Abstract
In Drosophila and several other metazoan organisms, there are two genes that encode related but distinct homologs of ADA2-type transcriptional adaptors. Here we describe mutations of the two Ada2 genes of Drosophila melanogaster. By using mutant Drosophila lines, which allow the functional study of individual ADA2s, we demonstrate that both Drosophila Ada2 genes are essential. Ada2a and Ada2b null homozygotes are late-larva and late-pupa lethal, respectively. Double mutants have a phenotype identical to that of the Ada2a mutant. The overproduction of ADA2a protein from transgenes cannot rescue the defects resulting from the loss of Ada2b, nor does complementation work vice versa, indicating that the two Ada2 genes of Drosophila have different functions. An analysis of germ line mosaics generated by pole-cell transplantation revealed that the Ada2a function (similar to that reported for Ada2b) is required in the female germ line. A loss of the function of either of the Ada2 genes interferes with cell proliferation. Interestingly, the Ada2b null mutation reduces histone H3 K14 and H3 K9 acetylation and changes TAF10 localization, while the Ada2a null mutation does not. Moreover, the two ADA2s are differently required for the expression of the rosy gene, involved in eye pigment production, and for Dmp53-mediated apoptosis. The data presented here demonstrate that the two genes encoding homologous transcriptional adaptor ADA2 proteins in Drosophila are both essential but are functionally distinct.
The ADA genes (encoding ADA1, ADA2, ADA3/NGG1, GCN5/ADA4, and SPT20/ADA5) were defined genetically in yeast (Saccharomyces cerevisiae) on the basis of the finding that mutations affecting them confer resistance to toxicity mediated by high-level expression of the GAL4-VP16 fusion protein (7). Yeast ADA proteins play a role in transcriptional initiation site selection, interact with basal transcription factors, and facilitate the acetylation of nucleosomal histones as components of ADA/GCN5 histone acetyltransferase (HAT) complexes. For yeast, at least three distinct ADA2-containing multisubunit complexes regulating transcriptional activation have been identified, namely, A2, ADA, and SAGA (15, 32). These complexes have multiple, distinct functions leading to gene-specific transcription activation. ADA2 has been suggested to have an essential role in these complexes by participating in the recruitment of basal transcription factors, the stabilization of interactions with acidic activation domains, and potentiation of the HAT activity of GCN5 (1, 3, 5). ada2 is not essential for yeast viability but is involved in rapid transcriptional responses to environmental signals (43). Yeast ada2 mutants grow slowly in minimal medium and are cold and heat sensitive.
Recently, several groups have reported that the Drosophila genome contains two distinct genes encoding ADA2 homologs (19, 24). Biochemical characterization of the two ADA2 proteins demonstrated that both of them interact with the HAT GCN5 and participate in transcription activation. On the other hand, ADA2a and ADA2b exhibit marked differences, e.g., they participate in distinct high-molecular-weight HAT-containing protein complexes, localize to different chromosomal loci, and have at least partly different partners of interaction (19, 24). Two different ADA2 homologs encoded by two distinct genes have been found not only in Drosophila but also in Arabidopsis and several vertebrate genomes (6, 24, 35). While Drosophila ADA2a and ADA2b are more similar to human Ada2a (hAda2a) and hAda2b, respectively, both ADA2s of Arabidopsis resemble ADA2a of Drosophila more than ADA2b (24). T-DNA insertion mutations of one of the Ada2 genes (Ada2b) in Arabidopsis have been found to exert pleiotropic effects on plant growth and development, including dwarf size, aberrant root development, and short petals and stamens in the flowers. Furthermore, Ada2b mutant plants are tolerant of freezing (41). Functional differences between the two human ADA2 proteins, Ada2α and Ada2β, have recently been reported (6). Ada2α was found to be a stable component of the human PCAF-containing HAT complex. In contrast, Ada2β appeared not to be a stable component of PCAF- or Gcn5-containing macromolecular complexes, but interacted with subunits of the Swi/Snf chromatin-remodeling complex (6).
ADA2-containing complexes of yeast and humans have been shown to participate in gene-specific transcription activation by several sequence-specific transcription factors (4, 5, 8, 23, 33). Among others, the transcription-activating activity of the tumor suppressor p53 requires the ADA2/ADA3/GCN5 adaptor complex in yeast (42). hADA3 and p53 interact physically in human cells, and an appropriate hADA3 function is essential for the full transcriptional activity of p53 and for p53-mediated apoptosis (42). A p53 homolog was recently identified in Drosophila (Dmp53) (10, 25). Although Dmp53 displays low similarity to its human counterpart in its amino acid sequence, structural similarity in the domain structures of human and Drosophila p53 proteins and functional homology between them clearly exist. An interaction between Dmp53 and the Drosophila ADA2b protein, but not ADA2a, has also been revealed by in vitro pull-down experiments (19). Dmp53 induces apoptosis in response to genotoxic stresses by transactivating proapoptotic target genes through specific response elements in their promoters. Interestingly, the response elements to which Dmp53 binds are similar or identical to those recognized by mammalian p53 (10). However, unlike its mammalian homolog, Dmp53 does not induce cell cycle arrest (25, 34). One of the best-characterized transcriptional targets of Dmp53 is the enhancer and promoter of the reaper (rpr) gene, which is highly responsive to Dmp53 activation (10, 34). In a number of cases, however, Dmp53-induced apoptosis has been demonstrated in the absence of or independently of rpr, and it is therefore reasonable to assume that proapoptotic targets other than rpr also play important roles in Dmp53 function (11, 17, 28).
In this paper, we describe mutations and transgenes which facilitate analyses of the functions of the individual Ada2 genes of Drosophila. An analysis of the Ada2 mutants shows that the two Ada2 genes of Drosophila are functionally distinct, and the ectopic expression of one cannot substitute for the loss of function of the other. Furthermore, the two Ada2 genes have different effects on nucleosomal H3 acetylation, TAF10 localization, eye pigment formation, and Dmp53-mediated processes. The data obtained by analyzing Ada2 mutants suggest that ADA2-containing HAT complexes contribute to both basal and activator-induced H3 acetylation.
MATERIALS AND METHODS
Drosophila stocks, P-element mobilization, and genetic crosses.
Fly stocks were raised and crosses were performed at 25°C on standard medium containing propionic acid. Stocks carrying a single P-element insertion in the Ada2b or Ada2a region of the third chromosome [EP(3)3412 and l(3)S096713, respectively] were kindly provided by the Szeged Drosophila Stock Center.
To mobilize P elements in the Ada2a and Ada2b regions, we crossed P-element-carrying stocks with a TM3 ryRK Sb1 Ser1 P(delta2-3)99B/Df(3R)C7 ry506 transposase source (Bloomington 1808). Within the F2 progeny, we scored for the loss of the miniwhite (w+mC) marker and identified lethal mutations on the basis of no complementation being shown with the deficiencies Df(3R)P14 and Df(3R)CA1, which cover the ADA2a and Ada2b regions, respectively (http://flybase.bio.indiana.edu). Lethal alleles affecting the Ada2a and Ada2b regions were sorted into complementation groups and characterized by molecular methods to find deletions in the Ada2a and Ada2b genes. One Ada2a (Ada2ad189) and three Ada2b (Ada2bd842, Ada2bd272, and Ada2bd52) alleles identified among the lethal jump-outs were used for the further analyses described here. The mutant alleles were balanced over TM3 Sb Ser or TM6c Tb Sb. For an explanation of the genetic symbols, see reference 22 or the FlyBase website (http://flybase.bio.indiana.edu).
Ada2a Ada2b double mutants were constructed by recombining Ada2bd842 and Ada2ad189 into the same chromosome. The double mutants were selected on the basis of noncomplementation with either Df(3R)P14 or Df(3R)CA1. Eight independent P[Dtl+ Rpb4+] Ada2ad189 Ada2bd842/TM6C Tb Sb strains were used to test the phenotypes of double Ada2a Ada2b mutants. P[Dtl+ Rpb4+] is a transgene used to replace functions other than those of Ada2a which are removed in Ada2ad189 cells (see below). P[Dtl+ Rpb4+] Ada2ad189 Ada2bd842 homozygous larvae were selected on the basis of their Tb+ phenotype.
Germ line mosaics were generated by the pole-cell transplantation technique, as described earlier (38). Pole cells were collected from the F1 progeny of the cross P[Dtl+ Rpb4+] Ada2ad189/TM3 Sb Ser × Df(3R)P14/CxD. Host embryos were derived from a cross between wild-type females and Fs(1)K1237 males. Fs(1)K1237 (ovoD1) is a dominant female-sterile mutation that disrupts the function of the germ line cells (18, 27). Eclosing +/Fs(1)K1237 females were analyzed individually by crossing with white1118 males, and chimeras were isolated and kept for several days to identify the genotypes of the progeny.
To determine the roles of the Ada2a and Ada2b genes in imaginal disk and abdominal histoblast cells, somatic clones were generated (through X-ray-induced mitotic recombination) in both Ada2ad189/KiS and Ada2bd842/KiS larvae. Most of the mitotic recombinations in the Ada2ad189/KiS (or Ada2bd842/KiS) cells led to the formation of Ada2ad189/Ada2ad189 (or Ada2bd842/Ada2bd842) cells, which do not carry the KiS allele and (unless the lack of the Ada2a [or Ada2b] gene interferes with the cell function) develop normal bristles (13). The X-ray-irradiated larvae descended from a cross between Ada2ad189/TM3 Sb Ser (or Ada2bd842/TM3 Sb Ser) females and KiS/TM3 Sb Ser males. The Ada2ad189/KiS flies also carried the P[Dtl+ Rpb4+] transgene. The larvae were irradiated in the third instar, 74 to 98 h following egg-laying, by 1,500 R of X rays (150 kV; 0.5-mm Al filter; 1,000 R/min). For the detection of mosaic spots, the Ada2ad189/KiS (or Ada2bd842/KiS) adult females were mounted and analyzed for Ki+ clones (36).
The pigment contents of the eyes of adult flies were determined in P[w+mc GAL4-ninaE.GMR]12 (BL1104) (in short, P[gmr-GAL4])/+ +/P[UAS-P53.Ex]3 (BL8418), P[gmr-GAL4]/+ Ada2bd842/P[UAS-P53.Ex], P[gmr-GAL4]/P[Dtl+ Rpb4+] Ada2ad189/P[UAS-P53.Ex], +/+ Ada2bd842/+, and +/+ +/+ animals, as described previously (2).
Loss-of-heterozygocity (LOH) assays were carried out by scoring for the appearance of the recessive multiple wing hairs (Mwh) phenotype as reported earlier (34). The genotypes used were mwh p53/TM6C control, mwh p53/Ada2bd84, and P[Dtl+ rpb4+] mwh p53/Ada2ad189. Late-third-instar (wandering) larvae were X-ray irradiated with 250 R (150 kV; 0.5-mm Al filter; 1,000 R/min). Every genetic combination was tested in two independent experiments, with each involving 8 to 15 wings.
To determine radiation-induced apoptosis in the wing imaginal disks, third-instar wild-type (w1118), Ada2b mutant (Ada2bd842 and Ada2bd52) and Ada2a mutant (P[Dtl+ Rpb4+] Ada2ad189) larvae were X-ray irradiated at 4,000 R (150 kV; 0.5-mm Al filter; 1,000 R/min), left to recover for a 4-h period, and then dissected. Wing imaginal disks were incubated in a 1.6-μg/ml acridine orange (AO) solution for 5 min at room temperature, washed three times for 5 min each time with phosphate-buffered saline, and mounted in phosphate-buffered saline. The stained cells were counted under a fluorescence microscope. At least three independent experiments, each involving 8 to 12 disks, were performed for every genetic combination.
Transgene construction and other molecular biology techniques.
Ada2a+ and Ada2b+ transgenes were constructed by inserting cDNAs or genomic DNA fragments into pUAST (9) and pCaSpeR4 (39), respectively. Plasmid pCaSpeR4-Dtl+Ada2a+Rpb4+ carries a 7.1-kb KpnI-XbaI genomic fragment (from positions −3200 to +3919 with respect to the Ada2a translation start site) (Fig. 1). Three modified versions of the above plasmid are pCaSpeR4-Ada2a+Rpb4+, pCaSpeR4- Dtl+Ada2a+, and pCaSpeR4- Dtl+Rpb4+. The first has a 3.2-kb deletion in the Dtl region, generated by the elimination of an XbaI-NaeI fragment (from positions −3919 to −658). The second was generated by NarI digestion and religation, which resulted in deletion of the Rpb4 coding region between +2225 and +3053. In the third plasmid, the Ada2a open reading frame is interrupted by a four-nucleotide insertion into an SpeI site at +971, which results in a stop codon in Ada2a after amino acid 227 but does not interfere with RPB4 translation.
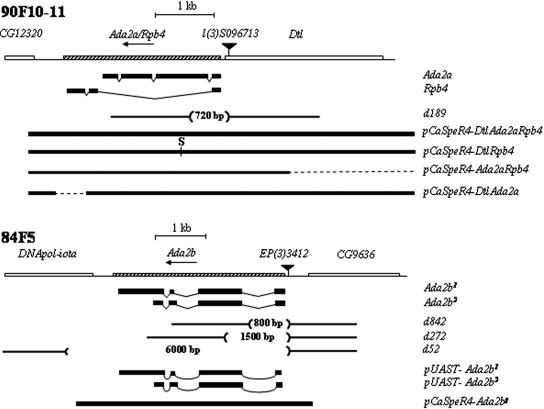
FIG. 1.
Drosophila 90F (Ada2a) and 84F (Ada2b) chromosomal regions. Both Ada2 genes are transcribed from right to left, and their exon-intron structures are indicated. The adjacent genes and the positions of P elements in the lines l(3)S096713 and EP(3)3412 are also shown. The positions and extensions of deletions generated by mobilization of these P elements are indicated in base pairs. Heavy lines indicate fragments used to generate transgenes for the phenotypic rescue of Ada2a and Ada2b mutants.
A 1,284-bp cDNA fragment encoding the ADA2b1 protein was obtained by reverse transcription-PCR (RT-PCR) on Drosophila total mRNA, using the primers A2bRI (at −10) (numbers refer to the translation start site of Ada2b) and A2bBHI (at +2519) (for the primer sequences, see Table 1). The PCR product was digested with EcoRI and BamHI and inserted into the EcoRI and BglII sites of pUAST to obtain pUAST-Ada2b1. From this plasmid, pUAST-Ada2b2 was constructed by replacing the Ada2b1 coding region between the codon for amino acid 297 and the stop codon with a PCR fragment corresponding to the third and fourth exons of Ada2b2 (generated with the primers AbNco [at +1670] and A2b2S [at +2967] (Table 1). pCaSpeR4-Ada2bg was constructed by insertion of the Ada2b genomic region from −593 to +3197 into the KpnI-NotI sites of pCaSpeR4. The Ada2b genomic fragment was obtained by PCR amplification using the primers A2bgenF (at −593), A2bgeneL (at +630), A2b3F (at +1716), and DNApol5R (at +3197) (Table 1).
TABLE 1.
Oligonucleotides used for this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| A2bRI | GCA TGA ATT CAT GAC CAC AAT CGC GGA TTT |
| A2bBHI | CGA TGG ATC CCC GAC AGC TAT CCA A |
| AbNco | CCA TAT GGC CAT GGC AAG |
| A2b2S | TTC AGT GGC TCA GCC AGC |
| A2bgenF | TTT AAT CCT GAC CAC CGC T |
| A2bgeneL | CAG GGT GGG TCG ATT ATG TTG |
| A2b3F | GTG CCG AAA TCG AAC TAA CCA |
| DNSpol5R | CAC AAT ATC TGA ATC CCG ACA TCT |
| st F | TTG AGC CCC TGA TAT TCG TC |
| st R | AAG TCG GCA CTC TCC TGA A |
| ry F | TAT CGA ATG GGT CGT CCT GT |
| ry R | GCT GCT GGT GGT AGT GTG TG |
| L17A F | GTG ATG AAC TGT GCC GAC AA |
| L17A R | CCT TAT TTC GCC CTT GTT G |
| rprFw | CCA GTT GTG TAA TTC CGA ACG A |
| rprRev | TCG CCT GAT CGG GTA TGT AGA |
| rpr probe | AAG AAA GAT AAA CCA ATG GCA GTG GCA |
| 18Fw | GCC AGC TAG CAA TTG GGT GTA |
| 18Rev | CCG GAG CCC AAA AAG CTT |
| 18 probe | TAT GGC TCT CTC AGT CG CTT CCG GG |
For the RT-PCR detection of rosy (ry), scarlet (st), and rpL17A transcripts, mRNAs were prepared from Ada2a, Ada2b, and wild-type wandering larvae and Ada2b and wild-type control animals 10 h after the white-pupa stage (P5), using Tri-reagent (Sigma). RT reactions were performed with Moloney murine leukemia virus reverse transcriptase (Sigma). The primers used for PCR amplifications were st F-st R (353 bp/467 bp), ry F-ry R (427 bp/708 bp), and L17A F-L17A R (268 bp/854 bp) (the sizes of the specific products from cDNA and genomic DNA templates, respectively, are given) (Table 1).
To detect mRNAs corresponding to Ada2b1 and Ada2b2, primers AbNco, AbRIBH, and A2bBHI (Table 1) were used, which resulted in amplification of the 290-bp and 224-bp products specific for Ada2b1 and Ada2b2, respectively. Descriptions of the PCR primers used to map deficiencies generated within the Ada2a and Ada2b regions are available upon request. Southern hybridizations were performed according to standard protocols. To detect changes in mRNA levels in wild-type versus Ada2bd842 mutants, total RNAs isolated from white pupae were analyzed by using Affymetrix Drosophila GeneChip arrays. Three independent pairs of samples of Ada2b and wild-type pupae were compared on six chips, and induction values relative to the basal levels are reported.
For the quantitative determination of rpr and rosy mRNAs, total RNAs were isolated with Tri-reagent (Sigma) according to the manufacturer's instructions. First-strand cDNAs were synthesized from 1 μg RNA, using TaqMan reverse transcription reagent (ABI). The relative abundance of rpr mRNA was quantified by quantitative RT-PCR (ABI Prism 7300), using TaqMan probes specific for rpr and for 18S rRNA as a control. The primers and probes for rpr mRNA and 18S rRNA were as follows: rprFw, rprRev, rpr probe, 18Fw, 18Rev, and 18 probe (Table 1). CT values were set against a calibration curve ranging over 2 orders of magnitude.
The extents of H3 and H4 acetylation in Ada2 mutant and wild-type animals were compared via the staining of polytene chromosomes obtained from the salivary glands of wandering larvae. Ac-H3 K14-, Ac-H3 K9-, and Ac-H4 K8-specific antibodies were purchased from Upstate. Mouse anti-Pol II (7G5) and anti-dTAF10 (TAF24) antibodies were raised against specific peptides as previously reported (14). The secondary antibodies were Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin and Alexa Fluor 555-conjugated goat anti-rabbit immunoglobulin. Polytene chromosome preparation and immunostaining were performed by previously published methods (29).
RESULTS
Characterization of Drosophila Ada2a and Ada2b mutations.
The Ada2a and Ada2b genes of Drosophila are located in the 90F10 and 84F5 cytological regions, respectively. Interestingly, both genes give rise to at least two protein products: Ada2a directs the synthesis of the ADA2a coactivator and the RNA Pol II subunit RPB4 mRNAs by alternative splicing (24), while Ada2b produces two related transcripts, encoding ADA2b1 and ADA2b2, which are identical in their N-terminal 330 amino acids but differ in their C-terminal ends (Fig. 1). Significant homology between ADA2a and ADA2b proteins exists only in their N-terminal regions, which are present in both ADA2b proteins. The RT-PCR detection of Ada2b messages by using primers permitting the detection of both forms indicates no apparent differences in the ratios of the two messages at different stages of Drosophila development (data not shown).
In order to facilitate functional studies of the two Ada2 genes of Drosophila, we generated Ada2a and Ada2b mutations by remobilizing P elements localized close to the 5′ ends of these genes. In the l(3)S096713 and EP(3)3412 lines, a P element is located 107 bp and 87 bp upstream of the transcription start sites of Ada2a and Ada2b, respectively (Fig. 1). The phenotype of l(3)S096713 homozygotes is larval lethal, while EP(3)3412 homozygotes are viable. By remobilizing the P element in l(3)S096713, we generated the deletion 189 (Ada2ad189), which removed nucleotides +107 to −613 of Ada2a (the numbering refers to the transcription start site of Ada2a). Ada2ad189 homozygotes and Ada2ad189/Df(3R)P14 (see Materials and Methods) heterozygotes are L1 lethal. Remobilization of EP(3)3412 resulted in the Ada2b alleles Ada2bd842, Ada2bd272, and Ada2bd52, which have deficiencies extending from the promoter region of Ada2b into the first intron, the second exon, and beyond the coding region into the adjacent gene, DNApol-iota, respectively (Fig. 1). The lethal phases of Ada2b mutants (Ada2bd842 and Ada2bd272 homo- or trans-heterozygotes and Ada2bd842/Ada2bd52 and Ada2bd272/Ada2bd52 mutants) are the late pupa (P5) and pharate-adult (pA) stages.
Since an imprecise jump-out of P elements might affect additional functions of Ada2a and Ada2b and since RPB4 is also produced from the Ada2a region via alternative mRNA splicing, we used transgenes to verify the Ada2 mutations and functionally dissect the Ada2a and Ada2b regions. A 7.1-kb genomic fragment (pCaSpeR4-Dtl+Ada2a+Rpb4+) (Fig. 1) completely restored the wild-type phenotype of Ada2ad189 and Ada2ad189/Df(3R)P14 animals. Although this indicated that the function(s) affected by the deficiency d189 is fully represented by this region, it still left open the question of whether the L1-lethal phenotype is related to Dtl, Ada2a, or Rpb4, all three located in the region, or a combination of them. To clarify this, we used further transgenes in which one of the three functions of the region was disabled (Fig. 1). Animals carrying the Dtl+Ada2a+ or Ada2a+Rpb4+ transgene in the Ada2ad189 background are L1 and L3 lethal, respectively, indicating that the loss of the Rpb4 transcript causes early larval lethality while the loss of the Dtl function results in late larval lethality. Ada2a mutants [Dtl+Rpb4+ transgene-carrier Ada2ad189 homozygotes or Ada2ad189/Df(3R)P14 animals] are either late-larval or early-pupal lethals. They follow a seemingly normal course of development until the L3 instar and survive for several days in the L3 stage, but a quarter of them (27%) never pupate. Three-quarters (73%) of Ada2a mutants form prepupae, which are similar to larvae in shape, are covered with a brownish cuticle, and die at this stage. Two transgenes, Dtl+Rpb4+ and Dtl+Ada2a+, together resulted in complete rescue, indicating that both the Rpb4 and Ada2a gene products are produced in sufficient amounts from a single-copy transgene. Consequently, from these data we concluded that the loss of the Ada2a function results in L3/early-pupa lethality.
Ada2b mutants (Ada2bd842 and Ada2bd272 homo- or trans-heterozygotes and Ada2bd842/Ada2bd52 and Ada2bd272/Ada2bd52 mutants) complete larval life and pupate like their heterozygous siblings. However, 77% of them perish at P5 and only 23% reach the pA stage, when a differentiated adult head, legs, and wings are clearly visible. The expression of either of the forms of Ada2b cDNAs or their combination resulted in partial rescue; 25% of the transgene-carrier animals also carrying an actin promoter-GAL4 driver completed normal development and emerged as adults. The rest died as pharate adults. A genomic fragment carrying the entire Ada2b coding region and 400 bp of its 5′ region (pCaSpeR-Ada2bg), however, resulted in complete rescue, implying that pupal lethality is a consequence of a lack of the Ada2b function. The penetrance of Ada2b mutations, similarly to that of Ada2a mutations, is 100%: no Ada2b or Ada2a animals emerged as adults during this study.
RT-PCR did not reveal either Ada2a-specific RNA in the Ada2ad189 P[Dtl+ Rpb4+] line or the Ada2b-specific transcript in Ada2bd842 and Ada2bd272/Ada2bd842 mutants (data not shown). Promoter mapping studies (26) indicated that both Ada2 genes have transcription regulatory regions extending <100 bp upstream from the transcription initiation sites. Therefore, we believe that the deletions d189, d842, and d272 d52 removed the promoters and parts of the 5′ regions of Ada2a and Ada2b, respectively, and can consequently be considered null alleles of the corresponding genes.
Ada2a Ada2b double mutants generated by recombination have a phenotype identical to that of the Ada2a mutants; most of the animals die in the third larval stage or several days later as prepupae. Neither the ectopic expression of the ADA2b protein in the Ada2a mutants nor the Ada2a+ transgene in the Ada2b mutants changed the phenotype, suggesting that the two proteins cannot substitute for each other; thus, the homologous ADA2 proteins have specific functions.
Analysis of Ada2 germ line and somatic mosaics.
Qi et al. (30) recently reported that the absence of Ada2b blocks oogenesis at an early stage. To determine whether the function of the Ada2a gene is also required in the female germ line, we transplanted germ line founder cells obtained from crosses producing, among others, Ada2ad189/Df(3R)P-14 eggs into ovoD1/+ host embryos (see Materials and Methods). The resulting adult females were analyzed for the ability to lay eggs and dissected to assess the differentiation of the egg primordia in them. Fourteen host embryos received germ line cells and developed into adult females that were able to lay eggs, but none of them had the Ada2ad189/Df(3R)P-14 germ line. The dissection of 42 more females which did not lay eggs revealed no signs of oogenesis progressing further than was characteristic for ovoD1/+ females (P < 0.05). This result clearly shows that the function of the Ada2a gene is required in the female germ line. To assess the requirements of ADA2a and ADA2b in somatic cells, we generated somatic mosaics by X-ray irradiation-induced mitotic recombination and recorded the appearance of the bristle morphology determined by the Ki+ marker (13). Mitotic recombination in Ada2ad189/Kis (or Ada2bd842/Kis) early-third-instar larvae leads to the formation of homozygous Ada2ad189 (or Ada2bd842) cells, which as a result of recombination have lost the Kis dominant marker and can therefore develop normal bristles (Ki+). We analyzed Ki+ clones at imaginal disk (head, notum, legs, and wing margin)- and histoblast-derived structures of the abdomen (tergites and sternites) since the two cell types display different proliferation dynamics (37). The results of the clonal analysis are summarized in Table 2. On disk-derived body parts, Ki+/Ki+, i.e., Ada2ad189/Ada2ad189 (or Ada2bd842/Ada2bd842), clones developed as frequently as those in the control, but they grew significantly smaller than those in the control (P < 0.01; Table 2). In contrast, on the abdomen the Ada2a and Ada2b clones were as large as those in the control (Table 2). Assuming that the difference seen between the body structures originated from imaginal cells and the histoblast is the result of the different division dynamics of the two cell types, the data indicate that the Ada2ad189/Ada2ad189 and Ada2bd842/Ada2bd842 cells do not die and can differentiate into normal bristles but cannot divide as many times as wild-type cells. Since the Ada2ad189/Ada2ad189 and Ada2bd842/Ada2bd842 cells originated from Ada2ad189/+ and Ada2bd842/+ cells, expression of the normal gene products in them at the time of clone induction made possible several rounds of cell divisions following mitotic recombination.
TABLE 2.
Analysis of Ada2a and Ada2b somatic mosaics
| Specimen | No. of flies | Parameters for imaginal disc derivatives
|
Parameters for abdomen
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Head
|
Notum
|
Leg
|
Wing margin
|
No. of clones | Sizea | Size of largest clonea | ||||||||||
| No. of clones | Sizea | Size of largest clonea | No. of clones | Sizea | Size of largest clonea | No. of clones | Sizea | Size of largest clonea | No. of clones | Sizea | Size of largest clonea | |||||
| Control | 90 | 9 | 2.7 ± 1.2 | 4 | 13 | 5.2 ± 3.8 | 16 | 21 | 8.4 ± 8.4 | 45 | 8 | 24.8 ± 14.9 | 46 | 79 | 2.8 ± 1.9 | 8 |
| Ada2a mutant | 36 | 2 | 1 ± 0 | 1 | 3 | 1.3 ± 0.5 | 2 | 12 | 1.2 ± 0.6 | 3 | 10 | 1.3 ± 0.7 | 3 | 22 | 2.0 ± 1.5 | 6 |
| Ada2b mutant | 48 | 3 | 1 ± 0 | 1 | 10 | 1.4 ± 0.7 | 3 | 10 | 2.2 ± 1.9 | 6 | 5 | 2.2 ± 0.8 | 3 | 44 | 2.6 ± 2.1 | 7 |
Bristle/cone.
Ada2a and Ada2b products play different roles in histone acetylation and exert different effects on TAF10 localization on polytene chromosomes.
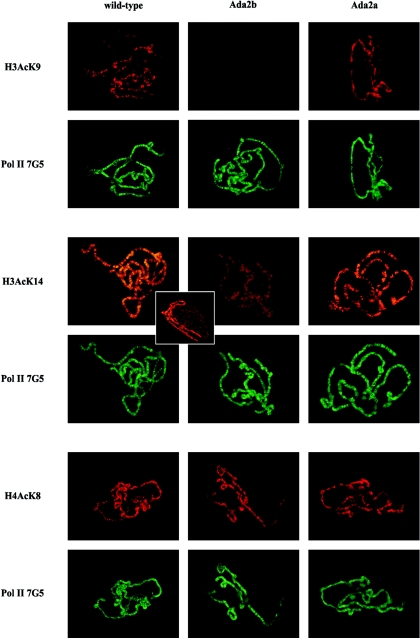
Yeast ADA2 participates in several chromatin-modifying complexes and potentiates GCN5 HAT activity. Since one of the primary targets of GCN5 is lysine 14 of nucleosomal H3 (16), we were interested in determining the effects of Ada2a and Ada2b mutations on H3 acetylation. The immunostaining of polytene chromosomes of late-third-instar larvae with an acetylated K14 H3-specific monoclonal antibody indicated a decreased level of H3 K14 acetylation in Ada2b mutants, while we could not detect any changes in H3 K14 acetylation in Ada2a null mutants (Fig. 2). The extensive role played by Ada2b in H3 K14 acetylation is clear from a comparison of the staining intensities of chromosomes obtained from Ada2b and wild-type animals (the latter can be easily identified from the morphology of the balancer chromosomes) that were stained and photographed together in mixed preparations (Fig. 2, inset). An overall decrease in the level of chromosome staining, without clear differences in the staining pattern of Ada2b chromosomes, is obvious compared with wild-type chromosomes. Similar to H3 K14 acetylation, H3 K9 acetylation is also decreased in Ada2b mutants (Fig. 2). In contrast, Ada2a mutations have no detectable effect on either H3 K14 or H3 K9 acetylation (Fig. 2), and anti-acetylated H4 lysine 8 antibody gives comparable signals on wild-type, Ada2a, and Ada2b polytene chromosomes (Fig. 2). It should be noted, however, that in Ada2a mutants the structure of the polytene chromosomes is altered and the banding pattern is often distorted (Fig. 3A). Whether the altered chromosome structure results from the loss of the Ada2a function specifically or is a more general effect of the retarded development of Ada2 animals at a late larval stage is unclear.
FIG.2.
Effects of Ada2a and Ada2b mutations on H3 K14, H3 K9, and H4 K8 acetylation on polytene chromosomes. Chromosomes immunostained with acetylated H3- and H4-specific polyclonal antibodies (H3AcK9, H3AcK14, and H4AcK8) and a Pol II-specific monoclonal antibody (Pol II 7G5) are shown. Genotypes are indicated at the top. The pictures indicating immunostaining in different mutants were obtained with identical data-recording settings. The inset shows Ada2b and wild-type polytene chromosomes immunostained in a mixed preparation with H3AcK14-specific polyclonal antibody. Chromosomes were identified on the basis of the structure of balancer chromosomes in wild-type animals.
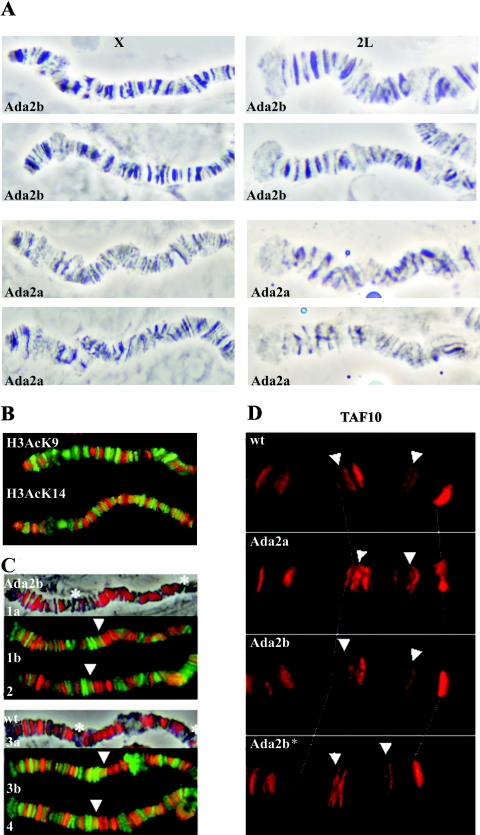
FIG. 3.
(A) Phase-contrast images of X and 2L polytene chromosome regions of Ada2b and Ada2a mutants. Note the distorted banding pattern and less condensed structure of Ada2a chromosomes. (B) Enlarged parts of wild-type X chromosomes coimmunostained for Pol II (green) and acetylated H3 K9 or H3 K14 (red) show identical distributions of H3 K9 and H3 K14 acetylation. (C) Enlarged parts of Ada2b (1a, 1b, and 2) andwild-type (3a, 3b, and 4) 2L chromosomes stained for H3AcK14 (red) and Pol II (1b, 2, 3b, and 4; green). H3AcK14 staining in both wild-type and Ada2b mutants is colocalized mostly, but not exclusively, with bands, while Pol II immunostaining is colocalized with interband regions. The asterisks and arrowheads indicate bands exhibiting weak staining for H3AcK14 and colocalization of H3AcK14 and Pol II, respectively. (In order to allow a comparison of the Ada2b mutant and the wild type, images of H3AcK14-specific antibody-stained chromosomes are color enhanced.) (D) Localization of TAF10 in the 3R region of wild-type and Ada2 polytene chromosomes. Genotypes are indicated in the pictures. Ada2b* is a chromosome from an Ada2b homozygote carrying the Ada2b+ transgene. Arrows point to locations where TAF10 localization in the Ada2b mutant is lost.
The loss of H3 K14/K9 acetylation in the Ada2b mutants is in accord with our biochemical separation and coimmunoprecipitation results, which indicated that ADA2b is part of the 1.8-MDa TFTC-like complex, together with the GCN5 HAT, while ADA2a is associated with a lower-molecular-weight complex (24).
Moreover, in the TFTC-like complex, ADA2b was found to be associated with TAF10, while in the ADA2a-containing complex, TAF10 was absent (24). Thus, we set out to determine whether in vivo mutation in either Ada2b or Ada2a would influence the binding of TAF10 to polytene chromosomes. A comparison of the immunolocalization patterns of TAF10 on polytene chromosomes of wild-type and Ada2a and Ada2b mutant third-instar larvae revealed the complete and reproducible loss of a few chromosomal bands (two to four per chromosome) in the Ada2b mutants (Fig. 3D). In addition, weaker labeling was observed in the Ada2b mutants at many other sites (data not shown, but see Discussion). The introduction of an Ada2b+ transgene into Ada2b animals restored a staining pattern of polytene chromosomes identical to that seen in wild-type animals, providing further evidence that the loss of TAF10 localization at these sites resulted from ADA2b depletion (Fig. 3D). In contrast, no significant changes were observed in Ada2a null mutants. Overall, these in vivo results indicate that Ada2a and -b exert different influences on the general histone acetylation observed on polytene chromosomes, and in agreement with the in vitro results, suggest that ADA2b, GCN5, and TAF10 can function together.
Ada2a and Ada2b differ in their effects on eye pigment formation.
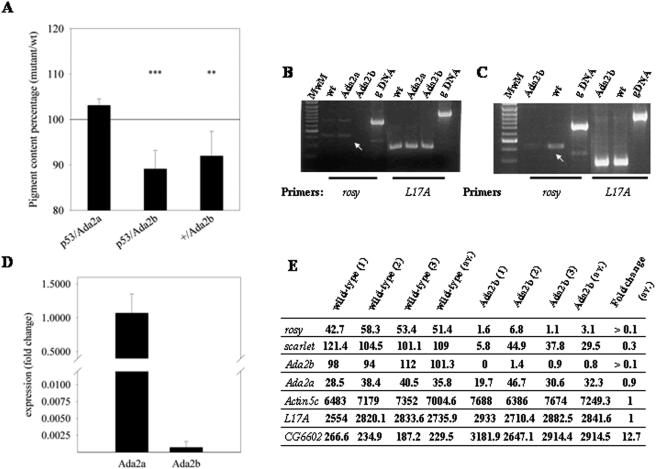
A functional difference between the two ADA2 proteins of Drosophila was suggested on the basis of their interactions with Dmp53, since in immunoprecipitation experiments ADA2b coprecipitated with Dmp53 while ADA2a did not (19). To extend these studies, we tested genetic interactions between the Ada2 genes and Dmp53. The ectopic expression of Dmp53 in the developing eye of Drosophila results in apoptosis, as observed with the rough eye phenotype of adult animals (21). The loss of function of factors which enhance or inhibit the Dmp53 function is expected to suppress or enhance the phenotype, respectively. To obtain a quantitative measure of the interplay of Dmp53 and ADA2a or -b, we determined the eye pigment contents of animals carrying one copy of a Dmp53+ transgene ectopically expressed in the eye in the Ada2a/+ and Ada2b/+ backgrounds. A comparison of the pigment contents of Ada2 heterozygotes and wild-type animals indicated that Ada2b mutation affected the pigment contents of the eyes of adult animals, while Ada2a mutation did not (Fig. 4A). Surprisingly, however, this effect was independent of Dmp53; the formation of red eye pigment in Ada2b heterozygotes was significantly reduced both in the presence and in the absence of ectopically expressed Dmp53. In concert with this, the mRNA level of rosy, a gene encoding xanthine dehydrogenase, which is involved in the formation of red eye pigments, exhibited a significant reduction in Ada2b mutant animals compared to wild-type animals (Fig. 4B to E), while the expression of rosy did not change in Ada2a null animals. In some experiments with Ada2b mutants, we also observed, though to a lesser extent, a decrease in the expression of scarlet, another gene involved in pigment formation (Fig. 4E and data not shown). This suggests that ADA2a and -b are differentially involved (directly or indirectly) in the transcriptional regulation of the rosy and scarlet genes. Indeed, preliminary data from chromatin immunoprecipitation experiments indicated that ADA2b is present at the promoter region of the rosy gene in S2 cells (data not shown).
FIG. 4.
Effects of Ada2a and Ada2b mutations on eye pigment formation. (A) Red eye pigment contents of Ada2a/Dmp53, Ada2b/Dmp53, and Ada2b/+ adult heterozygotes. The pigment contents of groups of 10 animals with the indicated genetic backgrounds were determined as described in Materials and Methods. Ratios of pigment contents of Ada2 mutants to those of the corresponding controls (p53/Ada2a versus p53/+, p53/Ada2b versus p53/+, and +/Ada2b versus +/+) are shown. The level of rosy mRNA is reduced in Ada2b mutants, as determined by RT-PCR (B and C), quantitative PCR (D), and DNA microarray analysis (E). The levels of rosy mRNA and L17A mRNA as a control were determined by RT-PCR with total RNA samples prepared from third-instar larvae (B) and 10-h pupae (C). The genotypes are indicated. wt, wild type; gDNA, genomic DNA control. (Note that Ada2a animals do not reach the pupa stage.) Arrows point to specific products present at a decreased level. (D) Level of rosy mRNA in Ada2a and Ada2b third-instar larvae compared to that in wild-type controls, as determined by quantitative RT-PCR. (E) Levels of selected mRNAs, including rosy, scarlet, Ada2a, and Ada2b, in wild-type and Ada2b mutant white pupae determined on three pairs of microarrays.
Ada2a and Ada2b differentially affect Dmp53 function.
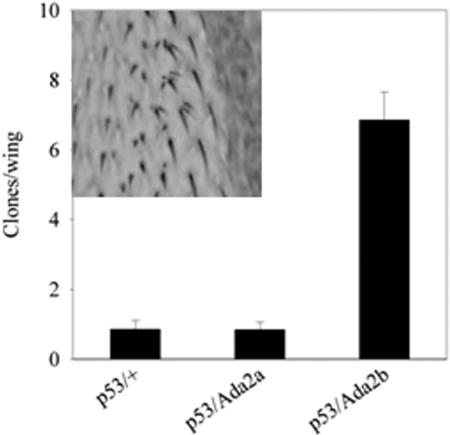
The rough eye phenotype arising from induced apoptosis is only one of a number of possibilities permitting the testing of Dmp53 functions in Drosophila. LOH assays are established means of demonstrating the role played by Dmp53 in preserving genome stability. We carried out LOH assays based on detection of the recessive multiple wing hairs (Mwh) phenotype revealed by the loss of the wild-type copy of mwh. Late-third-instar larvae heterozygous for Dmp53 and the mwh mutation and either wild-type or heterozygous for Ada2a or Ada2b were irradiated with a low-level X-ray dose (250 R). Cells which have lost the wild-type copy of mwh display the recessive Mwh phenotype, which is easily recognizable and can be scored on the wings. The numbers of mwh clones obtained following X-ray treatment were strikingly different for Dmp53+/+ Ada2a and Dmp53+/+ Ada2b animals (Fig. 5). For Ada2a heterozygotes, the frequency of clones was similar to that seen with wild-type animals. In contrast, Ada2b heterozygotes exhibited substantially elevated numbers of mwh cells under conditions that had no effect on wild-type flies or Ada2a heterozygotes. This result clearly demonstrates that ADA2b, unlike ADA2a, is involved in the pathway induced by Dmp53 to preserve genomic stability in response to DNA damage induced by low-level X-ray radiation.
FIG. 5.
Ada2b function is required for the preservation of genomic stability in response to a genotoxic effect. The numbers of mwh clones found on the wings of wild-type, Ada2a, and Ada2b animals following low-level X-ray irradiation at the L3 stage are shown in the graph. The inset shows an mwh clone in the wild-type background. Error bars denote the standard errors of the means.
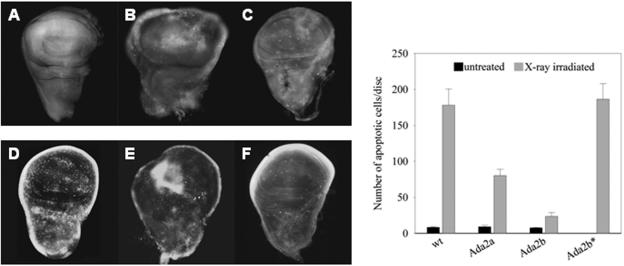
The Dmp53 function is also essential for radiation-induced apoptosis. DNA damage leads to Dmp53 activation, which through the transcriptional activation of proapoptopic factors brings about apoptosis. In order to test whether Ada2 functions are required for this process, we X-ray-irradiated wild-type and Ada2 mutant third-instar larvae, and after a 4-h recovery period, dissected wing imaginal disks and subjected them to staining with the vital stain AO. Staining of disks from larvae ectopically expressing Dmp53 in the eye disks showed that under the conditions used, AO specifically stained cells undergoing apoptosis (data not shown). Radiation with 4,000 R induced apoptosis in the disks of wild-type animals. For Ada2a mutants, and more extensively for Ada2b mutants, the number of cells undergoing apoptosis was significantly lower than that in wild-type animals (Fig. 6). For both tested Ada2b alleles (Ada2bd842 and Ada2bd52), the number of AO-stained cells was significantly decreased. The introduction of the Ada2bg transgene into Ada2b null animals restored apoptosis to the level seen in the wild type (Fig. 6), indicating that the decreased level in Ada2b mutants was a result of ablation of the Ada2b function. To test whether Ada2b mediates Dmp53 functions by altering rpr transcription (see the introduction), we compared the rpr mRNA levels in wild-type and Ada2 mutants. Interestingly, high-dose (4,000 R) X-ray irradiation, which resulted in a decrease in apoptosis in wing imaginal disks, induced the rpr message level similarly (a three- to fourfold increase compared with the nonirradiated controls) in wild-type and Ada2b larvae. Surprisingly, we could not detect rpr mRNA induction under the same conditions in Ada2a mutants. Taken together, these data not only suggest a different involvement of the ADA2 proteins in Dmp53-mediated processes but also indicate that ADA2b exerts its effect on a Dmp53-related pathway(s) other than those including rpr induction.
FIG. 6.
The level of radiation-induced apoptosis is decreased in Ada2 mutants. Wing imaginal disks of wild-type (D), Ada2a (E), and Ada2b (F) L3 larvae were irradiated at 4,000 R, left to recover for 4 h, and dissected, and the disks were stained with acridine orange. Controls (A, B, and C) were treated similarly, but without irradiation. (Right panel) Average number of AO-stained cells per wing disk, as determined in three independent experiments, each involving 10 disks or more. Ada2b* indicates Ada2b homozygotes carrying the Ada2b+ transgene.
DISCUSSION
Transcriptional coactivator complexes have recently received a great deal of attention, and their roles in several aspects of gene-specific transcription regulation are well documented. The SAGA complex of yeast, for example, is known to participate in activator interaction, histone acetylation, and TATA-binding protein interaction. Coactivator complexes of higher eukaryotes can be expected to exhibit an even broader functional versatility, underscored by their structural diversity (40). Biochemical studies of ADA2-containing Drosophila and human HAT complexes support this assumption and suggest functional differences between complexes built up from similar, but often distinct, building blocks or modules (19, 24). However, a full exploration of the extent of functional diversity of coactivator complexes resulting from the subtle alterations in their compositions requires a combination of powerful in vivo assays with biochemical and genetic approaches. As a step toward this goal, we report here the functional in vivo analysis of the two Ada2 genes of Drosophila and their corresponding protein products.
The two ADA2 proteins of Drosophila melanogaster are encoded by genes located at different regions of the third chromosome. Our results did not indicate a concerted regulation of the expression of the two Ada2 genes. It is noteworthy that both Ada2 genes have short 5′ regulatory regions with no obvious similarities which would suggest the presence of binding sites for common transcriptional activators. Interestingly, both Ada2 genes give rise to a number of protein products. One of the mRNAs of Ada2a encodes the fourth largest subunit of RNA Pol II (RPB4). An RPB4 mutation in S. cerevisiae results in a cold-sensitive phenotype, while an RPB4 deficiency in Schizosaccharomyces pombe is lethal (31). In Drosophila, the lack of RPB4 causes early larval lethality, as indicated by animals carrying mutations in the Ada2a and Rpb4 genes and a transgene providing Ada2a function. The losses of ADA2a and RPB4 have sharply different consequences, as demonstrated by the first-larval and third-larval lethal phenotypes of the corresponding null mutants, despite the fact that the two mRNAs arise from one transcription unit regulated by the same promoter (24). This suggests that a posttranscriptional regulatory mechanism(s) plays a role in governing ADA2a and RPB4 mRNA production. In light of numerous independent observations that RPB4 is involved in several steps of transcription and mRNA processing (for a review, see reference 12), it is reasonable to assume that the loss of the function(s) performed by this polymerase subunit compromises life in an earlier stage of development than in the case of the loss of ADA2a. On the other hand, exhaustion of the maternal pools of ADA2a and RPB4 by different stages of development might also lead to the different phenotypes. In either case, the observation that Ada2a/Rpb4 cotranscription leads to different levels of mRNAs and expression of the two transcription regulatory factors raises a number of interesting questions, which can be tackled with the help of the mutants we describe here.
Ada2b of Drosophila also produces at least two mRNAs, which are translated into proteins with identical N-terminal but different C-terminal regions. Upon assessing their presence at different stages of Drosophila development by RT-PCR, we did not find alterations in the levels of mRNAs leading to ADA2b1 and ADA2b2 production (data not shown). The functional equivalence of the two forms of ADA2b protein is indicated convincingly by the observation that ectopically expressed transgenes carrying cDNA fragments corresponding to either the Ada2b1 or Ada2b2 mRNA are equally effective at rescuing Ada2b mutations, and the two transgenes together produce a result identical to that produced by either alone. It is noteworthy that neither of the cDNAs containing Ada2b transgenes resulted in full rescue of the Ada2b mutant phenotype. Accordingly, it is possible that some function of Ada2b is missing from both cDNAs used. On the other hand, it is also conceivable that the GAL4-responsive promoter-driven pUAST transgenes produced the protein(s) at a level insufficient for full rescue. In contrast, a genomic fragment with the authentic Ada2b promoter resulted in complete rescue.
The Ada2a and Ada2b alleles we obtained by the remobilization of P elements inserted in the 5′ regions of the corresponding genes are apparently null alleles. Improper P-element excision in the Ada2ad189, Ada2bd842, Ada2bd272, and Ada2bd52 lines removed the promoter regions of the genes and substantial parts of the coding regions.
In contrast with the yeast ada2 mutation, which has a conditional phenotype (7), both Ada2 genes of Drosophila are essential. Ada2a and Ada2b mutants have well-defined and characteristic lethal phenotypes at specific stages of development, demonstrating the functional difference that exists between them. Since both ADA2 proteins of Drosophila have been shown to interact with GCN5, it is not surprising that Gcn5 mutations result in a phenotype similar to that furnished by Ada2a mutations (our unpublished observation; C. Antoniewsky [Pasteur Institute, France], personal communication). The phenotype of Ada2a Ada2b double mutants is identical to the more severe phenotype observed with the Ada2a null mutation, and ectopic expression of either of the ADA2 proteins does not change the phenotype observed in the absence of the other, indicating that they are unable to substitute for each other functionally. In light of these data, it is unlikely that the different consequences of Ada2a and Ada2b mutations are the results of different expression patterns of functionally identical proteins. The two ADA2 proteins of Arabidopsis are also believed to be functionally distinct, as the ectopic overexpression of Arabidopsis Ada2a did not complement Ada2b mutation (41). Plants carrying a mutation of Ada2b display pleiotropic effects at several stages of plant development and infertility. Mutation of the Ada2a gene of Arabidopsis, however, has so far not been described.
An analysis of somatic mosaics indicated that both Ada2 functions of Drosophila are required for normal cell division. Ada2a or Ada2b homozygous cells do not die but cannot divide as many times as normal cells. The differences seen in the number and size of homozygous mosaics on different body parts can be explained by the different proliferation dynamics of imaginal disk and histoblast (abdominal precursor) cells: imaginal disk cells proliferate throughout larval life, while histoblast cells grow in size, accumulate cell components, and divide only after the initiation of pupation. Consequently, the long persistence of normal Ada2 gene products permits normal proliferation of the Ada2a and Ada2b cells over the abdomen. An analysis of germ line mosaics indicated that Ada2a is indispensable in the female germ line. Qi et al. (30) recently reported that Ada2b homozygous germ cells are also arrested at an early stage of oogenesis.
Ada2a and Ada2b mutations affect H3 K14 and H3 K9 acetylation differently. Mutations of Ada2b which cause lethality in the late pupal stage result in a significant reduction of H3 K14/K9 acetylation (Fig. 2), while a mutation of Ada2a which has a late-larval-early-pupal lethal phenotype has no obvious effect on this type of histone modification (Fig. 2). Taken together, these observations are noteworthy for several reasons: they clearly demonstrate that the two ADA2 proteins have distinct functions, but with no indication that either ADA2 can substitute for the function of the other, and they further demonstrate that a reduced level of H3 K14/K9 acetylation (in the absence of ADA2b) does not interfere with development until the late larval stage. It is interesting that the loss of ADA2b, which is believed to be a component of various complexes with HAT activity, e.g., the SAGA/TFTC-like multiprotein complex(es), results in a later lethal phase than the loss of ADA2a, which participates in a smaller multiprotein complex(es). It is unlikely that the H3 K14/K9 acetylation observed in the late larval stage of Ada2b animals was due to a residual fraction of maternal ADA2b protein. It may rather be assumed that either HAT complexes other than SAGA/TFTC (which do not require ADA2b) or SAGA/TFTC-like complexes (but with reduced activity in the absence of ADA2b) are responsible for H3 K14/K9 acetylation in Ada2b mutants. Recently, Qi et al. (30) reported an analysis of Ada2b mutants generated by remobilization of the same P element that we used for this study. With respect to the effect on H3 K9, H3 K14, and H4 K8 acetylation, the phenotypes of Ada2b mutants Qi et al. studied and those described here are identical.
The decreased level of H3 K14/K9 acetylation in Ada2b mutants does not affect the localization of RNA Pol II profoundly; polytene chromosomes of Ada2a, Ada2b, and wild-type animals display similar staining intensities with Pol II-specific antibody (Fig. 2). The distributions of K9 and K14 acetylated H3 along the polytene chromosome seem to be very similar, if not identical (Fig. 3B). Fig. 3C demonstrates that Ac-H3 K14 colocalizes with bands, while Pol II is localized mostly to the interbands. In general, the staining pattern of Ac-H3 K14 parallels the banding pattern of unstained chromosomes, as if the distribution of Ac-H3 K14 were proportional to the DNA concentration (assuming that stronger bands represent highly compacted DNA). However, a careful comparison of native and stained chromosome preparations reveals that while the density and staining intensity correlate at many sites, in numerous cases they do not, and at some sites strong bands do not demonstrate intense staining with Ac-H3 K14 antibody (Fig. 3C). Furthermore, the colocalization of Ac-H3 K14 and Pol II can be found in other regions, both in bands and at less condensed areas (Fig. 3C). The band-interband distribution of Ac-H3 K14 and transcribed genes has also been observed by others (20, 29). From these data, in view of the Ac-H3 K14 localization and the phenotype of Ada2b mutation, it should be assumed that ADA2b-containing HAT complexes have multiple roles, which are exerted through participation in generally distributed “maintenance” and transcriptional activation-induced Ac-H3 K14/K9 acetylation. Interestingly, in contrast with the effect of Ada2b mutation, Ada2a deficiency does not change either histone 3 K9/K14 or histone 4 K8 acetylation to an extent detectable by the immunostaining technique we used.
Ada2a and Ada2b mutants also differ in that TAF10 localization at some, but not all, specific bands is lost in Ada2b null mutants. In contrast, the localization of TAF10 is not affected in Ada2a null mutants (Fig. 3D). This is in agreement with the previous observation that ADA2b, but not ADA2a, participates in a SAGA/TFTC-type multiprotein complex(es) together with TAF10 and GCN5 (24). This suggests that in certain complexes the presence of ADA2b is required for the incorporation of TAF10. In some cases, we observed that the labeling of TAF10 on the polytene chromosomes of Ada2b null mutants became weaker but did not disappear completely (data not shown). Since TAF10 was identified in both TFIID and TFTC complexes (14, 24), it is conceivable that at these chromosomal locations the binding of TFTC-type complexes, but not that of TFIID complexes, has been abolished. The complete disappearance of some TAF10-labeled sites in the Ada2b null mutants, however, suggests that at these loci only the TFTC-type complexes play a role in chromatin remodeling and/or transcription initiation. Overall, these in vivo results further corroborate our in vitro result that ADA2b, GCN5, and TAF10 can function together in the same complex. In Ada2a mutants, neither H3 K14/K9 acetylation nor TAF10 localization is changed considerably, as indicated by an assessment of the staining of Ada2a polytene chromosomes compared with that of wild-type chromosomes. Ada2a mutation, however, greatly affects the general structure of polytene chromosomes (Fig. 3A). We noted that Ada2a chromosomes are more fragile and that their banding pattern is often distorted, while we observed smaller puffs reflecting intensely transcribed regions on them. The cause of these morphological changes in the Ada2a chromosomes is at present unclear. It is interesting in this respect that the ADA2a protein has been shown to colocalize mostly with RNA Pol II in intensely transcribed interband regions (19). The phenotypes of Ada2 mutations and their effects on histone modification and TAF10 localization corroborate previous biochemical data indicative of functional differences between the two ADA2 proteins (19, 24). Since Drosophila is the first organism for which mutations have been obtained in both Ada2 genes, the question arose of whether functional differences between the two ADA2 proteins can also be seen in specific transcriptional activator-mediated processes. To address this issue, we chose genetic assays enabling us to follow alterations in the function of Dmp53, the tumor suppressor known to be involved in apoptosis induction in Drosophila (25). The rationale for monitoring the Dmp53 function in an Ada2 mutant background was the previous detection of a Dmp53 interaction with ADA2b, but not with ADA2a, by coprecipitation and an in vitro pull-down assay (19) and the fact that hADA3, which participates in coactivator complexes together with ADA2s, is required for human p53 activity (42).
Using an assay suitable for the detection of Dmp53-induced apoptosis in the eye, we found that the production of red eye pigment is significantly reduced in Ada2b heterozygotes, whereas it is not affected in Ada2a mutants. In accord with this, in Ada2b mutants we detected decreased mRNA levels of the rosy gene, and to a lesser extent, the scarlet gene, which are known to be involved in pigment formation/transport. However, this effect of Ada2b is independent of the Dmp53 status of the cells. Although these data did not establish a functional link between either ADA2a or -b and Dmp53, they do indicate that ADA2b, but not ADA2a, participates directly or indirectly in the transcription of these genes, pointing to another functional difference between the two ADA2 proteins. The observation that the effect of Ada2b on eye pigment formation was independent of the Dmp53 status of the cells did not argue against a functional link between ADA2b and Dmp53. It is possible that the assay we employed to detect dominant interactions was not sufficiently sensitive. Alternatively, since Dmp53 is assumed to have several proapoptotic targets (10, 11, 17), it could be that the transcription of some of them is affected by Ada2b while that of others is not. Mutations of the two Ada2 genes had different effects on Dmp53-mediated processes, which we monitored with the well-established LOH test and by the detection of radiation-induced apoptosis in imaginal disks. The significant increase in the level of mwh clones in an Ada2b mutant background and the decrease in the number of cells undergoing apoptosis in the wing disks in the absence of Ada2b suggest that the ADA2b protein participates in some of the Dmp53 functions. In both tests, a reduced level or lack of ADA2b suppressed the Dmp53-mediated functions. In contrast, Ada2a mutation exerted an appreciably weaker effect on the number of apoptotic cells in the disks and did not display any effect in the LOH assay. To ascertain whether either Ada2 gene is involved in the activation of rpr, a proapoptotic target of Dmp53, we compared the extent of radiation induction of rpr mRNA by quantitative RT-PCR for wild-type and mutant Ada2 animals. The levels of rpr message induction—three- to fourfold—in the wild-type and Ada2b mutants were identical, while for Ada2a mutants we repeatedly did not detect rpr mRNA induction following irradiation. Although this result might at first seem surprising, in light of numerous data indicating that Dmp53 can induce apoptosis through proapoptotic factors other than rpr it can be interpreted as Ada2b being involved in pathways which do not include rpr.
Qi et al. (30) recently reported the generation and analysis of Ada2b mutant alleles in Drosophila. Most of our results, such as the decreased levels of H3 acetylation and rpr induction in the absence of ADA2b, are in good agreement with their data. However, unlike us, they found that gamma irradiation led to an increase in the number of cells undergoing apoptosis in the disks of Ada2b animals. The explanation of the different results could lie in the conditions and techniques used to induce DNA damage. Most importantly, however, both we and Qi et al. (30) reached the conclusion that Ada2b plays a role in the DNA damage-induced Dmp53-dependent pathway. Our data additionally demonstrate that in this respect, and also as concerns many other functions that we tested in vivo, including eye pigment formation, H3 K14 and K9 acetylation, and TAF10 localization, ADA2b and ADA2a behave differently. The functional differences between the two homologous Drosophila ADA2 proteins described here on the basis of the results of in vivo studies are in full accord with previous data obtained from this and other laboratories by the in vitro biochemical separation of ADA2-containing complexes (19, 24). Thus, we believe that Ada2a and Ada2b mutants can serve as valuable new tools for the in vivo analysis of ADA2-containing complexes, which despite their structural and functional similarities, also exhibit noteworthy differences, particularly in higher eukaryotes.
Acknowledgments
We thank Katalin Ökrös and Bettina Nagy for their expert technical help and Christophe Antoniewsky for antibodies and for sharing unpublished results.
This work was supported by grants from the Hungarian Science Fund (OTKA T046414) and the Hungarian Ministry of Health (ETT 078/2003) to I.B., by funds from INSERM, CNRS, Hopital Universitaire de Strasbourg, Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Font Nationale de La Science ACI, INTAS (01-0211), and AICR (03-084) to L.T., and by grants from the European Community RTN (HPRN-CT-2004-504228) and STREP (LSHG-CT-2004-502950) to L.T. and I.B.
REFERENCES
- 1.Anafi, M., Y. F. Yang, N. A. Barlev, M. V. Govindan, S. L. Berger, T. R. Butt, and P. G. Walfish. 2000. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 14:718-732. [DOI] [PubMed] [Google Scholar]
- 2.Asburner, M. 1989. Drosophila laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 4.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 6.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23:6944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik, S. R., and M. R. Green. 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370:445-454. [DOI] [PubMed] [Google Scholar]
- 9.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 10.Brodsky, M. H., W. Nordstrom, G. Tsang, E. Kwan, G. M. Rubin, and J. M. Abrams. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103-113. [DOI] [PubMed] [Google Scholar]
- 11.Brodsky, M. H., B. T. Weinert, G. Tsang, Y. S. Rong, N. M. McGinnis, K. G. Golic, D. C. Rio, and G. M. Rubin. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24:1219-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choder, M. 2004. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 29:674-681. [DOI] [PubMed] [Google Scholar]
- 13.Gausz, J., H. Gyurkovics, and J. Szabad. 1987. KiS, a new cell marker mutation. Drosophila Inform. Serv. 66:190. [Google Scholar]
- 14.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 16.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 17.Jassim, O. W., J. L. Fink, and R. L. Cagan. 2003. Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. EMBO J. 22:5622-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komitopoulou, K., M. Gans, L. M. Margaritis, F. C. Kafatos, and M. Masson. 1983. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics 105:897-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labrador, M., and V. G. Corces. 2003. Phosphorylation of histone H3 during transcriptional activation depends on promoter structure. Genes Dev. 17:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. H., E. Lee, J. Park, E. Kim, J. Kim, and J. Chung. 2003. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 550:5-10. [DOI] [PubMed] [Google Scholar]
- 22.Lindsley, D. L., and G. G. Zimm. 1992. The genome of Drosophila melanogaster. Academic Press, Inc., San Diego, Calif.
- 23.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollmann, M., L. M. Young, C. J. Di Como, F. Karim, M. Belvin, S. Robertson, K. Whittaker, M. Demsky, W. W. Fisher, A. Buchman, G. Duyk, L. Friedman, C. Prives, and C. Kopczynski. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91-101. [DOI] [PubMed] [Google Scholar]
- 26.Pápai, G., O. Komonyi, Z. Tóth, T. Pankotai, S. Muratoglu, A. Udvardy, and I. Boros. 2005. Intimate relationship between the genes of two transcriptional coactivators, ADA2a and PIMT, of Drosophila. Gene 348:13-23. [DOI] [PubMed] [Google Scholar]
- 27.Perrimon, N., L. Engstrom, and A. P. Mahowald. 1984. The effects of zygotic lethal mutations on female germ-line functions in Drosophila. Dev. Biol. 105:404-414. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, C., G. E. Carney, B. J. Taylor, and K. White. 2002. Reaper is required for neuroblast apoptosis during Drosophila development. Development 129:1467-1476. [DOI] [PubMed] [Google Scholar]
- 29.Pile, L. A., and D. A. Wassarman. 2002. Localizing transcription factors on chromatin by immunofluorescence. Methods 26:3-9. [DOI] [PubMed] [Google Scholar]
- 30.Qi, D., J. Larsson, and M. Mannervik. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24:8080-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai, H., H. Mitsuzawa, M. Kimura, and A. Ishihama. 1999. The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol. 19:7511-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 33.Silverman, N., J. Agapite, and L. Guarente. 1994. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc. Natl. Acad. Sci. USA 91:11665-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sogame, N., M. Kim, and J. M. Abrams. 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA 100:4696-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockinger, E. J., Y. Mao, M. K. Regier, S. J. Triezenberg, and M. F. Thomashow. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szabad, J. 1978. Quick preparation of Drosophila for microscopic analysis. Drosophila Inform. Serv. 53:215. [Google Scholar]
- 37.Szabad, J., and P. J. Bryant. 1982. The mode of action of “discless” mutations in Drosophila melanogaster. Dev. Biol. 93:240-256. [DOI] [PubMed] [Google Scholar]
- 38.Szabad, J., and J. Szidonya. 1980. Developmental analysis of fs(1)1867, an egg resorption mutation of Drosophila melanogaster. Basic Life Sci. 16:95-108. [DOI] [PubMed] [Google Scholar]
- 39.Thummel, C. S., and V. Pirrotta. 1991. New PCaSpeR P element vectors. Drosophila Inform. Newsl. 71:50. [Google Scholar]
- 40.Timmers, H. T., and L. Tora. 2005. SAGA unveiled. Trends Biochem. Sci. 30:7-10. [DOI] [PubMed] [Google Scholar]
- 41.Vlachonasios, K. E., M. F. Thomashow, and S. J. Triezenberg. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, T., T. Kobayashi, R. Takimoto, A. E. Denes, E. L. Snyder, W. S. el-Deiry, and R. K. Brachmann. 2001. hADA3 is required for p53 activity. EMBO J. 20:6404-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, M., L. Newcomb, and W. Heideman. 1999. Regulation of gene expression by glucose in Saccharomyces cerevisiae: a role for ADA2 and ADA3/NGG1. J. Bacteriol. 181:4755-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]