Abstract
Mutation of BRG1, hBRM, and their associated factors, INI1 and BAF57, in primary human tumors has suggested that inactivation of human SWI/SNF (hSWI/SNF) complexes may be involved in neoplastic transformation. BT549 is an invasive human breast carcinoma cell line that lacks expression of BAF57, a key hSWI/SNF subunit that mediates interaction with transcriptional activators and corepressors. In this study we investigated the role of BAF57 in suppressing tumorigenesis by establishing BT549 stable cell lines that expresses full-length BAF57 protein. BT549 clones expressing BAF57 demonstrated marked phenotypic changes, slow growth kinetics, and restoration of contact inhibition. Altered growth was found to be due in part to cell cycle arrest and induction of apoptosis. Furthermore, microarray analysis revealed that BAF57-mediated cell death was associated with up-regulation of proapoptotic genes including the tumor suppressor familial cylindromatosis (CYLD), which was found to be a direct target of BAF57 as determined by chromatin immunoprecipitation analysis. Increased expression of CYLD in BT549 cells induced apoptosis, while its suppression by small interfering RNA inhibited cell death in BAF57 expressing BT549 cells. These findings demonstrate the importance of BAF57 in cell growth regulation and provide a novel link between hSWI/SNF chromatin remodelers and apoptosis.
Transcriptional regulation in normal and neoplastic tissues is a complex and dynamic process that is in part dependent on multisubunit protein complexes that modulate higher-order chromatin structure. The tightly compacted nature of chromatin makes DNA generally inaccessible to the transcription machinery. Thus, ATP-dependent protein complexes are required to modify nucleosomal structure to allow for the regulation of transcriptional activation and repression. The human SWI/SNF (hSWI/SNF) complex is a ubiquitous 2-MDa multimeric complex that regulates gene expression by remodeling nucleosomal structure in an ATP-dependent manner (29, 40, 48). hSWI/SNF complexes include either the BRG1 or hBRM DNA-dependent ATPase and their associated factors BAF250, BAF170, BAF155, BAF60, BAF57, BAF53, and BAF45. The role of each BAF has not been completely elucidated, but it is clear that BRG1 or hBRM in combination with BAF170, BAF155, and BAF45 can reconstitute chromatin remodeling, while other BRG1- or hBRM-associated factors (BAFs) such as BAF60 and BAF57 mediate interaction with transcriptional activators or repressors (5, 19, 22, 33, 34). Recent studies have shown that BRG1- and hBRM-based hSWI/SNF complexes can be found in association with enzymes involved in transcriptional regulation of genes that have been implicated in the control of cell growth and transformation (32, 33, 41, 45). Biochemical characterization of BRG1 and hBRM complexes revealed that mSin3A/HDAC2, which can deacetylate histones H3 and H4, and the type II protein arginine methyltransferase PRMT5, which can symmetrically methylate histones H3 and H4, can be found in complex with hSWI/SNF subunits (32, 33, 41). Furthermore, protein-protein interaction studies showed that BAF57 can mediate interaction with transcriptional activators such as c-MYC and estrogen receptor, as well as mSin3A/B and PRMT5 transcriptional repressors (5, 33). However, little is known about the genes regulated by BAF57.
Mutation of the hSWI/SNF subunits, BRG1, hBRM, BAF57, and BAF45, has been found to be associated with a wide variety of tumors (12, 36, 47, 50). Recent work utilizing in vivo models has suggested that an intact hSWI/SNF complex is critical for maintenance of normal tissue homeostasis and that disruption of this complex may contribute to neoplasia and transformation to a malignant phenotype (10, 16, 38, 39). While homozygous inactivation of Brg1 is lethal early in development, deletion of Brm does not appear to affect viability, suggesting that there might be some level of redundancy in the ATPase function of hSWI/SNF complexes (10, 37). However, there appears to be a strict requirement for Brg1 and BAFs including BAF45 and BAF155, whose homozygous inactivation leads to early embryonic lethality (16, 23, 38). Mice with Brg1 or Ini1 haploinsufficiency are prone to development of subcutaneous and malignant rhabdoid tumors, respectively (10, 16, 24, 38). In addition, tumors that arise in haploinsufficient Ini1 mice exhibit loss of heterozygosity and are more aggressive, with 100% of animals developing lymphomas within 11 weeks (39). Loss of heterozygosity is not observed in tumors from haploinsufficient Brg1 mice, although inactivating somatic mutation cannot be ruled out. Despite the fact that these studies implicate BRG1 and BAF45 in tumorigenesis and suggest that both SWI/SNF subunits function as tumor suppressors, it is uncertain whether other genetic alterations are involved in inducing cancer in Brg1 or Ini1 heterozygotic mice.
To fully define the role played by Brg1 and Ini1 during development and to elucidate their role in tumor induction, conditional knockout mice were developed. In the case of Brg1, tissue-specific inactivation brought about a block in T-cell development, which was marked by thymic atrophy and a reduced number of mature thymocytes (15). Conditional inactivation of Ini1 affected hematopoiesis and resulted in aggressive CD8+ T-cell lymphomas, indicating that Ini1 is essential for normal T-cell development (39). These remarkable in vivo data are further supported by findings that show loss of specific components of the SWI/SNF complex in primary human tumors. Specific inactivating mutations of INI1 have been described in aggressive rhabdoid tumors arising in pediatric patients, and BRG1/hBRM deletions within non-small-cell lung cancers have been associated with a particularly poor prognosis (36, 47). Finally, direct interaction between BRG1- and hBRM-associated factors and key regulators of the cell cycle including the retinoblastoma and BRCA1 tumor suppressor proteins lends further support to the notion that BRG1- and hBRM-based hSWI/SNF complexes affect cell growth and proliferation by interacting with and probably controlling tumor suppressor pathways (6, 8, 13, 18, 43, 46, 52).
The BT549 cell line is an invasive human ductal epithelial breast cancer line that lacks expression of BAF57, which can bind DNA via its high-mobility group-like domain and mediate interaction with transcriptional activators as well as transcriptional repressors (5, 33, 49). Recent reports describe the importance of BAF57-containing complexes in transcriptional repression of neuron and T lymphocyte-specific genes, as well as tumor suppressor genes such as BRCA1 (3, 4, 11). Therefore, restoring BAF57 activity in tumorigenic cells that lack this BRG1- and hBRM-associated factor might provide clues about its function in promoting normal cell growth. To investigate the importance of BAF57 expression as it relates to the transformed phenotype, we utilized retroviral transduction to reintroduce BAF57 into BT549 cells. We demonstrate that reexpression of BAF57 results in reversal of the transformed phenotype via cell cycle arrest and induction of apoptosis. We also show that reexpression of BAF57 induces a wide variety of genes including the proapoptotic tumor suppressor gene CYLD. These findings indicate that BAF57-containing BRG1 and hBRM chromatin remodeling complexes affect multiple activities involved in the control and maintenance of cell survival and proliferation.
MATERIALS AND METHODS
Plasmid constructions.
A retroviral vector for BAF57 expression was generated by inserting a 1.3-kbp EcoRI DNA fragment, which was PCR amplified using pBS(KS+)/BAF57 (49), into EcoRI-linearized pBABE-puromycin. The 5′ primer (5′-GGAATTCCCGAAGCCGGAGCTGCGGGCGCT-3′) was modified to include an EcoRI site (underlined), while the 3′ primer (5′-GGAATTCCGGATCCGTTATTTGTCATCGTCGTCCTTGTAGTCTTCTTTTTTCTCATCTTCTGGTATGGGATCTGTTGG-3′) included a flag-tagged sequence followed by a stop codon and an EcoRI site (underlined). Plasmids pCDNA3-neomycin/Flag-CYLD and pCDNA3-neomycin/Flag-CYLD(1-932) (residues 1 to 932 of CYLD) for expression of flag-tagged wild-type and mutant cylindromatosis were previously described (44). Vectors expressing a short hairpin RNA (shRNA) against CYLD (pSUPER/CYLD) or a mutated shRNA (pSUPER/MutCYLD) were provided by Hannah Farmer and Alan Ashworth (The Breakthrough Breast Cancer Research Center, London, United Kingdom).
Establishment of stable cell lines and transient transfection assays.
BT549 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and human insulin (0.023 U/ml). To establish BT-pBABE and BT-BAF57 cell lines, 80 to 90% confluent plates were washed with 1× phosphate-buffered saline and incubated with 5 ml of high-titer retroviral supernatant, which was harvested from Bing cells transfected with either pBABE-puromycin or pBABE-puromycin/Flag-BAF57 in the presence of 20 μg/ml prolybrene. After a 6-h incubation, each plate was supplemented with 5 ml of medium, and cells were allowed to grow for an additional 48 h before 2.5 μg/ml puromycin was added. Drug-resistant colonies were expanded in the presence of puromycin. Several clones were examined by Western blot analysis, and BT-BAF57 clones 3 and 6 were selected for further studies. To generate BT549 cell lines that express either wild-type or mutant CYLD, and BT-BAF57 cells that express CYLD small interfering RNA (siRNA) or nonspecific (NS) siRNA, approximately 1 × 106 cells were seeded in 10-cm plates and transfected for 5 to 6 h with 2 μg of either control or expression plasmids using Lipofectamine reagent (Invitrogen, Inc.). In the case of BT-BAF57 cells, transfections were carried out in the presence of 0.5 μg of pBABE-hygromycin. Two days after transfection, BT549 cells expressing wild-type or mutant CYLD were selected using 200 μg/ml neomycin, while BT-BAF57 cells expressing CYLD siRNA or NS siRNA were selected in the presence of 426 μg/ml hygromycin.
To assess the effects of BAF57 on CYLD expression by transient transfection assays, normal human mammary epithelial cells (HMEC) (Cambrex Inc.) and transformed breast epithelial cells (BT 549, HCC1937, and MCF7) were electroporated using the Amaxa Nucleofector II. HMEC were grown in mammary epithelial cell growth medium, which was supplemented with 52 μg/ml bovine pituitary extract, 5 μg/ml insulin, 10 ng/ml human recombinant epidermal growth factor, and 0.5 μg/ml hydrocortisone. HCC1937 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, while MCF7 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% FBS. Approximately 5 × 105 HMEC or 2 × 106 transformed breast epithelial cells were electroporated with 5 μg of pBABE/Fl-BAF57 plasmid using 100 μl of the appropriate Amaxa solution, and 48 h posttransfection total RNA was isolated and transcript levels were measured by reverse transcription-PCR (RT-PCR).
Proliferation, colony formation, and cell cycle assays.
Stable BAF57 cell lines or control cells were seeded at a density of 5 × 104 cells per 10-cm plate in triplicate and allowed to grow for various periods of time. Next, cells were harvested, and the number of viable cells was determined by trypan blue dye exclusion. Absolute cell numbers were determined in three separate experiments under identical culture conditions, and the results were plotted as the mean absolute number of cells recovered. To measure contact inhibition, approximately 2 × 103 cells were plated and allowed to grow for 2 weeks before they were fixed with 10% buffered formalin solution and stained with 0.1% crystal violet stain. To analyze the DNA content of each cell line, approximately 5 × 104 cells were seeded in 15-cm tissue culture plates and allowed to grow before they were fixed and stained with propidium iodide solution (20 μg/ml propidium iodide, 200 μg/ml RNase A). Cells were incubated for 30 min at room temperature in the dark before they were analyzed on a FACSCalibur flow cytometer (Becton Dickinson). CellQuest software (Becton Dickinson) was used for statistical and flow cytometric data analysis.
Immunoprecipitation and Western blot analysis.
Approximately 200 μg of nuclear extract from either BT549 or BT-BAF57 cells was precleared with 30 μl of a 50% slurry of protein A agarose beads for 2 h at 4°C. Precleared nuclear extracts were incubated with 20 μl of either preimmune or immune anti-BAF57 antibodies for an additional 2 h at 4°C before the addition of 40 μl of preblocked protein A agarose beads. Next, samples were incubated overnight at 4°C and washed four times with 1 ml of buffer A (40 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% NP-40), and bound proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and analyzed by Western blotting. To examine flag-tagged BAF57 expression, 20 μg of nuclear extract from each cell line was loaded on an 8% SDS-polyacrylamide gel, transferred onto nitrocellulose membrane, and detected by enhanced chemiluminescence reagents according to the manufacturer's recommendations (Amersham Pharmacia Biotech, Inc.). Antibodies specific for hSWI/SNF subunits have been previously described (33, 41, 42). Anti-Flag M2 (sc-807), anti-MAD1 (sc-222), anti-p16 (sc-468), anti-p21 (sc-397), anti-p27 (sc-528), anti-CDC25A (sc-7157), anti-cyclin A (sc-751), anti-cyclin D1 (sc-718), anti-cyclin E1 (sc-481), anti-caspase 3 (sc-7148), anti-BAX (sc-7480), and anti-CYLD (H-419) antibodies were purchased from Santa Cruz; anti-cyclins B and C were purchased from GeneTex; anti-HDAC2 was purchased from Zymed; and anti-BCL-2 was purchased from Pharmingen.
Reverse transcriptase PCR and microarray analysis.
Total RNA was isolated from BT549, BT-pBABE, and BT-BAF57 cell lines using Trizol reagent according to the manufacturer's instructions (Invitrogen, Inc.). Reverse transcription was performed using 10 to 20 μg of total RNA in a 20-μl reaction containing 20 pmol of specific 3′ primer, 3.5 mM MgCl2, a 1 mM concentration of each deoxynucleoside triphosphate, 1× Taq Pol buffer (Invitrogen, Inc.), 15 U of AMV-RT (Promega, Inc.), and 2.5 U of RNasin. Either 0.2 μl or 2 μl of the reverse transcriptase reaction mixture was PCR amplified using specific primers in a 50-μl reaction mixture containing 2.5 U of Taq polymerase (Invitrogen, Inc.) and 2 μCi of [α-32P]dCTP. Amplified fragments were separated from nonspecific products by electrophoresis on a 5% polyacrylamide gel and quantitated with a Molecular Dynamics PhosphorImager. Specific primer pairs were used to amplify the following genes (regions): CYLD (+19 to + 348), CDC25A (+337 to + 771), CYCLIN E1 (+262 to + 659), BAX (+39 to + 369), BCL-2 (+32 to + 365), and GAPDH (+178 to + 578).
Microarray analysis was performed using 10 μg of total RNA isolated from either BT549 or BT-BAF57 cells. Affymetrix human genome U133 (HG-U133A and HG-133B) high-density expression array chips, which include more than 39,000 human transcripts, were used to identify genes whose expression was altered upon reexpression of BAF57. The analysis was performed by the Ohio State University Comprehensive Cancer Center microarray facility (www.osuccc.osu.edu/microarrays). GENECHIP 3.3 software (Affymetrix) was utilized to normalize signals and compare the data generated from BT549 and BT-BAF57, with BT549 defining the baseline. Relative changes in expression levels, indicating the relative change in expression between BT-BAF57 and BT549 baseline targets, were used to identify genes differentially expressed. A file containing genes demonstrating at least a twofold difference was produced, and statistical analysis was performed as previously described (21). Results of microarray expression analysis were further confirmed by RT-PCR as described above.
Chromatin immunoprecipitation assay.
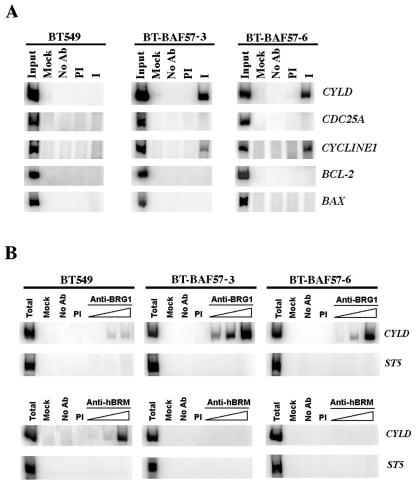
Cross-linked chromatin was prepared as described (14). Briefly, cells were treated with 1% formaldehyde and harvested in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.1], 100 mM NaCl, 5 mM EDTA, 0.5% SDS, protease inhibitors). Cells were collected by centrifugation and resuspended in 250 μl of immunoprecipitation buffer (100 mM Tris-HCl [pH 8.6], 5 mM EDTA, 0.3% SDS, 1.7% Triton X-100, protease inhibitors). Chromatin was then solubilized to a bulk size of 0.25 to 2 kbp by sonication with a microtip Branson sonifier 450 and immunoprecipitated in the presence of either anti-BAF57, anti-BRG1, or anti-hBRM antibodies and 40 μl of preblocked protein A Sepharose beads overnight at 4°C. Bound nucleoprotein complexes were washed and eluted as described previously (33). Eluted DNA was PCR amplified in the presence of 100 pmol of specific 5′ and 3′ primers in a 50-μl reaction mixture containing 2 μCi of [α-32P]dCTP for 35 cycles. Primer pairs used to amplify genomic sequences spanned promoter regions of CYLD (−190 to + 65), CDC25A (−185 to + 43), CYCLIN E1 (−246 to + 38), BCL-2 (−250 to + 88), BAX (−211 to + 136), and ST5 (−252 to + 34).
RESULTS
Establishment of a stable BT549 cell line that expresses BAF57.
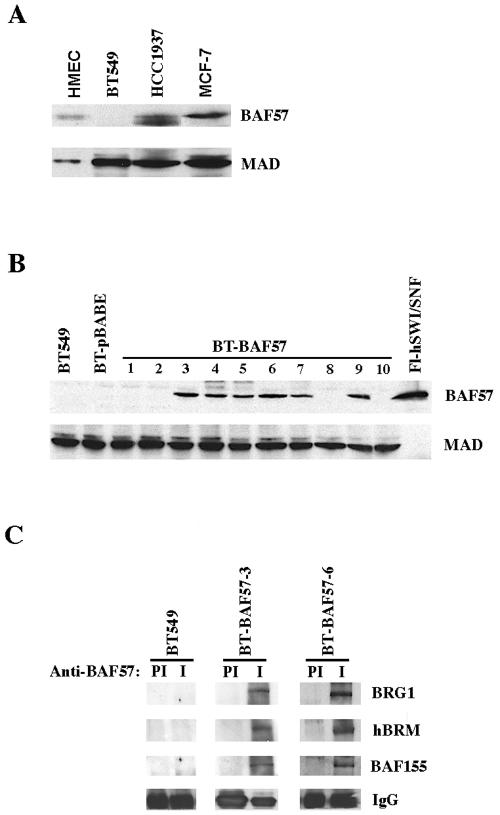
Several studies have shown that reintroduction of BRG1 and BAF45 in cells that lack these hSWI/SNF subunits leads to cell cycle arrest (2, 6, 13, 18, 46, 52). Inhibition of cell growth results from either up-regulation of cyclin-dependent kinase inhibitors and or repression of cyclins. Since BAF57 has been shown to be missing in the highly invasive BT549 breast cancer cell line (12), we reexamined its expression in normal as well as transformed mammary epithelial cells (Fig. 1A). Western blot analysis using whole cell extracts from normal HMEC and the transformed breast cancer cell lines BT549, HCC1937, and MCF7 confirmed the low levels of BAF57 in HCC1937 and the lack of BAF57 expression in BT549 cells. To determine whether reexpression of BAF57 could trigger similar changes as those caused by BRG1 and INI1, we used retroviral transduction to establish BT549 stable cell lines that express BAF57. Following infection of BT549 cells with retroviral particles, which were packaged using either empty vector or vector containing BAF57 cDNA, puromycin-resistant clones were selected and BAF57 expression was examined by Western blot analysis using anti-BAF57 antibodies. BAF57 was detected in several BT549 clones but not in BT549 or control BT549 cells transduced with empty vector (Fig. 1B). As a control, the same Western blot was probed with anti-MAD antibodies, and equal amounts of protein were found within each sample. We selected BT-BAF57 clones 3 and 6 for all subsequent experiments because BAF57 expression was less toxic, and cells were able to proliferate despite the delay in their doubling time (Fig. 2).
FIG. 1.
Retrovirus-mediated expression of BAF57 in BT549 cells. (A) Whole-cell extracts (20 μg) from normal HMEC or transformed BT549, HCC1937, and MCF7 breast cancer cell lines were analyzed by Western blotting using anti-BAF57 antibody. As a control, MAD levels were also measured by reprobing the same blot with anti-MAD antibody. (B) Analysis of ectopically expressed BAF57 in stable BT-BAF57 cell lines. Approximately 20 μg of nuclear extract isolated from either BT549, BT549/pBABE (BT-pBABE), or BT549/BAF57 (BT-BAF57) cells and 100 ng of immunopurified flag-tagged human SWI/SNF (Fl-hSWI/SNF) were analyzed by Western blotting using anti-BAF57 antibody. Anti-MAD was used as a control to show equal loading. (C) Ectopically expressed BAF57 is incorporated into BRG1- and hBRM-based hSWI/SNF complexes. Nuclear extracts from BT549 and BT-BAF57 clones 3 and 6 were immunoprecipitated with either preimmune (PI) or immune (I) anti-BAF57 antibodies, and after extensive washing the retained proteins were analyzed by Western blotting using anti-BRG1, anti-hBRM, and anti-BAF155 antibodies.
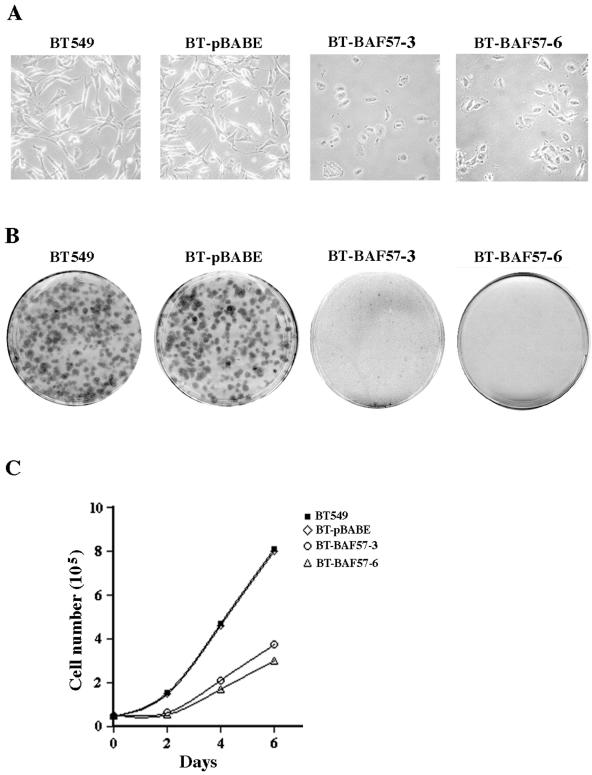
FIG. 2.
Reexpression of BAF57 induces flat-cell morphology and inhibits transformation and cell proliferation. (A) An equal number (5 × 104) of BT549, BT-pBABE, BT-BAF57 clone 3, and BT-BAF57 clone 6 cells was seeded into 10-cm plates, and after 3 days pictures were taken at ×40 magnification. (B) Approximately 2 × 103 BT549, BT-pBABE, and BT-BAF57 clones 3 and 6 cells were grown for 14 days before anchorage-dependent colonies were visualized by crystal violet staining. This experiment was performed three times, and representative plates are shown. (C) Proliferation was measured by plating an equal number (5 × 104) of cells from the indicated cell lines and performing cell counts every 2 days for 6 days. The number of viable cells was determined by trypan blue exclusion. This experiment was repeated three times, and the data points represent the average counts from nine plates.
Next, we tested whether ectopically expressed BAF57 was incorporated into endogenous BRG1- and hBRM-based hSWI/SNF complexes by performing immunoprecipitation experiments (Fig. 1C). Following immunoprecipitation with either preimmune or immune anti-BAF57 antibodies, the retained proteins were analyzed by Western blotting. As expected, we were unable to detect hSWI/SNF subunits in immunoprecipitates from the BAF57-deficient cell line BT549. In contrast, Western blot analysis of immune complexes from BT-BAF57 cells revealed that the hSWI/SNF subunits BRG1, hBRM, and BAF 155 coimmunoprecipitate with retrovirally expressed BAF57. Preimmune immunoprecipitates were devoid of any hSWI/SNF subunit. Collectively, these results demonstrate that ectopically expressed BAF57 is capable of associating with endogenous BRG1 and hBRM complexes.
Reexpression of BAF57 results in altered cell morphology and reduced proliferation.
We noted a marked difference in cellular morphology between the parental BT549 cell line and the retrovirally transduced BT549 clones expressing BAF57 including BT-BAF57 clones 3 and 6 (Fig. 2A). Unlike other BAF57-expressing clones, which could not be expanded due to their low viability, cells of BT-BAF57 clones 3 and 6 were able to proliferate despite their altered size and morphology. BT-BAF57 cells appeared larger and less extended and showed fewer dendritic projections (Fig. 2A). Wild-type and control BT549 cells transduced with empty vector (BT-pBABE) grown under the same conditions did not exhibit any change in their morphology, suggesting that BAF57 reexpression induces these phenotypic changes.
Because BT-BAF57 cells displayed a flat phenotype, we compared their growth characteristics to BT549 and BT-pBABE cell lines (Fig. 2B). BAF57-expressing cells were unable to form colonies, as determined by crystal violet staining. However, BT549 and BT-pBABE cells formed visible colonies as early as 6 days after the tissue culture plates were seeded, suggesting that reexpression of BAF57 inhibits BT549 cell growth and proliferation. To determine the cause of this apparent loss of transformation, we compared the proliferation rate of BT-BAF57 cells to that of BT549 and control BT-pBABE cell lines by performing repeated absolute counts of viable cells placed in culture (Fig. 2C). We found that BT549 and BT-pBABE cells grew two- to threefold faster than BT-BAF57 cells. Thus, it appears that BAF57 expression triggered a significant decrease in BT549 proliferation. Taken together, these experiments indicate that reexpression of BAF57 can alter growth of the highly invasive and transformed breast cancer BT549 cell line.
Restoration of BAF57 expression is associated with cell cycle arrest and induction of apoptosis.
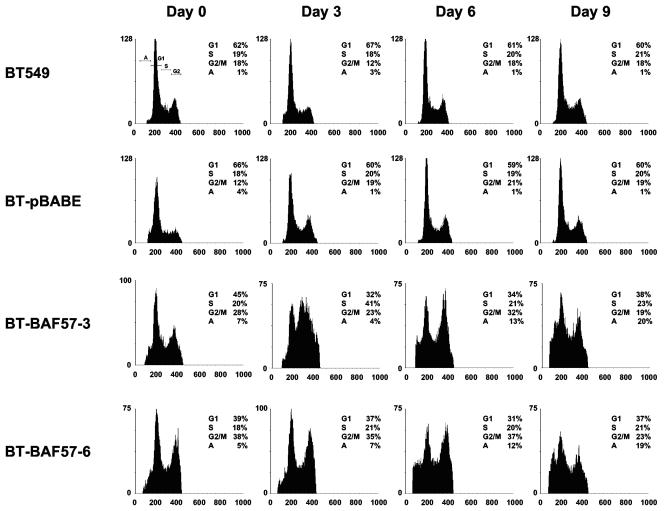
Reduced cell growth and proliferation might be related to cell cycle arrest and induction of programmed cell death. To determine whether BT-BAF57 cells were undergoing cell cycle block and/or apoptosis, we performed cell cycle analysis by propidium iodide staining and flow cytometric analysis (Fig. 3). Both parental BT549 and control BT-pBABE cell lines showed similar cell cycle profiles at day 0, whereas reexpression of BAF57 in BT549 resulted in a higher percentage of cells accumulating in phases G2 to M of the cell cycle. When cells were allowed to grow further, we noticed that there was an increase in the proportion of BT-BAF57 clone 3 cells in S phase, while BT-BAF57 clone 6 cells did not exhibit any change in their cell cycle profiles. We also remarked that by day 6 there was a greater percentage of BT-BAF57 cells undergoing apoptosis (12 to 13%). This trend continued to day 9, when we observed a further increase in the subdiploid fraction in BT-BAF57 cells but not in the BT549 and BT-pBABE cell lines. These results show that reexpression of BAF57 brings about cell cycle changes that are consistent with the growth characteristics of BT-BAF57 cells.
FIG. 3.
FACS analysis of wild-type and BAF57-expressing BT549 cell lines. An equal number (5 × 104) of BT549, BT-pBABE, BT-BAF57 clone 3, or BT-BAF57 clone 6 cells was seeded into 15-cm plates and allowed to grow for the indicated times. Next, cells were fixed, stained with propidium iodide, and analyzed on a FACSCalibur flow cytometer. The cell cycle profile of each cell line was determined at least three times, and representative histograms are shown.
BAF57-induced cell cycle arrest and death involve down-regulation of gene products critical for cell cycle progression and inhibition of apoptosis.
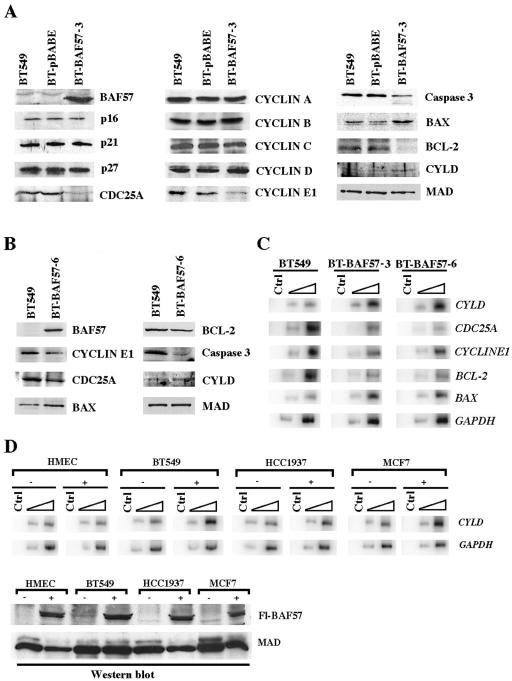
Cell cycle analysis showed evidence of BT-BAF57 cells accumulating in phases G2 to M and G1 to S between days 3 and 6, suggesting that cell cycle arrest was an important mechanism by which BAF57 inhibits BT549 cell growth and proliferation. Previous work has shown that reintroduction of BRG1 or INI1 in deficient cell lines causes cell cycle arrest that is marked by transcriptional repression of members of the cyclin family, including cyclins A and D, as well as induced expression of cyclin-dependent kinase inhibitors such as p15, p16, and p21 (6, 18, 46, 52). We therefore performed a series of Western blot analyses to examine the level of cyclins and cyclin-dependent kinase inhibitors in BT549, BT-pBABE, and BT-BAF57 cell lines at day 6 (Fig. 4A). These experiments showed that BAF57 expression had no effect on p16, p21, and p27. Similarly, there was no change in the level of members of the cyclin family except for cyclin E1, which showed a 2- to 2.5-fold decrease at the protein level and 1.8- to 2-fold decrease at the mRNA level (Fig. 4A and B).
FIG. 4.
Reexpression of BAF57 affects genes involved in cell cycle regulation and cell death. (A) Whole-cell extracts (20 μg) from BT549, BT-pBABE, and BT-BAF57 clone 3 cells were analyzed by Western blotting using antibodies specific for the indicated proteins. As a control, BAF57 levels were measured in BT549, BT-pBABE, and BT-BAF57 cell lines. (B) Genes affected by BAF57 reexpression were analyzed in BT-BAF57 clone 6 as described in panel A. (C) Total RNA from BT549 and BT-BAF57 clones 3 and 6 was used in RT-PCRs as described in Material and Methods. Different amounts of each RT reaction (0.2 and 2 μl) were PCR amplified in the presence of [α-32P]dCTP and gene-specific primers. Control (Ctrl) represents PCRs lacking 5′ primer. (D) Total RNA from untransfected (−) or transiently transfected (+) HMEC or BT549, HCC1937, and MCF7 cells was used to measure the levels of CYLD mRNA as described in panel C. Western blot analysis was performed to show that BAF57 was expressed at similar levels in the transiently transfected cells.
The fact that there was no change in the level of cyclins A and B suggested that there might be other mechanisms by which G2 to M arrest is induced. To identify target genes regulated by BAF57, we compared global gene expression in BT549 and BT-BAF57 cells by microarray analysis. We found that among the inhibited genes, CDC25A was down-regulated twofold (see Table S2 in the supplemental material). This result was also confirmed by RT-PCR analysis, which showed that CDC25A was reduced 3- to 3.6-fold (Fig. 4C). Unlike cyclin E1, which through its association with cyclin dependent kinase 2 (Cdk2) can regulate G1 to S phase transition, CDC25A phosphatase has recently been shown to be involved in regulating events both at the G1 to S and G2 to M phases of the cell cycle (27). CDC25A can activate cyclin E-Cdk2 prior to DNA replication and is involved in activating cyclin B-Cdk1, which is known to be essential for the transition from G2 to M (30, 31). Furthermore, CDC25A appears to be rate limiting in cells progressing through mitosis, because the addition of glutathione transferase-CDC25A to depleted cell extracts can restore cyclin B-Cdk1 activation (27). More importantly, siRNA interference experiments have shown that reducing CDC25A levels not only inhibits the transition from G1 to S but also delays progression from G2 to M (27). Therefore, we analyzed the levels of CDC25A in BT549, BT-pBABE, and BT-BAF57 by Western blotting (Fig. 4A and B). We found that CDC25A levels were reduced 2.2- to 3-fold in BT-BAF57 cells. These results suggest that BAF57 is involved in regulating cyclin E1 and CDC25A protein levels.
A large proportion of BT-BAF57 cells undergoes apoptosis as measured by fluorescence-activated cell sorter (FACS) analysis. To determine whether there are changes in the levels of proteins involved in inducing apoptosis, we analyzed the expression of major components of the apoptotic pathway at day 6 (Fig. 4A and B). Western blot analysis showed that the levels of the antiapoptotic protein BCL-2 were reduced 1.9- to 3.4-fold in BT-BAF57 cells, whereas the levels of the proapoptotic BAX protein were increased twofold in comparison to BT549 and BT-pBABE cells. When we measured the levels of caspase 3, we found that its levels were also reduced twofold. These results are in complete agreement with the FACS analysis, which shows an increase in the number of subdiploid cells by day 6. Since BCL-2 and BAX act upstream of the executioner caspase 3, we examined whether BAF57 could affect their expression at the transcriptional level. While BCL-2 mRNA was down-regulated threefold, BAX levels were induced 3- to 3.5-fold (Fig. 4C). These findings suggest that reexpression of BAF57 induces cell death by controlling expression of key apoptotic factors.
BAF57 affects transcriptional expression of the tumor suppressor CYLD.
Having found that BAF57 inhibits growth of BT549 cells by altering the level of proteins involved in the control of cell cycle progression and death, we sought to examine the effect of BAF57 on global gene expression. Total RNA from BT549 and BT-BAF57 cells was used to identify target genes whose expression is affected in the presence of BAF57 (see Tables S1 to S4 in the supplemental material). We found that 410 genes were up-regulated while 469 genes were down-regulated, suggesting that BAF57 reexpression can activate as well as repress transcription of target genes. Among the genes affected in BT-BAF57 cells, both cyclin E1 and CDC25A were down-regulated twofold. These results are consistent with the protein expression data, which show that BT-BAF57 cells have lower levels of cyclin E1 and CDC25A.
Because BT-BAF57 cells show increased apoptosis in comparison to control BT549 and BT-pBABE cells, we checked the list of differentially expressed genes for the presence of proapoptotic genes. We found that the familial cylindromatosis (CYLD) gene was up-regulated twofold in cells that express BAF57 (see Table S1 in the supplemental material). Recent work has shown that CYLD is a tumor suppressor gene involved in regulating apoptosis, and mutation of this locus gives rise to cylindromas, which develop mainly in hairy areas of the body including the scalp (7). Furthermore, functional studies have shown that the tumor suppressing properties of CYLD correlate with its ability to deubiquitinate tumor necrosis factor receptor-associated factor 2 (TRAF2), TRAF6, and NEMO, which transduce antiapoptotic signals through the NF-κB pathway (9, 25, 44). Therefore, we wanted to confirm the microarray results and examine the CYLD mRNA levels by RT-PCR analysis. As expected, CYLD mRNA levels were induced 4- to 4.5-fold in BT-BAF57 clones 3 and 6 (Fig. 4C), while CYLD protein levels were increased 2- to 2.5-fold (Fig. 4A and B). These results suggest that BAF57 affects BT549 cell growth by altering the expression of genes involved in regulating apoptosis.
To exclude the possibility that the observed increase in CYLD expression is due to clonal selection of BT-BAF57 cells, we transiently transfected normal and transformed epithelial cells with pBABE-BAF57 and analyzed the CYLD transcript levels (Fig. 4D). When total RNA from either HMEC or BT549, HCC1937, or MCF7 cells was used to measure CYLD mRNA by RT-PCR, we found that its levels were induced 1.9- to 2.8-fold in transformed human breast cancer cell lines. CYLD levels were unchanged in normal human mammary epithelial cells. To ensure that the changes in CYLD mRNA were not caused by fluctuations in the total amount of RNA, we measured the levels of GAPDH and determined that its levels were comparable in all samples tested (Fig. 4D). We also measured the levels of the flag-tagged BAF57 protein and found that it is expressed at similar levels in transiently transfected cells (Fig. 4D). Taken together, these findings indicate that reexpression of BAF57 induces CYLD expression in transformed breast cancer cell lines but not in normal mammary epithelial cells.
To determine whether BAF57 is directly involved in transcriptional regulation of target genes identified by Western blotting and microarray analysis, we analyzed recruitment of BAF57 to their promoter region (Fig. 5A). We found that while BAF57 is recruited to the promoter region of CYLD and cyclin E1, there was a lack of recruitment of BAF57 to CDC25A, BCL-2, and BAX, suggesting that BAF57 can alter gene expression by direct and indirect mechanisms. Since BAF57 can be found in association with BRG1- and hBRM-based hSWI/SNF complexes and since we have found that reintroduction of BAF57 in BT549 cells induces CYLD expression, we sought to examine recruitment of BRG1 and hBRM ATPases to the CYLD promoter. When cross-linked chromatin from BT549 and BT-BAF57 cells was immunoprecipitated with either anti-BRG1 or anti-hBRM antibodies, we were able to detect a four- to fivefold enrichment in BRG1 recruitment to the CYLD promoter in BT-BAF57 cells (Fig. 5B). In stark contrast, hBRM was detected at the CYLD promoter in BT549 but not in BT-BAF57 clones 3 and 6. These results suggest that BRG1- and hBRM-based hSWI/SNF complexes are differentially recruited to the CYLD promoter.
FIG. 5.
BAF57 is associated with the CYLD and CYCLIN E promoters. (A) Cross-linked chromatin from BT549, BT-BAF57 clone 3, and BT-BAF57 clone 6 cells was immunoprecipitated with either preimmune serum (PI) or anti-BAF57 antibodies (I), and the eluted genomic DNA was PCR amplified using promoter-specific primers for the indicated genes. Input represents 1/750 of total chromatin used in each reaction. (B) BRG1 and hBRM are differentially recruited to the CYLD promoter. Chromatin immunoprecipitation was conducted as described in panel A using either anti-BRG1 or anti-hBRM antibodies, and the eluted DNA (1, 2, or 5 μl) was PCR amplified using CYLD or suppressor of tumorigenicity 5 (ST5) promoter-specific primers. Mock, reaction without chromatin; No Ab, reaction without antibody.
Wild-type but not mutant CYLD associated with familial cylindromas can induce apoptosis in BT549 cells.
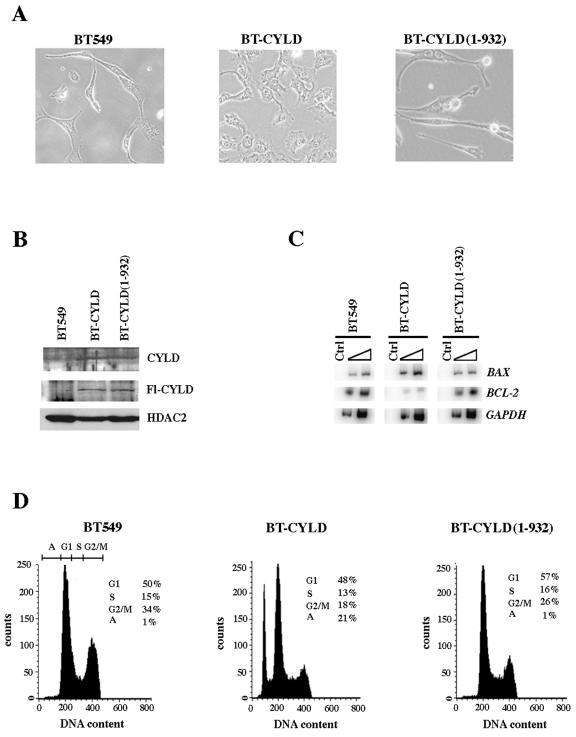
Recent studies have shown that loss or reduced expression of CYLD is antiapoptotic and that this effect is mediated through stimulation of the NF-κB pathway (9). To assess the contribution of CYLD to apoptosis, we transfected BT549 cells with either wild-type flag-tagged CYLD (Fl-CYLD) or C-terminally truncated flag-tagged CYLD [CYLD(1-932)] and examined the morphology and DNA content in drug-resistant polyclonal cell lines (Fig. 6). Expression of wild-type Fl-CYLD induced flat cell formation in BT-CYLD cells, whereas mutant Fl-CYLD-expressing BT549 cells [BT-CYLD(1-932)] did not exhibit any significant change in their morphology (Fig. 6A). To ensure that the observed changes were due to increased expression of wild-type Fl-CYLD, we analyzed whole cell extracts by Western blotting using anti-flag antibodies (Fig. 6B). Both flag-tagged wild-type and mutant CYLD proteins were expressed at similar levels. In contrast, BT549 cells transfected with vector alone showed no expression. We also measured the levels of endogenous CYLD and found that its levels were very low in BT549, while in transfected BT549 cells the CYLD levels were increased twofold. When we examined the DNA content of an equal number of BT549, BT-CYLD, or BT-CYLD(1-932) cells, we found that 21% of cells underwent apoptosis when wild-type Fl-CYLD was expressed (Fig. 6D). Neither BT549 nor BT-CYLD(1-932) showed any increase in the number of cells undergoing cell death.
FIG. 6.
CYLD induces apoptosis in BT549 cells. (A) An equal number (5 × 104) of cells from the BT-549, BT-CYLD, and BT-CYLD(1-932) cell lines was seeded into 10-cm plates, and pictures were taken at ×40 magnification. (B) Western blot analysis was conducted using 20 μg of whole-cell extract from the indicated stable cell lines. Both wild-type and mutant CYLD were detected using either anti-flag or anti-CYLD antibody. To show equal loading, the same blot was stripped and reprobed with anti-HDAC2 antibody. (C) Total RNA from BT549, BT-CYLD(1-932), and BT-CYLD was PCR-amplified using either BAX, BCL-2, or GAPDH gene-specific primers as described in the legend of Fig. 4C. (D) Approximately 5 × 104 cells from the BT549, BT-CYLD or BT-CYLD(1-932) cell lines were seeded into 10-cm plates and allowed to grow until plates became 80 to 90% confluent. Cells were then analyzed by FACS analysis. Cell cycle distribution of each cell line was measured at least three times, and representative histograms are shown.
Analysis of BAX and BCL-2 transcript levels in BT-BAF57 cells showed that although both genes are not direct targets of BAF57 (Fig. 5A), their levels fluctuate in a manner consistent with cells undergoing apoptosis (Fig. 4A to C). To verify whether expression of wild-type CYLD could induce similar changes in BAX and BCL-2 mRNA levels, we analyzed total RNA by RT-PCR (Fig. 6C). While BAX levels were induced two- to threefold, BCL-2 was inhibited four to fivefold in BT-CYLD, suggesting that expression of wild-type Fl-CYLD can influence both genes at the transcriptional level. Collectively, these results suggest that BAF57-induced apoptosis is mediated by the familial cylindromatosis tumor suppressor gene CYLD.
Suppression of CYLD inhibits BAF57-induced apoptosis.
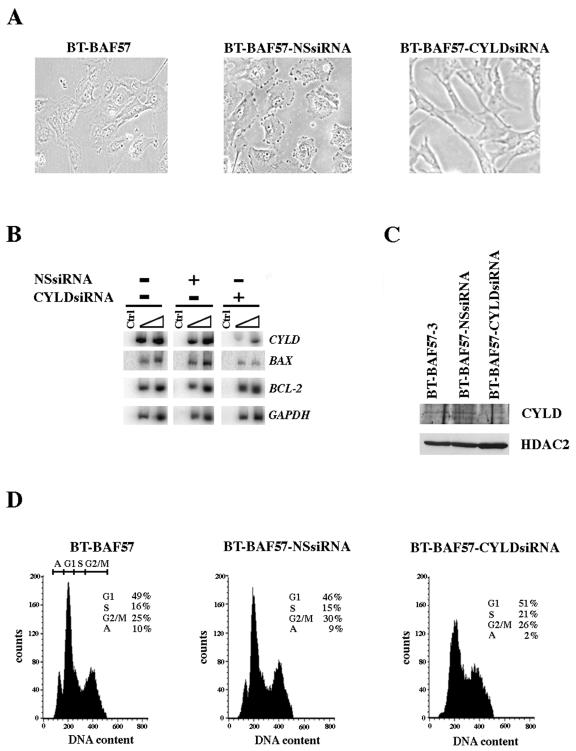
To unequivocally demonstrate that BAF57 induces apoptosis by up-regulating CYLD expression, we utilized RNA interference to knock down CYLD expression in BT-BAF57 cells (Fig. 7). BT-BAF57 cells were transfected with either vector alone, a construct designed to express human CYLD-specific siRNA, or an unrelated NS siRNA. After drug selection, we analyzed the morphology, CYLD mRNA and protein levels, and the DNA content of each cell line. Both control cell lines, BT-BAF57 and BT-BAF57-NSsiRNA, exhibited flat phenotype morphology; however, in BT-BAF57-CYLD siRNA cells where expression of CYLD was knocked down by more than 70% at the mRNA level and two- to threefold at the protein level (Fig. 7B and C), there was a complete loss of flat cell formation, and most cells looked elongated like the parental BT-549 cell line. More importantly, when we examined the cell cycle profile of all three cell lines by FACS analysis, we found that knocking down CYLD expression not only restores cell morphology but also reduces the number of apoptotic cells (Fig. 7C). This was supported by the RT-PCR analysis, which showed that BAX mRNA levels were reduced by 2- to 3.7-fold, while BCL-2 levels were unaffected (Fig. 7B). Taken together, these results indicate that BAF57 induces apoptosis by increasing the levels of CYLD and that reduced expression of CYLD is antiapoptotic.
FIG. 7.
Knock-down of CYLD by siRNA restores BT549 cell morphology and inhibits apoptosis. (A) An equal number (5 × 104) of BT-BAF57, BT-BAF57-NSsiRNA, and BT-BAF57-CYLDsiRNA cells was seeded into 10-cm plates, and pictures were taken at ×40 magnification. (B) To measure the mRNA levels of CYLD, BAX, and BCL-2, RT-PCR was conducted using total RNA from the indicated stable cell lines. As a control GAPDH levels were also analyzed. (C) To measure endogenous CYLD protein levels, Western blot analysis was conducted using whole-cell extracts from the indicated stable cell lines. As a control HDAC2 levels were also analyzed. (D) An equal number (5 × 104) of BT-BAF57, BT-BAF57-NSsiRNA, and BT-BAF57-CYLDsiRNA cells was seeded into 10-cm plates and analyzed by FACS analysis as described in the legend of Fig. 6D.
DISCUSSION
Here we demonstrate that restoration of BAF57 expression in the invasive breast carcinoma cell line BT549 results in flat cell morphology, cell cycle arrest, and induction of apoptosis. We show that these changes are accompanied by changes in expression of genes involved in the control of cell growth and survival. Retroviral-mediated expression of BAF57 resulted in stable, constitutive levels of BAF57 protein that associated with BRG1- and hBRM-based hSWI/SNF chromatin remodeling complexes. Given the role of BAF57 in mediating interaction with corepressor complexes, our observation demonstrating transcriptional repression of cyclin E and CDC25A, both of which are involved in controlling events at the G1/S and or G2/M, emphasizes the importance of BRG1- and hBRM-based hSWI/SNF complexes in maintaining cell growth regulation and homeostasis. Moreover, the involvement of BAF57 in transcriptional activation of the proapoptotic gene CYLD further extends the role played by hSWI/SNF chromatin remodeling complexes to pathways that are critical for normal cell growth and proliferation. Our findings are consistent with other work demonstrating that intact hSWI/SNF complexes are critical to cell growth and regulation and that deletion or inactivation of hSWI/SNF components results in loss of growth control and cellular transformation (10, 16, 36, 38, 47, 50).
Previous work has shown that cyclin E and CDC25A are critical for cells to progress through the cell cycle. Cyclin E is important for G1/S transition, while CDC25A appears to be required during G1/S as well as G2/M phases of the cell cycle. We have found that reexpression of BAF57 reduces the levels of cyclin E and CDC25A by inhibiting their transcription. In the case of CDC25A, it is clear that BAF57 exerts its effects in an indirect manner because chromatin immunoprecipitation assays showed that BAF57 is not recruited to the CDC25A promoter region. However, in the case of cyclin E, BAF57 appears to be directly involved in its regulation. We have shown that reexpression of BAF57 not only induces cell cycle arrest but also promotes cell death. Analysis of the microarray data revealed that the familial cylindromatosis tumor suppressor gene, CYLD, is induced in cells that express BAF57, a result that was also confirmed by RT-PCR analysis. Furthermore, chromatin immunoprecipitation assays demonstrated that BAF57 is directly involved in CYLD transcriptional induction. We have examined BRG1 and hBRM recruitment to the CYLD promoter region and found that both ATPases are differentially recruited in BT549 and BT-BAF57 cells (Fig. 5B). We have recently identified an association between MeCP2 and hBRM and shown that the MeCP2/hBRM complex is involved in transcriptional silencing of the fragile X mental retardation (FMR1) gene (17). Therefore, it is possible that hBRM might be involved in transcriptional silencing of CYLD, while BRG1 recruitment is associated with CYLD transcriptional activation. It is clear that more studies are required to elucidate the transcriptional mechanisms involved in regulating CYLD gene expression, and studies are under way to assess the ordered recruitment of chromatin remodeling complexes and epigenetic regulators to the CYLD promoter.
Further characterization demonstrated that transfected wild-type CYLD induces apoptosis in BT549 cells, whereas a mutant form of CYLD, which lacks the C-terminal sequences important for the deubiquitinating activity, was unable to bring about cell death. These results confirm the involvement of CYLD in BAF57-mediated apoptosis but do not rule out the involvement of other proapoptotic proteins. In order to verify the functional significance of CYLD in BAF57-mediated apoptosis, we used RNA interference to knock down CYLD expression in BT-BAF57 cells. CYLD-specific siRNA inhibited apoptosis in BT-BAF57 cells, whereas expression of an NS siRNA had no effect on CYLD expression and apoptosis. These results reinforce the notion that CYLD is the primary target through which BAF57 exerts its proapoptotic effects.
It is well established that overexpression of BCL-2 protects against apoptosis, while its down-regulation enhances cell death and increases the efficacy of chemotherapy (1, 26, 28). Similarly, BAX expression has been shown to be increased in cells undergoing apoptosis (20, 51). We have shown that BCL-2 and BAX are inversely affected by BAF57 expression. BCL-2 mRNA levels were reduced, while BAX levels were increased in BT-BAF57 cells, suggesting that BAF57-induced cell death might be initiated by altering the BCL-2 and BAX transcript levels. To verify whether BAF57 is directly involved in transcriptional regulation of BCL-2 and BAX, we conducted chromatin immunoprecipitation experiments, which revealed that BAF57 is not recruited to the promoter region of these cell death regulators. We have shown that wild-type CYLD induces apoptosis in BT549, in a manner similar to BAF57, by affecting BCL-2 and BAX transcript levels. These findings suggest that BCL-2 and BAX are downstream targets of BAF57 and CYLD and that, despite the fact that BAF57 is not directly recruited to the BCL-2 and BAX promoters, it is able to control their levels by increasing expression of CYLD, which can in turn trigger apoptosis.
Our results showing that reexpression of BAF57 can inhibit cellular transformation by inducing cell cycle arrest and apoptosis are consistent with previous work, which has shown that reintroduction of BRG1 or hBRM into SW13 cells can induce cell cycle arrest and flat cell morphology (13, 43). In this study, BRG1 and hBRM were shown to cooperate with the retinoblastoma family of tumor suppressor proteins to mediate the induction of flat-cell morphology. Recent studies have also shown that reexpression of BAF45 in malignant rhabdoid tumor cell lines causes growth suppression by inhibiting expression of cyclin A and p16INK4A (35). We have examined the level of both proteins, and we have found that there was no fluctuation in their levels when BAF57 was reexpressed in the invasive breast cancer cell line BT549. Therefore, it appears that there are multiple pathways affected by hSWI/SNF subunits.
BAF57 is a hSWI/SNF subunit that has been shown to mediate interactions with activator proteins such as c-MYC, as well as corepressor proteins including mSin3A/HDAC and PRMT5 (32, 33). Since BAF57 is directly involved in CYLD activation, future work will be focused on understanding the nature and composition of the chromatin remodeling complexes recruited at this promoter and on defining the histone modifications associated with its regulation. These studies will help elucidate the mechanism by which CYLD is regulated and will provide information that can be used to modulate its expression in cancer cells.
Supplementary Material
Acknowledgments
We thank Hannah Farmer and Alan Ashworth for providing the shRNA expression constructs and S. Ackerman for critical reading of the manuscript.
This work was supported by the Sidney Kimmel Scholar Award SKF-03-022, National Cancer Institute grant K01 CA89854, and American Cancer Society grant RSG-0418201-GMC to S.S. and by an International Scholarship from the Howard Hughes Medical Institute and a Leukemia and Lymphoma Society of America Scholar Award to G.M.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abou El Hassan, M. A., D. C. Mastenbroek, W. R. Gerritsen, G. Giaccone, and F. A. Kruyt. 2004. Overexpression of Bcl2 abrogates chemo- and radiotherapy-induced sensitisation of NCI-H460 non-small-cell lung cancer cells to adenovirus-mediated expression of full-length TRAIL. Br. J. Cancer 91:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ae, K., N. Kobayashi, R. Sakuma, T. Ogata, H. Kuroda, N. Kawaguchi, K. Shinomiya, and Y. Kitamura. 2002. Chromatin remodeling factor encoded by ini1 induces G1 arrest and apoptosis in ini1-deficient cells. Oncogene 21:3112-3120. [DOI] [PubMed] [Google Scholar]
- 3.Baker, K. M., G. Wei, A. E. Schaffner, and M. C. Ostrowski. 2003. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876-17884. [DOI] [PubMed] [Google Scholar]
- 4.Battaglioli, E., M. E. Andres, D. W. Rose, J. G. Chenoweth, M. G. Rosenfeld, M. E. Anderson, and G. Mandel. 2002. REST repression of neuronal genes requires components of the hSWI/SNF complex. J. Biol. Chem. 277:41038-41045. [DOI] [PubMed] [Google Scholar]
- 5.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betz, B. L., M. W. Strobeck, D. N. Reisman, E. S. Knudsen, and B. E. Weissman. 2002. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 21:5193-5203. [DOI] [PubMed] [Google Scholar]
- 7.Bignell, G. R., W. Warren, S. Seal, M. Takahashi, E. Rapley, R. Barfoot, H. Green, C. Brown, P. J. Biggs, S. R. Lakhani, C. Jones, J. Hansen, E. Blair, B. Hofmann, R. Siebert, G. Turner, D. G. Evans, C. Schrander-Stumpel, F. A. Beemer, A. van Den Ouweland, D. Halley, B. Delpech, M. G. Cleveland, I. Leigh, J. Leisti, and S. Rasmussen. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160-165. [DOI] [PubMed] [Google Scholar]
- 8.Bochar, D. A., L. Wang, H. Beniya, A. Kinev, Y. Xue, W. S. Lane, W. Wang, F. Kashanchi, and R. Shiekhattar. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102:257-265. [DOI] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., S. M. Nijman, A. M. Dirac, and R. Bernards. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424:797-801. [DOI] [PubMed] [Google Scholar]
- 10.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 11.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195-199. [DOI] [PubMed] [Google Scholar]
- 12.DeCristofaro, M. F., B. L. Betz, C. J. Rorie, D. N. Reisman, W. Wang, and B. E. Weissman. 2001. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell. Physiol. 186:136-145. [DOI] [PubMed] [Google Scholar]
- 13.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 14.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebuhr, T. C., G. I. Kovalev, S. Bultman, V. Godfrey, L. Su, and T. Magnuson. 2003. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 198:1937-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidi, C. J., A. T. Sands, B. P. Zambrowicz, T. K. Turner, D. A. Demers, W. Webster, T. W. Smith, A. N. Imbalzano, and S. N. Jones. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harikrishnan, K. N., M. Z. Chow, E. K. Baker, S. Pal, S. Bassal, D. Brasacchio, L. Wang, J. M. Craig, P. L. Jones, S. Sif, and A. El-Osta. 2005. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37:254-264. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks, K. B., F. Shanahan, and E. Lees. 2004. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell. Biol. 24:362-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao, P. W., C. J. Fryer, K. W. Trotter, W. Wang, and T. K. Archer. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23:6210-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, X. L., T. Olsson, I. M. Johansson, T. Brannstrom, and P. Wester. 2004. Dynamic changes of the anti- and pro-apoptotic proteins Bcl-w, Bcl-2, and Bax with Smac/Diablo mitochondrial release after photothrombotic ring stroke in rats. Eur. J. Neurosci. 20:1177-1188. [DOI] [PubMed] [Google Scholar]
- 21.Huang, Y., M. Prasad, W. J. Lemon, H. Hampel, F. A. Wright, K. Kornacker, V. LiVolsi, W. Frankel, R. T. Kloos, C. Eng, N. S. Pellegata, and A. de la Chapelle. 2001. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc. Natl. Acad. Sci. USA 98:15044-15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, T., M. Yamauchi, M. Nishina, N. Yamamichi, T. Mizutani, M. Ui, M. Murakami, and H. Iba. 2001. Identification of SWI/SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 276:2852-2857. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. K., S. O. Huh, H. Choi, K. S. Lee, D. Shin, C. Lee, J. S. Nam, H. Kim, H. Chung, H. W. Lee, S. D. Park, and R. H. Seong. 2001. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21:7787-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovalenko, A., C. Chable-Bessia, G. Cantarella, A. Israel, D. Wallach, and G. Courtois. 2003. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424:801-805. [DOI] [PubMed] [Google Scholar]
- 26.Letai, A., M. D. Sorcinelli, C. Beard, and S. J. Korsmeyer. 2004. Antiapoptotic BCL-2 is required for maintenance of a model leukemia. Cancer Cell 6:241-249. [DOI] [PubMed] [Google Scholar]
- 27.Mailand, N., A. V. Podtelejnikov, A. Groth, M. Mann, J. Bartek, and J. Lukas. 2002. Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 21:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahta, R., and F. J. Esteva. 2003. Bcl-2 antisense oligonucleotides: a potential novel strategy for the treatment of breast cancer. Semin. Oncol. 5:143-149. [DOI] [PubMed] [Google Scholar]
- 29.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 30.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21-32. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, I., and I. Hoffmann. 2000. Cell cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 4:107-114. [DOI] [PubMed] [Google Scholar]
- 32.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24:9630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 35.Reincke, B. S., G. B. Rosson, B. W. Oswald, and C. F. Wright. 2003. INI1 expression induces cell cycle arrest and markers of senescence in malignant rhabdoid tumor cells. J. Cell. Physiol. 194:303-313. [DOI] [PubMed] [Google Scholar]
- 36.Reisman, D. N., J. Sciarrotta, W. Wang, W. K. Funkhouser, and B. E. Weissman. 2003. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 63:560-566. [PubMed] [Google Scholar]
- 37.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, C. W., S. A. Galusha, M. E. McMenamin, C. D. Fletcher, and S. H. Orkin. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97:13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, C. W., M. M. Leroux, M. D. Fleming, and S. H. Orkin. 2002. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2:415-425. [DOI] [PubMed] [Google Scholar]
- 40.Sif, S. 2004. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell. Biochem. 91:1087-1098. [DOI] [PubMed] [Google Scholar]
- 41.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strober, B. E., J. L. Dunaief, Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trompouki, E., E. Hatzivassiliou, T. Tsichritzis, H. Farmer, A. Ashworth, and G. Mosialos. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793-796. [DOI] [PubMed] [Google Scholar]
- 45.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 46.Versteege, I., S. Medjkane, D. Rouillard, and O. Delattre. 2002. A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene 21:6403-6412. [DOI] [PubMed] [Google Scholar]
- 47.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 48.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes promoter targeting of chromatin-modifying complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G. R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 51.Yoshida, T., I. Tomioka, T. Nagahara, T. Holyst, M. Sawada, P. Hayes, V. Gama, M. Okuno, Y. Chen, Y. Abe, T. Kanouchi, H. Sasada, D. Wang, T. Yokota, E. Sato, and S. Matsuyama. 2004. Bax-inhibiting peptide derived from mouse and rat Ku70. Biochem. Biophys. Res. Commun. 321:961-966. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Z. K., K. P. Davies, J. Allen, L. Zhu, R. G. Pestell, D. Zagzag, and G. V. Kalpana. 2002. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 22:5875-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.