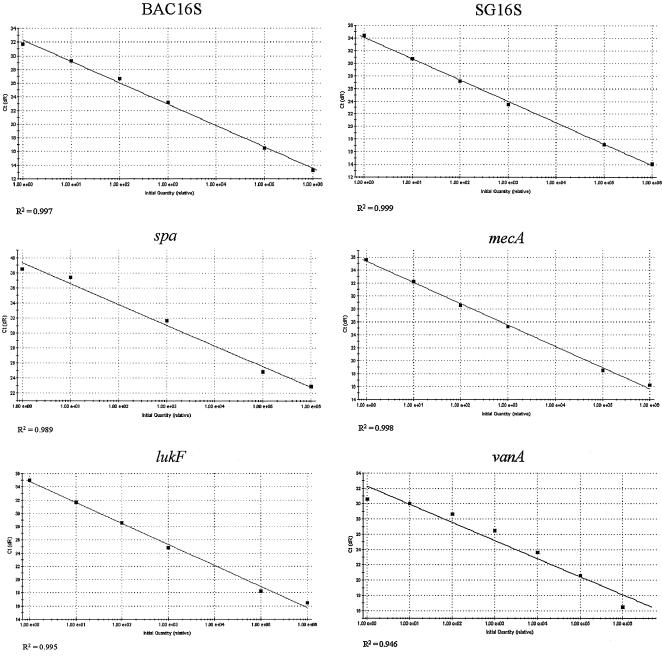
DNA isolated from PVL-positive MRSA and a VRE were serially diluted and probed with the molecular beacons to determine the sensitivity of the assay (Fig. 3). The MRSA DNA was probed with molecular beacons for BAC16S, SG16S, mecA, spa, and lukF individually, and VRE DNA was probed for vanA. As few as 13 genome copies of MRSA could be detected when probed for each target, and as few as three genome copies could be detected when VRE DNA was probed with the vanA molecular beacon.
FIG. 3.
Sensitivity of molecular beacon probes. DNA samples from MRSA and VRE were serially diluted and probed with BAC16S, SG16S, spa, mecA, and lukF for MRSA and vanA for VRE to determine the limit of detection for each probe. For MRSA, each molecular beacon used was able to detect 13 genomes of MRSA DNA, and for VRE, the vanA probe was able to detect three genomes of VRE DNA. Ct, cycle threshold. dR, first derivative of the fluorescence changes with respect to temperature.
DISCUSSION
In the clinical microbiological laboratory, the identification and susceptibility testing of a bacterial isolate remains the principal focus of the routine diagnostic work. Phenotypic assays in automated formats generate the majority of the reported microbiologic data, and turnaround times are still measured in days, not hours.
While the identification of an MRSA in a blood culture taking as long as 4 days to finalize after the time of sampling may be considered acceptable practice (17), the accelerated rise in antibiotic resistance, the lack of new antibiotics in development, and the emergence of community-acquired resistant strains are all undermining the empirical approach that broad-spectrum antibiotics provided such patients in the past.
The four recent examples of the vanA operon transferring to MRSA on a plasmid demonstrates that the continued ability to rely on vancomycin as the most effective anti-staphylococcal antibiotic for the treatment of methicillin-resistant strains is being compromised (1, 4, 23). The fact that these strains were not all routinely detected in the microbiological laboratory using conventional automated systems only highlights the urgent need for improved diagnostics (23).
The goal of the present study was to develop a diagnostic platform, for use in the clinical setting and that ultimately could be applied to primary specimens, that is rapid, specific, and able to answer multiple biological questions in a single-tube assay. The assay was based on the identification of confirmed targets, the presence of which is predictive for known biological properties; the probes against these targets were combined in a multiplex format and assessed for their compatibility. As a result of its clinical importance, an S. aureus strain, specifically an MRSA strain, was selected as the prototypical organism to demonstrate the simplicity, speed, accuracy, and flexibility of this new diagnostic platform.
Genotypic approaches to the identification of MRSA have each justifiably focused on the mecA gene target as the predictor for methicillin resistance. However, because mecA is also present in coagulase-negative staphylococci, the commonly isolated methicillin-resistant S. epidermidis also needs to be considered in the development of a diagnostic assay. One method, described by Francois and colleagues (6), details the use of an immunocapture method to segregate S. aureus from other bacterial isolates; the S. aureus isolates are then tested for the presence of mecA. This approach improves specificity but increases expenditures of time, labor, and money. The recent real-time PCR assay introduced by Huletsky and coworkers (8) takes advantage of the unique chromosomal location of the mecA-harboring cassette, the Staphylococcus chromosomal cassette (SCCmec), and its juxtaposition to orfX. The latter is a conserved gene in S. aureus and S. epidermidis, but it has unique sequences in S. aureus that can be specifically targeted using a molecular beacon probe. This approach may be limited by the promiscuity of the SCCmec type IV and V elements (9) and by the creation of genetic polymorphisms at the PCR priming sites.
The multiplex platform presented in our studies provides a format in which various molecular beacon hybridization probes may be interchanged so as to create panels for differing diagnostic assays. Each probe is designed to answer a specific “Yes/No” question, for which a positive result correlates with the presence of a predictive biological trait. The ability to test four different distinct targets simultaneously means that both MRSA isolates and the rare vancomycin-resistant S. aureus strains can be identified using probes for mecA and vanA. We have also designed a beacon for the lukF gene, a predictive target for the PVL determinant and a marker for CA-MRSA strains.
Although amplification of the current targets in our assay enables us to accurately identify MRSA and to distinguish it from methicillin-resistant S. epidermidis isolates when in pure culture, it is clear that these current targets would not discriminate MRSA from a mixture of MSSA and methicillin-resistant S. epidermidis or distinguish VMRSA from a mixture of MRSA and VRE. In an attempt to overcome this limitation, we have identified an S. epidermidis-specific target that, together with spa, mecA, and vanA, would provide a robust assay to not only discriminate MRSA from other organisms but also identify those samples that are contaminated with one or more colonizing or infecting strains. Development of an Enterococcus-specific probe would enable detection of VRE contamination of MRSA. Recent developments in RT-PCR technology have led to the production of several spectrofluorometric thermal cyclers that can detect up to six fluorophores in a single reaction. With this technology, our assay could be expanded to include two additional targets, which could be used to determine species specificity and the detection of a known virulence determinant.
Finally, the recent advances in DNA extraction methods that enable the purification of high-quality nucleic acid directly from clinical samples, including blood, sputum, cerebrospinal fluid, urine, stool, and nasal, rectal, and vaginal swabs, now provides the capability for this rapid RT-PCR assay to be used to identify bacterial pathogens directly from the specimen in a manner that we believe will dramatically affect clinical decisions and treatment and infection control patient management (13, 17, 21, 25).
Acknowledgments
This study was funded by a research grant from the Health Care Foundation of New Jersey to B.N.K.
We thank Barun Mathema for a critical reading of the manuscript.
REFERENCES
- 1.Anonymous. 2004. Brief report: vancomycin-resistant Staphylococcus aureus —New York. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 2.Anonymous. 2003. National Nosocomial Infection Surveillance (NNIS) System report, data summary, from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control 31:481-498. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990 through May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 4.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 5.Elsayed, S., B. L. Chow, N. L. Hamilton, J. D. Gregson, J. D. Pitout, and D. L. Church. 2003. Development and validation of a molecular beacon probe-based real-time polymerase chain reaction assay for rapid detection of methicillin resistance in Staphylococcus aureus. Arch. Pathol. Lab. Med. 127:845-849. [DOI] [PubMed] [Google Scholar]
- 6.Francois, P., D. Pittet, M. Bento, B. Pepey, P. Vaudaux, D. Lew, and J. Schrenzel. 2003. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisold, A. J., E. Leitner, G. Muhlbauer, E. Marth, and H. H. Kessler. 2002. Detection of methicillin-resistant Staphylococcus aureus and simultaneous confirmation by automated nucleic acid extraction and real-time PCR. J. Clin. Microbiol. 40:2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage φPVL carrying Panton-Valentine leukocidin genes. Gene 215:57-67. [DOI] [PubMed] [Google Scholar]
- 11.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreiswirth, B. N., S. M. Lutwick, E. K. Chapnick, J. D. Gradon, L. I. Lutwick, D. V. Sepkowitz, W. Eisner, and M. H. Levi. 1995. Tracing the spread of methicillin-resistant Staphylococcus aureus by Southern hybridization using gene-specific probes of mec and Tn554. Microb. Drug Resist. 1:307-313. [DOI] [PubMed] [Google Scholar]
- 13.Louie, L., J. Goodfellow, P. Mathieu, A. Glatt, M. Louie, and A. E. Simor. 2002. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 40:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 15.Mongkolrattanothai, K., S. Boyle, M. D. Kahana, and R. S. Daum. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 37:1050-1058. [DOI] [PubMed] [Google Scholar]
- 16.Patel, J. B. 2001. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6:313-321. [DOI] [PubMed] [Google Scholar]
- 17.Peters, R. P., M. A. van Agtmael, S. A. Danner, P. H. Savelkoul, and C. M. Vandenbroucke-Grauls. 2004. New developments in the diagnosis of bloodstream infections. Lancet Infect. Dis. 4:751-760. [DOI] [PubMed] [Google Scholar]
- 18.Reischl, U., H.-J. Linde, M. Metz, B. Leppmeier, and N. Lehn. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J. Clin. Microbiol. 38:2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Said-Salim, B., B. Mathema, and B. N. Kreiswirth. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect. Control Hosp. Epidemiol. 24:451-455. [DOI] [PubMed] [Google Scholar]
- 20.Said-Salim, B., B. Mathema, K. Braughton, S. Davis, D. Sinsimer, W. Eisner, F. DeLeo, and B. N. Kreiswirth. 2005. Differential distribution and expression of Panton-Valentine leukocidin among community-acquired methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 43:3373-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrestha, N. K., M. J. Tuohy, G. S. Hall, C. M. Isada, and G. W. Procop. 2002. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J. Clin. Microbiol. 40:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 23.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 25.Warren, D. K., R. S. Liao, L. R. Merz, M. Eveland, and W. M. Dunne, Jr. 2004. Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J. Clin. Microbiol. 42:5578-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]