Abstract
The RASSF1A isoform of RASSF1 is frequently inactivated by epigenetic alterations in human cancers, but it remains unclear if and how it acts as a tumor suppressor. RASSF1A overexpression reduces in vitro colony formation and the tumorigenicity of cancer cell lines in vivo. Conversely, RASSF1A knockdown causes multiple mitotic defects that may promote genomic instability. Here, we have used a genetic approach to address the function of RASSF1A as a tumor suppressor in vivo by targeted deletion of Rassf1A in the mouse. Rassf1A null mice were viable and fertile and displayed no pathological abnormalities. Rassf1A null embryonic fibroblasts displayed an increased sensitivity to microtubule depolymerizing agents. No overtly altered cell cycle parameters or aberrations in centrosome number were detected in Rassf1A null fibroblasts. Rassf1A null fibroblasts did not show increased sensitivity to microtubule poisons or DNA-damaging agents and showed no evidence of gross genomic instability, suggesting that cellular responses to genotoxins were unaffected. Rassf1A null mice showed an increased incidence of spontaneous tumorigenesis and decreased survival rate compared with wild-type mice. Irradiated Rassf1A null mice also showed increased tumor susceptibility, particularly to tumors associated with the gastrointestinal tract, compared with wild-type mice. Thus, our results demonstrate that Rassf1A acts as a tumor suppressor gene.
Loss of heterozygosity (LOH) at chromosome 3p21.3 is extremely common in human carcinomas of the lung and breast (14) and is believed to be a critical early event in the pathogenesis of lung cancer (11, 42). Comparative analysis of deletions in this region in lung and breast cancer cell lines has identified a critical deletion interval of 120 kb containing eight genes (14). While none of the genes in this deletion region are mutated at a significant frequency in cancer (14), one of these genes, RASSF1, is a strong candidate as the major tumor suppressor gene on the basis of its frequent epigenetic silencing and LOH in lung cancers (6).
The two major isoforms of RASSF1, A and C, are produced by alternative promoter usage and mRNA splicing (2, 6). Transcripts from both isoforms share four common exons (exons 3 to 6), which encode a Ras association (RA) domain, PEST sequences, and a consensus phosphorylation site for ataxia telangiectasia-mutated (ATM) kinase (6). Since the RASSF1 RA domain can associate with Ras oncoproteins in vitro (21) and ATM kinases play an essential role in DNA damage, RASSF1A/C may serve as a link between these two signaling pathways. Overexpression and knockdown studies of RASSF1 have implicated RASSF1A in regulating cell cycle progression and RASSF1C in Ras-mediated cellular activities. Both isoforms localize to the microtubules and the mitotic spindle, suggesting a possible involvement in regulating microtubule dynamics (39). Microtubules are protected against nocodazole-induced depolymerization by RASSF1A, and to some extent by RASSF1C, overexpression (39). The RASSF1 isoforms have been reported to play a role in mitotic progression; overexpression of RASSF1A but not RASSF1C causes a prometaphase arrest by preventing activation of the anaphase-promoting complex/cyclosome (APC/C), whereas short interfering RNA-mediated depletion of RASSF1A accelerates mitotic progression and causes mitotic defects (33). Furthermore, overexpression of RASSF1A has been shown in numerous studies to result in reduction of in vitro colony formation and anchorage-independent growth and reduction of the tumorigenicity of cancer cell lines in vivo (2, 6, 9). In contrast, only one study has reported that overexpression of RASSF1C resulted in growth inhibition in vitro and in vivo, and then only in the prostate and renal cell carcinoma cell lines LNCaP and KRC/Y, respectively, not the lung cancer line U2020 (15). These results suggest that RASSF1A function is distinct from RASSF1C and that RASSF1A may function as a tumor suppressor gene. Indeed, epigenetic inactivation of RASSF1A is frequently reported in human cancers (reviewed in reference 7) and in some cases has been shown to correlate with poor prognosis (17, 41, 43). In contrast, only one study has reported loss of expression of RASSF1C in cancer (38).
Here, we have generated Rassf1A null and conditional mice in order to dissect the functions attributed to the RASSF1 isoforms. We reexamine the role of RASSF1A in protecting microtubule stability and cell cycle progression and specifically examine the role of RASSF1A as a tumor suppressor gene.
MATERIALS AND METHODS
Construction of the Rassf1A targeting vector and generation of Rassf1A mutant mice.
To generate the Rassf1A targeting vector (pAH19), DNA fragments for the 5′, conditional, and 3′ homology arms were amplified from AB2.2 genomic DNA by PCR using Platinum PCR Supermix High Fidelity (Invitrogen, San Diego, CA) for 10 to 18 cycles. The homology arms consisted of a 4.5-kb 5′ arm (containing the entire BLU gene [GenBank accession number AF333027] flanked by AscI sites), a 1.7-kb conditional arm (containing exon 1α flanked by HindIII sites), and a 4.4-kb 3′ homology arm (containing exons 1βγ, 2αβ, and 2γ flanked by NotI sites), which were cloned into pGEM T-Easy (Promega, Madison, WI), sequenced, and then subcloned into the AscI, HindIII, and NotI sites of pFlexible (36), respectively. Ten micrograms of the PvuI-linearized pAH19 was electroporated into 107 AB2.2 embryonic stem (ES) cells (from mouse strain 129S5/SvEvBrd) (24) and grown under 3-μg/ml puromycin selection. Cells were cultured on a lethally irradiated SNL76/7 feeder layer (18). ES clones were picked into 96-well plates after 9 days of drug selection and expanded, and targeted clones were identified by Southern blot analysis using PCR-amplified probes on NdeI- or SacII-digested genomic DNA with a 5′ or 3′ external probe, respectively. pAS3 is the 254-bp 5′ external probe (forward primer, 5′-TCT GGA GGG TGA GAC TCG GTT TTG TAA A-3′; reverse primer, 5′-TAC ATA GTG AGA CTT TGT CTC AGA AAA ATC AAC CTC CTT C-3′), and pAT1 is the 579-bp 3′ external probe (forward primer, 5′-GCT GAG ACT GAG CAG AAA ATC AAG GAC TAC AAT GG-3′, and reverse primer, 5′-CTT GGC CAT GCC GTT CAG TTC GCT CAA AGA GTG C-3′). The correctly targeted allele was termed Rassf1ABrdm1 (m1). To generate null and conditional alleles, a cell line with the m1 allele was electroporated with 10 μg of a Cre-expressing plasmid (pTURBO-Cre; GenBank accession number AF334827) or an Flpe-expressing plasmid (pFLPe) (27), respectively. After 3 days of culture, the cells were trypsinized, reseeded at 3 × 105 cells/10-cm-diameter plate, and cultured in 0.2 μM 2-deoxy-2-fluoro-β-d-arabinofuranosyl-5-iodouracil (FIAU) for 5 days. After this time, the cells were grown for a further 3 days in drug-free medium before the resulting colonies were trypsinized, pooled, and analyzed by Southern blotting using the pAS3 probe on KpnI-digested genomic DNA or using the pR20P probe (a 1.2-kb PstI fragment from the conditional arm) on HindIII-digested genomic DNA. The allele in the Flp-transfected clones showing loss of the 12.1-kb KpnI fragment but not the 1.7-kb HindIII fragment was termed Rassf1ABrdc1 (c1) (“conditional” allele). The allele generated with Cre, which showed a 9.1-kb KpnI fragment and loss of the 1.7-kb HindIII fragment, was termed Rassf1ABrdm2 (m2) (“null” allele). ES cell clones for the null and conditional alleles were transmitted through the germ line and bred to homozygosity (maintained on a mixed 129/Sv-C57BL/6 background). Mice were housed in accordance with Home Office regulations (United Kingdom). For tumor induction studies mice were whole body irradiated with a sublethal dose of 3.5 Gy at 4 weeks of age.
Genotyping of Rassf1A mutant mice by genomic PCR.
Genomic DNA was amplified in 50-μl reaction mixtures using 45 μl Platinum PCR Supermix (Invitrogen) and 100 ng of each primer pair (RSF-5/RSF-3 or RSF-C/RSF-3). The primers were as follows: RSF-3 (reverse primer), 5′-CCA GGC TTC CTT CTC ACT CCT CTG CCG C-3′; RSF-5 (forward primer), 5′-CTC GCC CCT GTC AGA CCT CAA TTT CCC-3′; and RSF-C (forward primer), 5′-AAG GAG AAA GAA AGC TGC TCT GGG GTT CT-3′. The PCR cycle profile was as follows: 1 cycle at 94°C for 2 min followed by 30 cycles at 94°C for 30 s, 65°C for 1 min, and 72°C for 30 s, with a final cycle of 72°C for 10 min.
Reverse transcriptase PCR (RT-PCR).
RNA was extracted from +/+, c1/c1, and m2/m2 mouse lung (RNAqueous; Ambion, Austin, TX), and 1.5 μg of total RNA was used to generate cDNA (RETROscript; Ambion). Specific primers were as follows: for Rassf1A exons 1α, 1βγ, and 2αβ (330-bp product), 5′-ATG TCG GCG GAG CCA GAA CTC ATT GAA CTA-3′ and 5′-CAC GTT CGT ATC CCG CTC TAG TGC AGA GT-3′; for Rassf1C exons 2γ and 3 (220-bp product), 5′-ATG GGC GAG GCT GAA ACA CCT TCC TTC GAA-3′ and 5′-CAG GCT CAT GAA GAG GTT GCT GTT GAT CT-3′; for Rassf1 exons 3 and 4 (372-bp product), 5′-GAT GAG GCT GTA GAG CGG GAG ACA CCC G-3′ and 5′-CTT GGC CAT GCC GTT CAG TTC GCT CAA AG-3′; and for β-actin exons 2 and 3 (215-bp product), 5′-ACC AAC TGG GAC GAT ATG GAG AAG A-3′ and 5′-TAC GAC CAG AGG CAT ACA GGG ACA A-3′. The cDNA was amplified in 50-μl reaction mixtures using 45 μl Platinum PCR Supermix (Invitrogen) and 100 ng of each primer pair with the following PCR cycle profile: 1 cycle at 94°C for 2 min followed by 30 cycles at 94°C for 30 s, 65°C for 1 min, and 72°C for 30 s, with a final cycle of 72°C for 10 min. All PCR products were cloned into pGEM T-Easy and sequenced to confirm their identity.
Quantitative RT-PCR.
RNA was extracted from +/+ and m2/m2 mouse liver (Trizol; Invitrogen), and 5 μg of total RNA was used to generate cDNA (Superscript II reverse transcriptase; Invitrogen). Quantitative PCR (qPCR) was carried out using the relative quantitation comparative CT method in separate tubes on the ABI 7900HT sequence detection system in accordance with the manufacturer's instructions (ABI user bulletin no. 2; Applied Biosystems, Foster City, CA). The target sequence was designed to exons 1 and 2 of Rassf1C (5′-AGC ACA ACC AGC AGT GGC TA-3′ and 5′-TTG ATT TTC TGC TCA GTC TCA-3′; 167-bp product), and the reference PCR product was designed to exons 3 and 4 of β-actin (5′-GAT CTG GCA CCA CAC CTT CT-3′ and 5′-GGG GTG TTG AAG GTC TCA AA-3′; 138-bp product). Both PCR products were tested to confirm the presence of a single clean product by gel electrophoresis and melt curve analysis on the ABI 7900HT sequence detection system. A validation experiment was carried out to confirm that the PCR efficiencies of the two products were sufficiently similar. qPCRs were performed with the QuantiTect Syber Green PCR kit (QIAGEN, Valencia, CA) in accordance with the manufacturer's instructions, using a 50-μl PCR volume, 5 ng cDNA, and 125 ng (2.5 ng/μl) primer. Five replicates of all PCRs were carried out, and the final quantification was determined relative to RNA from a wild-type mouse. The PCR products after qPCR were checked by melt curve analysis and gel electrophoresis to confirm the presence of a single clean product. The data are represented as the mean relative quantities of Rassf1C mRNA ± standard deviations (SD) (using liver RNA samples from four mice of each genotype).
Cell cycle and CD antigen analysis of thymocytes.
Thymuses and spleens were excised from 6- to 7-week-old wild-type and Rassf1A null (m2/m2) mice and were gently crushed through 100-μm cell strainers (BD Falcon, Bedford, MA) in phosphate-buffered saline (PBS). After centrifugation and washing with PBS/2% fetal bovine serum, cells were divided into two groups for analysis. For analysis of CD antigen expression, cells were stained with CD2-fluorescein isothiocyanate (FITC), CD4-phycoerythrin (PE), CD8-FITC, and/or CD45R (B220)-FITC (Pharmingen, Palo Alto, CA) and analyzed by flow cytometry (20,000 cells examined per sample). For cell cycle analysis, cells (1.5 × 106) were fixed with 70% ethanol for 30 min at 4°C, RNase A treated (2 μg/ml; Sigma, St. Louis, MO) for 30 min at room temperature, and stained with propidium iodide (50 μg/ml; Sigma), and the DNA content was analyzed by flow cytometry. All fluorescence-activated cell sorter (FACS) data analysis was performed using WinMDI version 2.8 software.
Micronucleus assay.
Blood was collected from wild-type and Rassf1A null 6- to 7-week-old and 7-month-old mice by cardiac puncture during terminal anesthesia and immediately fixed in methanol overnight at −80°C. DNA, CD71 antibody staining, and FACS analysis were performed using MicroFlow reagents from Litron Laboratories (Rochester, NY) as described by the manufacturer. Fifty thousand red cells were examined from each animal. For irradiation studies, 6- to 7-week-old mice were whole body irradiated at 0.7 Gy and blood was collected 48 h later.
Histology.
Tissues from wild-type and Rassf1A null mice were collected and fixed in 10% neutral buffered formalin. Tissue samples were processed routinely and embedded in paraffin, sectioned at 4 μm, stained with hematoxylin and eosin, and analyzed by light microscopy.
Cell growth and proliferation of MEFs.
Mouse embryonic fibroblasts (MEFs) were derived from 13.5-day-old embryos as described previously (29). All studies were performed on four to seven independently collected batches of MEFs, which were cultured up to passage 7 (P7). For cell growth assays MEFs were seeded in triplicate at a density of 3 × 105 cells/6-cm-diameter plate at day 0. Cells were counted daily using a Coulter counter (Beckman Coulter, Fullerton, CA). Data were collected from cell counts of four independent experiments.
Survival assays.
Cytotoxicity was evaluated using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay, in which 4 × 103 wild-type or Rassf1A null MEFs in 200 μl of complete medium were placed into each well of a 96-well plate and 24 h later were exposed to a DNA-damaging agent (methyl methanosulfonate [MMS], mitomycin C [MMC], hydroxyurea [HU], ionizing radiation, or UV) or microtubule-binding agent (nocodazole, vinblastine, colchicine, or taxol). Exceptions include MMS treatment, in which cells were exposed to the drug for only 1 h, and ionizing radiation, in which cells were irradiated prior to plating. After 5 days, viability was assessed using CellTiter reagent (Promega), according to the manufacturer's recommendations. A minimum of eight wells were analyzed and averaged for each data point at each dose from four independent experiments. Results are expressed as means ± SD. All DNA-damaging and microtubule-binding chemicals were purchased from Sigma.
Spectral karyotyping.
Spectral karyotyping analysis was carried out as previously described (31) on metaphase spreads prepared from wild-type and Rassf1A null MEFs (at P1) using the mouse SkyPaint probe mixture supplied by Applied Spectral Imaging (Vista, CA). Spectral images were captured using a Nikon Eclipse E800 epifluorescence microscope equipped with an SD200 SpectraCube system (Applied Spectral Imaging). At least 20 raw spectral image files were acquired for each genotype using Spectral Imaging version 2.3 software (Applied Spectral Imaging) and analyzed using SkyView version 1.6.1 software (Applied Spectral Imaging).
Microtubule stability analysis.
To evaluate microtubule stability in MEFs, cells were seeded onto polylysine-coated coverslips (Becton Dickinson) 24 h prior to treatment with nocodazole or colcemid for 30 min at 37°C. Coverslips were washed twice with cold PBS and were stained with mouse monoclonal anti-α-tubulin antibody at 1:200 (Molecular Probes, Eugene, OR) for 1 h at room temperature. After washing with PBS, Alexa Fluor 568-conjugated anti-mouse secondary antibody (Molecular Probes, Eugene, OR) was used and DNA was counterstained with Hoechst (Sigma). At each drug concentration, 100 to 150 cells were scored for microtubule integrity. Untreated cells have an extensive network of microtubule fibers that fills the cytoplasm. Local absence of defined fibers was scored as partial microtubule depolymerization, and the absence of any fibers was indicative of complete microtubule depolymerization. The micrographs shown are representative of >75% of the cells in terms of microtubule integrity at each concentration. Images were obtained with a Zeiss confocal microscope and processed using Photoshop 7.0 (Adobe Systems, San Jose, CA).
Immunofluorescence.
MEFs were grown on polylysine-coated coverslips and pulsed with 40 μM bromodeoxyuridine (BrdU) for 1 h prior to fixation in 4% paraformaldehyde. Cells were permeabilized in 80% ethanol at −20°C and briefly in 0.1% Triton X-100/0.02% sodium dodecyl sulfate in PBS. Cells were blocked in 1% bovine serum albumin and stained with anti-BrdU and nuclease (Amersham Biosciences, Piscataway, NJ), rabbit anti-PH3 at 1:400 (Upstate Biotechnology, Waltham, MA), or rabbit anti-γ-tubulin 1:1,000 (Sigma). After washing with detergent, cells were incubated with Alexa Fluor 568-conjugated anti-mouse secondary antibody or Alexa Fluor 568-conjugated anti-rabbit secondary antibody (Molecular Probes). Hoechst (Sigma) was used to visualize DNA. At least 300 cells were scored for each experiment, and experiments were carried out with three to seven different batches of MEFs. When scoring cells for centrosome numbers, binucleate cells were excluded from the analysis. It was not possible to accurately determine the number of centrosomes in P7 MEFs as >80% of the cells contained no centrosomes (had been dispersed or degraded).
Statistics.
For most comparisons, the Student t test (unpaired) was used to generate a P value. For the Kaplan-Meier curves, the log rank test was used.
RESULTS
Generation of Rassf1A null and conditional mice.
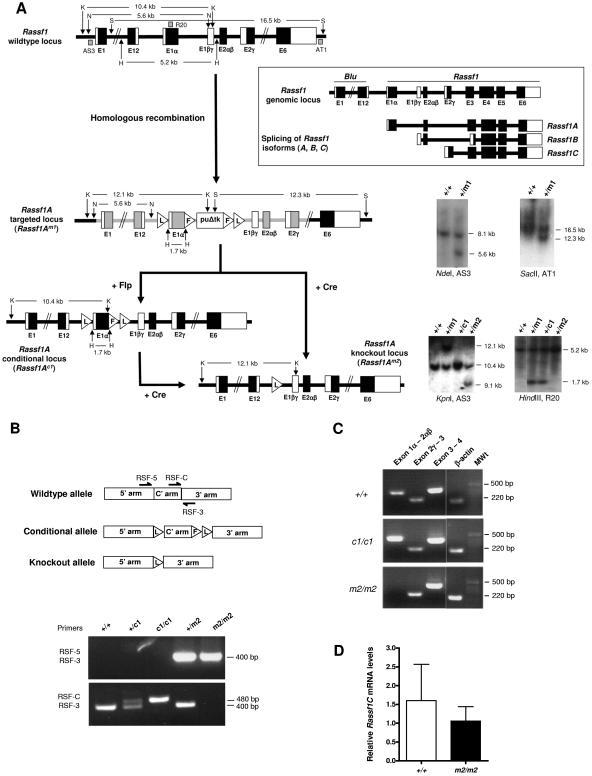
To assess the physiologic role of RASSF1A, we used gene targeting to generate mice lacking Rassf1A. In contrast to a previous study in which Rassf1 and hence all the isoforms were deleted (16) we specifically ablated the Rassf1A isoform by targeted deletion of exon 1α (Fig. 1A). Furthermore, we used a conditional approach to target Rassf1A, whereby the targeted locus contained loxP sites flanking exon 1α and FRT sites flanking the selection cassette (Fig. 1A). The conditional targeting vector was introduced into ES cells and correctly targeted clones, Rassf1A+/Brdm1 (+/m1), were identified by Southern blotting (Fig. 1A). +/m1 ES cell clones were then exposed to either Flpe (to delete the selection cassette) or Cre (to delete both exon 1α and the selection cassette) to generate the ES cell lines with the Rassf1A+/Brdc1 (+/c1) and Rassf1A+/Brdm2 (+/m2) alleles, respectively (Fig. 1A). These ES cell clones were used to generate chimeric mice that transmitted the mutated allele through the germ line. All offspring were genotyped by PCR using the strategy shown in Fig. 1B, which can distinguish between the five possible genotypes (+/+, +/c1, c1/c1, +/m2, and m2/m2).
FIG. 1.
Generation of null and conditional alleles of Rassf1A. A. (Box) Through the use of alternative promoter selection and alternative mRNA splicing of the RASSF1 transcript, three isoforms have been described, namely, RASSF1A, RASSF1B, and RASSF1C (RASSF1A mRNAs also come in multiple tissue-related splice forms, including heart-specific RASSF1D [AF102771] and pancreas-specific RASSF1E [AF102772]) (2). However, the two major isoforms are the ubiquitously expressed RASSF1A and RASSF1C, as RASSF1B is hematopoietic cell specific. RASSF1A is a six-exon gene that initiates transcription at exon 1α. Thus to specifically ablate the Rassf1A isoform, LoxP (L) sites were positioned to allow Cre-mediated excision of exon 1α and generate a null knockout (m2) allele. The puroΔtk cassette, flanked by FRT (F) sites, can be excised using Flpe to generate a conditional (c1) allele. The regions of homology used in the targeting vector (pAH19) are shown in gray. Blu is a gene which lies adjacent to Rassf1. Southern blot analysis of the Rassf1A alleles was performed using three probes: pAS3 and pAT1, which are 5′ and 3′ of the targeted allele, respectively, and pR20, which is on the conditional allele. Restriction enzyme sites and fragment sizes are as indicated. H, HindIII; K, KpnI; N, NdeI; S, SacII. B. PCR genotyping of tail genomic DNA, using a combination of two primer pairs (either RSF-5/RSF-3 or RSF-C/RSF-3), can distinguish between all five genotypes (+/+, +/c1, c1/c1, +/m2, and m2/m2) based on the presence and size of the product(s), which can be resolved on a 2% agarose gel. C. RT-PCR was performed to confirm deletion of the floxed exons in Rassf1A null mice. RT-PCR performed on RNA extracted from +/+ and c1/c1 mice shows a 330-bp product corresponding to Rassf1A exons 1α, 1βγ, and 2αβ. In contrast, no such product is seen in RNA from m2/m2 mice. RNA from +/+, c1/c1, and m2/m2 mice all show 220- and 372-bp products corresponding to exons 2γ and 3 and 3 and 4 of Rassf1C and Rassf1 (common to all isoforms), respectively. β-actin was used as a positive control for cDNA production. D. Quantitative RT-PCR was performed to confirm there was no difference in the level of mRNA expression of the Rassf1C isoform between wild-type and Rassf1A null mice. Data are presented as mean relative levels of Rassf1C mRNA ± SD (n = 4).
Rassf1A null and conditional mice are viable, healthy, and fertile.
Interbreeding of heterozygous (+/c1 or +/m2) mice produced homozygote conditional (c1/c1) and null (m2/m2) offspring at the expected Mendelian ratios. These mice were indistinguishable from littermate controls in terms of growth, development, and reproductive abilities. No histological differences could be seen between wild-type +/+, c1/c1, and m2/m2 mice in all tissues examined (heart, lung, spleen, brain, thymus, kidney, stomach, intestine, pancreas, and testis; data not shown). RT-PCR performed on m2/m2 lung cDNA confirmed the absence of Rassf1A exon 1α, while the other major isoform, Rassf1C, remained unaffected (Fig. 1C), as confirmed by quantitative RT-PCR (Fig. 1D). RT-PCR on c1/c1 lung cDNA showed that the conditional allele was spliced appropriately, the loxP site in the intron following exon 1α did not interfere with splicing from exon 1α to exon 2αβ, and, given that m2/m2 mice showed no obvious abnormalities, the c1 allele was not characterized further. However, conditional inactivation of Rassf1A using the c1/c1 mice will make it possible to evaluate the role of Rassf1A in specific tissues.
Lymphocyte development is normal in Rassf1A null mice.
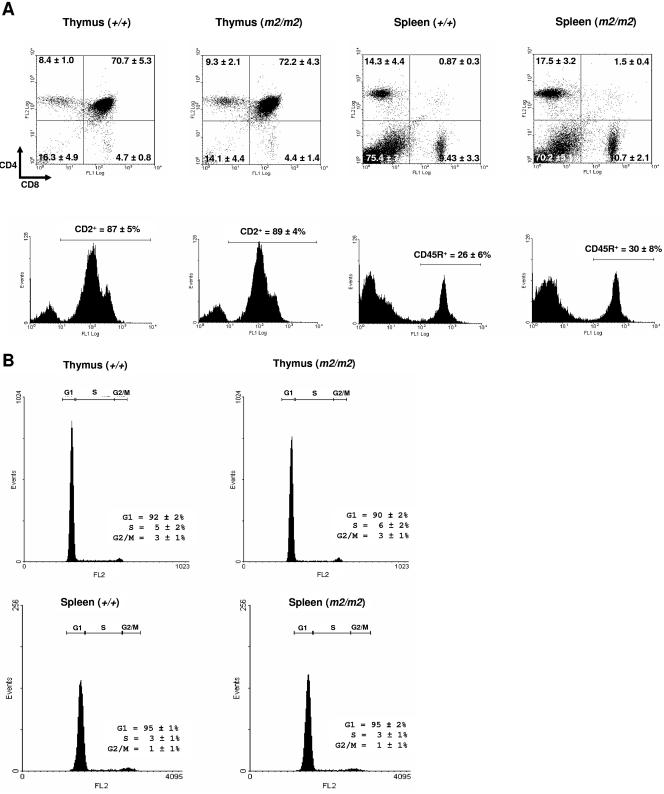
Rassf1A has been hypothesized to function as a negative regulator of cell cycle progression and cell proliferation (32). Given that tight cell cycle control is important for lymphocyte proliferation, we investigated the lymphocytic composition of thymuses and spleens from Rassf1A null mice. Loss of Rassf1A did not grossly affect thymic or splenic weight at either 6 to 7 weeks of age or 7 months of age (data not shown). Analysis of thymuses from Rassf1A null mice showed a thymocyte population comprising mainly double-positive (CD4+ CD8+) T cells, nearly identical to that of wild-type mice (Fig. 2A). Furthermore, similar proportions of mature CD4+ and CD8+ T cells were present in the spleens of wild-type and Rassf1A null mice (Fig. 2B). FACS analysis also demonstrated that the percentage of cells in the thymus and spleen that expressed CD2 (T-cell lineage) or CD45R (B220) (B-cell lineage) antigens, respectively, were unchanged in Rassf1A null mice (Fig. 2A). Thus, loss of Rassf1A gene function does not affect the lymphocytic component of the immune system.
FIG. 2.
Analysis of thymic and splenic lymphocytes in Rassf1A null and wild-type animals. A. Spleens and thymuses were taken from adult animals and stained with CD4-PI/CD8-FITC, CD2-FITC, or CD45R-FITC. A minimum of 10,000 events were analyzed from each organ. Data are presented as means ± SD (n = 7). B. Propidium iodide cell cycle analysis on thymic and splenic lymphocytes. A minimum of 10,000 events were analyzed from each organ. Data are presented as means ± SD (n = 7).
Rassf1A null MEFs have no gross cell cycle defects.
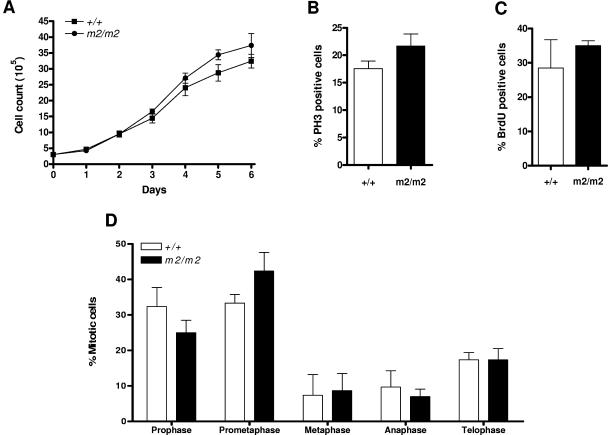
Numerous studies performed using overexpression and short interfering RNA-mediated knockdown of RASSF1A in vitro have implicated RASSF1A in playing a role in regulating cell cycle and mitotic progression (5, 25, 32, 33, 39). However, FACS analysis of propidium iodide-stained Rassf1A null and wild-type thymocytes, splenocytes, and MEFs did not show any accumulation of cells in a particular phase of the cell cycle (Fig. 2B and data not shown). There was also no difference in the growth curves of Rassf1A null and wild-type MEFs (Fig. 3A). Consistent with this, confocal microscopy of asynchronous Rassf1A null MEFs also showed no difference in the proportion of G2/M phase cells identified by phosphorylated histone H3 (4) (Fig. 3B) or in S phase cells identified by BrdU incorporation (Fig. 3C). Transient knockdown of Rassf1A has been previously shown to decrease the time spent in mitosis, presumably by premature APC/C activation and faster progression toward anaphase (33). In Rassf1A null MEFs, cells in each mitotic phase were scored and no significant difference was found in the proportion of premetaphase to postmetaphase cells (Fig. 3D). A slight increase in prometaphase cells was detected in Rassf1A null MEFs, which may reflect difficulties in spindle assembly in the absence of Rassf1A. Thus the absence of Rassf1A does not result in overt misregulation of the cell cycle or mitotic progression.
FIG. 3.
Analysis of the cell cycle and mitotic properties of wild-type and Rassf1A null MEFs. A. Growth curves of wild-type (+/+) and Rassf1A null (m2/m2) MEFs. Cell counts were performed in triplicate at intervals of 24 h. Data represent means ± SD of four independently collected batches of MEFs. B. PH3 staining of wild-type and Rassf1A null MEFs to analyze the proportion of the population in G2/M phase. At least 300 cells were scored for each experiment, and the data are presented as means ± SD (n = 3). C. BrdU staining of wild-type and Rassf1A null MEFs to analyze the proportion of the population in S phase. At least 300 cells were scored for each experiment, and the data are presented as means ± SD (n = 3). D. Analysis of the proportion of wild-type and Rassf1A null MEFs in each stage of mitosis. At least 200 cells were scored for each experiment, and the data are presented as means ± SD (n = 3).
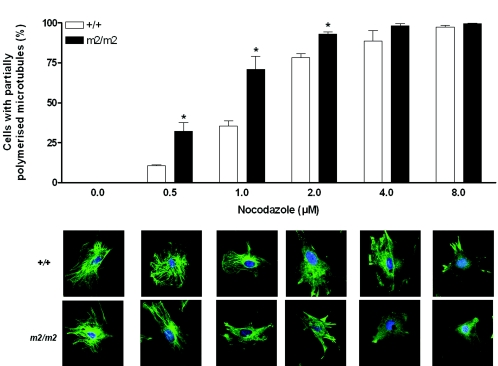
Rassf1A null MEFs show an increased sensitivity to microtubule-depolymerizing agents.
Protection of microtubules by Rassf1 has been shown previously (5, 16, 39), and we demonstrate here that at least a portion of this activity can be attributed to the Rassf1A isoform. Rassf1A null MEFs showed more extensive depolymerization of the microtubule network at low concentrations of nocodazole compared to wild-type MEFs (32.2% ± 10.7% versus 10.5% ± 1.1% of cells depolymerized at 0.5 μM, respectively; Fig. 4). Microtubule fibers could still be detected at 1 μM nocodazole in wild-type cells but could hardly be seen in Rassf1A null MEFs at this concentration. Similar results were found with another microtubule-depolymerizing agent, colcemid (data not shown).
FIG. 4.
Rassf1A null MEFs are more sensitive than wild-type MEFs to nocodazole-induced microtubule depolymerization. Wild-type and Rassf1A null (m2/m2) MEFs were incubated with nocodazole at the indicated concentrations for 30 min before staining with anti-α-tubulin antibody. At total of 100 to 150 cells were scored for each experiment, and the data are presented as means ± standard errors of the means (n = 4), with representative images shown for each concentration. Asterisk, P < 0.05 for +/+ versus m2/m2 at the concentration indicated.
Rassf1A null mice show no evidence of genomic stability.
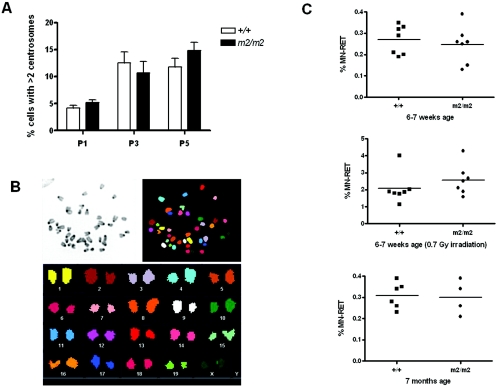
Transient knockdown of Rassf1A in foreskin fibroblasts has been shown to cause mitotic abnormalities including multiple centrosomes and lagging chromosomes that were postulated to contribute to genomic instability (33). However, Rassf1A null MEFs did not display lagging chromosomes (data not shown) or show distinct aberrations in centrosome numbers (P1 to P3), although at later passages (P5) a larger percentage of cells with more than two centrosomes was noticed than in wild-type cells (though not significant by Student's t test analysis; Fig. 5A). A complete analysis of the total chromosome content using spectral karyotyping (SKY) of metaphase spreads from wild-type and Rassf1A null MEFs showed no evidence of gross chromosomal rearrangements or aneuploidy (Fig. 5B). To confirm the absence of aneuploidy in Rassf1A null MEFs, we used the highly sensitive flow-cytometric micronucleus assay, which provides a quantitative measure of in vivo chromosome damage (10). Micronuclei can arise from acentric chromosome fragments or whole chromosomes that have not been incorporated in the main nuclei at cell division (20). As shown in Fig. 5C, Rassf1A null mice at either 6 to 7 weeks (0.24% ± 0.08%, n = 7) or 7 months of age (0.30% ± 0.08%, n = 4) did not show higher levels of micronuclei than wild-type littermates (0.27% ± 0.06%, n = 7 and 0.31% ± 0.06%, n = 6, respectively). Since irradiation can stimulate micronucleus formation (1, 30), Rassf1A null mice at 6 to 7 weeks of age were irradiated with 0.7 Gy and blood was collected 48 h later. Still, irradiated Rassf1A null mice (2.5% ± 0.8%, n = 7) did not show significantly higher levels of micronuclei than irradiated wild-type littermates (2.0% ± 0.9%, n = 7; Fig. 5C). Taken together, these data suggest that the absence of Rassf1A does not result in gross genomic instability.
FIG. 5.
Rassf1A null MEFS do not show evidence of increased genomic instability. A. Quantitation of centrosome numbers determined by γ-tubulin staining of wild-type and Rassf1A null (m2/m2) MEFs at P1, P3, and P5. At least 300 cells were scored for each experiment, and the data are presented as means ± SD (n = 7). B. SKY analysis. A SKY hybridization and detection kit was used to visualize the 19 autosomal and 2 sex chromosomes in colors. At least 20 raw spectral image files were acquired for each genotype, and images were analyzed using SkyView software. C. Micronucleus assays on blood from Rassf1A null and wild-type mice at 6 to 7 weeks of age (top), 6 to 7 weeks of age 48 h after irradiation with 0.7 Gy (center), and 7 months of age (bottom). Peripheral blood was stained with anti-CD71-FITC antibody and propidium iodide and analyzed by flow cytometry. A minimum of 50,000 events were analyzed for each sample. Data represent analysis of blood from four to seven animals of each genotype.
Rassf1A deficiency does not affect sensitivity to DNA-damaging or microtubule-binding agents.
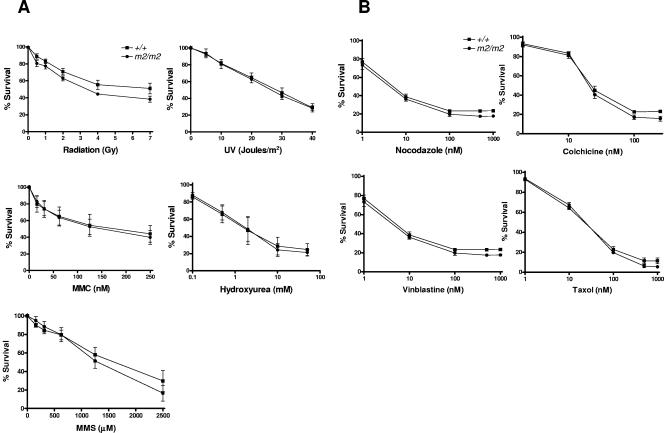
To examine the effect of Rassf1A deficiency on cell growth and viability after DNA damage, we performed an MTT assay on Rassf1A null MEFs exposed to a variety of DNA-damaging agents, including irradiation to induce DNA double-strand breaks, UV to induce nucleotide alterations, and HU to inhibit DNA synthesis, as well as the DNA cross-linking agent MMC and the alkylating agent MMS. As shown in Fig. 6, Rassf1A null MEFs did not show increased sensitivity compared to wild-type MEFs to the induced forms of DNA damage or microtubule poisons. This phenotype was confirmed in another cell type, by performing clonogenic assays on double-targeted (m2/m2) ES cells (data not shown). Thus Rassf1A deficiency does not impair checkpoint responses triggered by DNA damage or failure to correctly assemble mitotic spindles.
FIG. 6.
Rassf1A null MEFs cells do not show an enhanced cytotoxicity to DNA damaging- and microtubule-binding agents. The cytoxicity of DNA-damaging and microtubule-binding agents on Rassf1A null MEFs was examined using the MTT assay in a 96-well format (as described in Materials and Methods). Treatments include the DNA-damaging agents irradiation, UV, MMS, MMC, and HU (A) and the microtubule-binding agents nocodazole, colchicine, vinblastine, and taxol (B). Data were normalized to untreated cells of the same genotype, and results are expressed as % survival (representing the means of four to six independent experiments ± standard errors of the means).
Rassf1A deficiency plays a role in tumorigenesis.
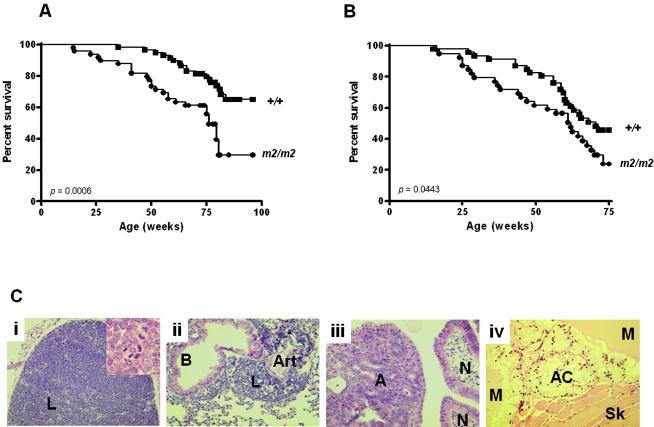
Using our Rassf1A null mice, we addressed the key question of whether loss of Rassf1A is relevant in tumorigenesis. As shown in Fig. 7A, Rassf1A null mice showed a significantly reduced rate of survival compared to wild-type littermates. In comparison to the death of 30% of the wild-type mice, 50% of the Rassf1A null mice died during the course of the study. In both cohorts the predominant cause of death was cancer, in particular lymphoma (Table 1). In addition to the deaths shown in Table 1, two Rassf1A null mice showed no tumors at the time of death (both died from cardiac failure and pneumonia). To accelerate tumorigenesis, Rassf1A null mice were irradiated with 3.5 Gy at 4 weeks of age (Fig. 7B). Irradiated Rassf1A null mice showed an increased incidence of tumorigenesis and decreased survival rate compared with wild-type mice, with 27 of the 39 Rassf1A null mice dying (69%) compared to 24 of the 46 wild-type mice (52%). It is noteworthy that, although the majority of all tumors that developed were lymphomas, often with metastatic spread to many organs, Rassf1A null mice showed more tumor formation (adenomas and adenocarcinoma) in the gastrointestinal (GI) tract than wild-type mice (Fig. 7C).
FIG.7.
Rassf1A null mice develop tumors with and without irradiation. Kaplan-Meier plots show the percentages of survival of wild-type (+/+) and Rassf1A null (m2/m2) mice in the (A) nonirradiated and (B) irradiated cohorts. C. Hematoxylin- and eosin-stained sections (×180 magnification) showing tumor formation. (i) thymic lymphoma (L), high grade, confined to thymus (m2/m2, irradiated; inset: high-grade lymphoma cells with many mitotic figures shown at high-power view [×360]); (ii) metastatic lymphoma (L) in lung, with high-grade lymphoma surrounding a bronchus (B) and artery (Art) (m2/m2, irradiated); (iii) small intestinal adenoma (A) (left) with adjacent normal intestinal villi (N) (right) (m2/m2, irradiated); (iv) metastatic mucinous adenocarcinoma (AC) producing pale-yellow-staining mucin (M) and invading skeletal muscle (Sk) (bottom right) of groin (m2/m2, nonirradiated).
TABLE 1.
Tumors found in wild-type and Rassf1A null micea
| Type and site of tumor | No. of mice
|
|||
|---|---|---|---|---|
| Spontaneous
|
Irradiated
|
|||
| +/+ | m2/m2 | +/+ | m2/m2 | |
| Lymphoma/leukemia | 14 | 17 | 18 | 20 |
| Carcinoma | ||||
| Adenocarcinoma | ||||
| Ovary | 1 | |||
| Lung | 1 | 2 | 4 | 1 |
| Gl tract | 1 | |||
| Squamous carcinoma of skin | 1 | |||
| Sarcoma | ||||
| Angiosarcoma | 1 | 2 | 1 | 2 |
| Osteogenic | 1 | 1 | ||
| Benign | ||||
| Adenoma | ||||
| Adrenal glands | 1 | |||
| Lung | 1 | |||
| Gl tract | 1 | 2 | ||
| Lipoma | 1 | |||
| Myxoma | 1 | |||
| Pheochromocytoma | 1 | |||
| Sex cord tumor (ovary) | 1 | |||
This table represents the different tumor types that were found; hence mice that developed multiple tumors are represented more than once. For the spontaneous cohort, 24 of the 48 Rassf1A null (m2/m2) mice died (50%) compared to 18 of the 59 wild-type mice (30%). For the irradiated cohort, 27 of the 39 Rassf1A null (m2/m2) mice died (69%) compared to 24 of the 46 wild-type mice (52%).
DISCUSSION
RASSF1A methylation has been reported in almost all types of human cancer (reviewed in references 7 and 22). However, how RASSF1A functions as a tumor suppressor gene remains largely unknown. Recent work has suggested that RASSF1A might provide the crucial link between tumor suppression and the cell cycle (16, 33) by controlling the stability of microtubules and the activity of APC/C. To delineate the role of RASSF1A in the cell cycle and tumorigenesis, we have used a genetic approach and generated Rassf1A knockout mice. Due to concerns that the homozygous null mice may be embryonically lethal, we also generated a conditional Rassf1A allele. However, Rassf1A null mice unexpectedly proved to be viable, fertile, and healthy. Most surprisingly, loss of Rassf1A did not grossly impair cell cycle progression in vivo. Lymphocytes from Rassf1A null mice were identical to those from wild-type littermates in terms of proportions of T and B lymphocytes and showed no difference in the proportion of cells in each cell cycle phase. The growth kinetics and cell cycle profiles of Rassf1A null MEFs were indistinguishable from wild-type MEFs. A closer, but stationary, analysis of the mitotic phases revealed a subtle difference between Rassf1A null and wild-type MEFs in the prometaphase population, the stage during which spindle assembly occurs. Protection of microtubules by Rassf1 has been shown previously (5, 39), and we demonstrate that at least a portion of this activity can be attributed to the Rassf1A isoform. This is consistent with the recent finding that RASSF1C can bind tubulin but is far less effective than RASSF1A at stabilizing microtubules (39).
The viability of the Rassf1A null mouse and the absence of any obvious cell cycle defects are perhaps not altogether that surprising, as the traditional view of the mammalian cell cycle depicts many components as fulfilling essential steps that dictate the sequential order of cell cycle events, yet Cyclin E, Cyclin D, Cdk2, Cdk4, and Cdc25C knockout mice have all been shown to be viable (reviewed in reference 34). Reasons for this could include compensation by other proteins with similar activity or the fact that some of these gene products have major roles but only in specific cell lineages. Other reasons could lie in the fact that most of the studies that declare genes to be essential for regulation of the cell cycle stem from analysis of cultured cells, many of which have defects in other key regulatory genes. This is particularly relevant for RASSF1A, as studies on the role of RASSF1A in the cell cycle have largely been performed using transformed immortalized cells such as HeLa cells, which have an abrogated Rb checkpoint due to human papillomavirus E7 expression, or H1299 cells, which are p53 null. Therefore, these cells lack many of the normal growth control regulations that would be imposed on a cell in vivo.
Previous in vitro observations using RASSF1A mutant cDNAs or transient knockdown of RASSF1A have led to the hypothesis that an absence of RASSF1A results in genomic instability (16, 33, 39). However, Rassf1A null mice did not display increased genomic instability or abrogation of DNA repair pathways. This lack of susceptibility to aneuploidy is unlikely to be due to functional compensation by Rassf1C, as Rassf1 null MEFs have also been reported to show no gross chromosomal aberrations (16). A more likely explanation is that, in order for a cell with such severe chromosomal abnormalities to be able to exist, defects in other key checkpoint (tumor suppressor) genes would also need to be present.
We addressed the key question of whether loss of Rassf1A is relevant in tumorigenesis. Rassf1A null mice showed a significantly increased incidence of tumorigenesis compared to wild-type littermates. Irradiated Rassf1A null mice also showed a reduced rate of survival and increased incidence of tumorigenesis, compared to irradiated wild-type mice. Irradiated heterozygous Rassf1A mice (+/m2) did not show an increased incidence of tumorigenesis or decreased rate of survival compared to wild-type mice; however, given the small size of this cohort (3 of the 12 mice [25%] developed tumors) the possibility of a haploinsufficiency effect cannot be excluded. In both the irradiated and nonirradiated cohorts, although the predominant tumor type was lymphoma, Rassf1A null mice showed additional tumors associated with the gastrointestinal tract, not commonly seen in the wild-type mice. Well-differentiated endocrine carcinomas of gastric origin have been shown to have frequent alterations in 3p21 (23), and epigenetic transcriptional silencing of RASSF1A is a frequent event in gastric tumorigenesis and might play an important role in the malignant progression of gastric adenocarcinomas (3, 12, 35). In addition, RASSF1A methylation has also been reported in colorectal cancer and hepatocellular carcinomas (13, 26, 28, 37, 40, 44). Interestingly, these are the same tumor types in which promoter methylation of the APC gene (adenomatous polyposis coli [APC]) has also been reported (13). Given that APC stimulates microtubule assembly and bundling (19, 45) and that lack of microtubule binding associated with cancer-associated mutations of APC are believed to contribute to defects in the assembly of mitotic spindles and lead to missegregation of chromosomes (8), it is tempting to speculate that aberrant APC function may play a role in the neoplastic progression of gastrointestional tract tissues with methylated or inactivated RASSF1A.
Acknowledgments
L.V.D.W. and D.J.A. are recipients of Australian NHMRC postdoctoral research fellowships (CJ Martin/RG Menzies Fellowship and CJ Martin Fellowship, respectively). K.K.T. is supported by an MRC predoctoral fellowship, M.A.G. is supported by an MRC clinical research training fellowship, and M.J.A. is supported by Cancer Research UK. This research was supported by the Wellcome Trust.
We thank Ron Laskey for helpful discussions, Ian Roberts for assistance with cytogenetic analysis, Graham Bignell for assistance with quantitative RT-PCR experiments, Evelyn Grau for microinjection of the targeted ES cell lines, and the personnel of Team 83 for care and monitoring of the animals.
REFERENCES
- 1.Bhilwade, H. N., R. C. Chaubey, and P. S. Chauhan. 2004. Gamma ray induced bone marrow micronucleated erythrocytes in seven strains of mouse. Mutat. Res. 560:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Burbee, D. G., E. Forgacs, S. Zochbauer-Muller, L. Shivakumar, K. Fong, B. Gao, D. Randle, M. Kondo, A. Virmani, S. Bader, Y. Sekido, F. Latif, S. Milchgrub, S. Toyooka, A. F. Gazdar, M. I. Lerman, E. Zabarovsky, M. White, and J. D. Minna. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J. Natl. Cancer Inst. 93:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byun, D. S., M. G. Lee, K. S. Chae, B. G. Ryu, and S. G. Chi. 2001. Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res. 61:7034-7038. [PubMed] [Google Scholar]
- 4.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatiotemporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallol, A., A. Agathanggelou, S. L. Fenton, J. Ahmed-Choudhury, L. Hesson, M. D. Vos, G. J. Clark, J. Downward, E. R. Maher, and F. Latif. 2004. RASSF1A interacts with microtubule-associated proteins and modulates microtubule dynamics. Cancer Res. 64:4112-4116. [DOI] [PubMed] [Google Scholar]
- 6.Dammann, R., C. Li, J. H. Yoon, P. L. Chin, S. Bates, and G. P. Pfeifer. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 25:315-319. [DOI] [PubMed] [Google Scholar]
- 7.Dammann, R., U. Schagdarsurengin, M. Stunnikova, M. Rastetter, C. Seidel, L. Liu, S. Tommasi, and G. P. Pfeifer. 2003. Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol. Histopathol. 18:665-677. [DOI] [PubMed] [Google Scholar]
- 8.Dikovskaya, D., I. P. Newton, and I. S. Nathke. 2004. The adenomatous polyposis coli protein is required for the formation of robust spindles formed in CSF Xenopus extracts. Mol. Biol. Cell 15:2978-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreijerink, K., E. Braga, I. Kuzmin, L. Geil, F. M. Duh, D. Angeloni, B. Zbar, M. I. Lerman, E. J. Stanbridge, J. D. Minna, A. Protopopov, J. Li, V. Kashuba, G. Klein, and E. R. Zabarovsky. 2001. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc. Natl. Acad. Sci. USA 98:7504-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heddle, J. A. 1973. A rapid in vivo test for chromosomal damage. Mutat. Res. 18:187-190. [DOI] [PubMed] [Google Scholar]
- 11.Hung, J., Y. Kishimoto, K. Sugio, A. Virmani, D. D. McIntire, J. D. Minna, and A. F. Gazdar. 1995. Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. JAMA 273:558-563. [PubMed] [Google Scholar]
- 12.Kang, G. H., S. Lee, J. S. Kim, and H. Y. Jung. 2003. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab. Investig. 83:519-526. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S., H. J. Lee, J. H. Kim, H. S. Lee, J. J. Jang, and G. H. Kang. 2003. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am. J. Pathol. 163:1371-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lerman, M. I., and J. D. Minna. 2000. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung CancerChromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 60:6116-6133. [PubMed] [Google Scholar]
- 15.Li, J., F. Wang, A. Protopopov, A. Malyukova, V. Kashuba, J. D. Minna, M. I. Lerman, G. Klein, and E. Zabarovsky. 2004. Inactivation of RASSF1C during in vivo tumor growth identifies it as a tumor suppressor gene. Oncogene 23:5941-5949. [DOI] [PubMed] [Google Scholar]
- 16.Liu, L., S. Tommasi, D. H. Lee, R. Dammann, and G. P. Pfeifer. 2003. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene 22:8125-8136. [DOI] [PubMed] [Google Scholar]
- 17.Maruyama, R., S. Toyooka, K. O. Toyooka, K. Harada, A. K. Virmani, S. Zochbauer-Muller, A. J. Farinas, F. Vakar-Lopez, J. D. Minna, A. Sagalowsky, B. Czerniak, and A. F. Gazdar. 2001. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 61:8659-8663. [PubMed] [Google Scholar]
- 18.McMahon, A. P., and A. Bradley. 1990. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62:1073-1085. [DOI] [PubMed] [Google Scholar]
- 19.Munemitsu, S., B. Souza, O. Muller, I. Albert, B. Rubinfeld, and P. Polakis. 1994. The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 54:3676-3681. [PubMed] [Google Scholar]
- 20.Nusse, M., B. M. Miller, S. Viaggi, and J. Grawe. 1996. Analysis of the DNA content distribution of micronuclei using flow sorting and fluorescent in situ hybridization with a centromeric DNA probe. Mutagenesis 11:405-413. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz-Vega, S., A. Khokhlatchev, M. Nedwidek, X. F. Zhang, R. Dammann, G. P. Pfeifer, and J. Avruch. 2002. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 21:1381-1390. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer, F. P., J. H. Yoon, L. Liu, S. Tommasi, S. P. Wilczynski, and R. Dammann. 2002. Methylation of the RASFF1A gene in human cancers. Biol. Chem. 383:907-914. [DOI] [PubMed] [Google Scholar]
- 23.Pizzi, S., C. Azzoni, D. Bassi, L. Bottarelli, M. Milione, and C. Bordi. 2003. Genetic alterations in poorly differentiated endocrine carcinomas of the gastrointestinal tract. Cancer 98:1273-1282. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez-Solis, R., P. Liu, and A. Bradley. 1995. Chromosome engineering in mice. Nature 378:720-724. [DOI] [PubMed] [Google Scholar]
- 25.Rong, R., W. Jin, J. Zhang, M. S. Sheikh, and Y. Huang. 2004. Tumor suppressor RASSF1A is a microtubule-binding protein that stabilizes microtubules and induces G2/M arrest. Oncogene 23:8216-8230. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto, N., T. Terai, Y. Ajioka, S. Abe, O. Kobayasi, S. Hirai, O. Hino, H. Watanabe, N. Sato, T. Shimoda, and H. Fujii. 2003. Frequent hypermethylation of RASSF1A in early flat-type colorectal tumors. Oncogene 23:8900-8907. [DOI] [PubMed] [Google Scholar]
- 27.Schaft, J., R. Ashery-Padan, F. van der Hoeven, P. Gruss, and A. F. Stewart. 2001. Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31:6-10. [DOI] [PubMed] [Google Scholar]
- 28.Schagdarsurengin, U., L. Wilkens, D. Steinemann, P. Flemming, H. H. Kreipe, G. P. Pfeifer, B. Schlegelberger, and R. Dammann. 2003. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene 22:1866-1871. [DOI] [PubMed] [Google Scholar]
- 29.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 30.Shima, N., S. A. Hartford, T. Duffy, L. A. Wilson, K. J. Schimenti, and J. C. Schimenti. 2003. Phenotype-based identification of mouse chromosome instability mutants. Genetics 163:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shing, D. C., C. A. Morley-Jacob, I. Roberts, E. Nacheva, and N. Coleman. 2002. Ewing's tumour: novel recurrent chromosomal abnormalities demonstrated by molecular cytogenetic analysis of seven cell lines and one primary culture. Cytogenet. Genome Res. 97:20-27. [DOI] [PubMed] [Google Scholar]
- 32.Shivakumar, L., J. Minna, T. Sakamaki, R. Pestell, and M. A. White. 2002. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell. Biol. 22:4309-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, M. S., S. J. Song, N. G. Ayad, J. S. Chang, J. H. Lee, H. K. Hong, H. Lee, N. Choi, J. Kim, H. Kim, J. W. Kim, E. J. Choi, M. W. Kirschner, and D. S. Lim. 2004. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat. Cell Biol. 6:129-137. [DOI] [PubMed] [Google Scholar]
- 34.Su, T. T., and J. Stumpff. 9 March 2004, posting date. Promiscuity rules? The dispensability of cyclin E and Cdk2. Sci. STKE 224: pe11. [Online.] http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;stke.2242004pe11. [DOI] [PMC free article] [PubMed]
- 35.To, K. F., W. K. Leung, T. L. Lee, J. Yu, J. H. Tong, M. W. Chan, E. K. Ng, S. C. Chung, and J. J. Sung. 2002. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int. J. Cancer 102:623-628. [DOI] [PubMed] [Google Scholar]
- 36.van der Weyden, L., D. J. Adams, L. W. Harris, D. Tannahill, M. J. Arends, and A. Bradley. 2005. Null and conditional Semaphorin 3B alleles using a flexible puroΔtk LoxP/FRT vector. Genesis 41:171-178. [DOI] [PubMed]
- 37.van Engeland, M., G. M. Roemen, M. Brink, M. M. Pachen, M. P. Weijenberg, A. P. de Bruine, J. W. Arends, P. A. van den Brandt, A. F. de Goeij, and J. G. Herman. 2002. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene 21:3792-3795. [DOI] [PubMed] [Google Scholar]
- 38.Vos, M. D., C. A. Ellis, A. Bell, M. J. Birrer, and G. J. Clark. 2000. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J. Biol. Chem. 275:35669-35672. [DOI] [PubMed] [Google Scholar]
- 39.Vos, M. D., A. Martinez, C. Elam, A. Dallol, B. J. Taylor, F. Latif, and G. J. Clark. 2004. A role for the RASSF1A tumour suppressor in the regulation of tubulin depolymerisation and genomic instability. Cancer Res. 64:4244-4250. [DOI] [PubMed] [Google Scholar]
- 40.Wagner, K. J., W. N. Cooper, R. G. Grundy, G. Caldwell, C. Jones, R. B. Wadey, D. Morton, P. N. Schofield, W. Reik, F. Latif, and E. R. Maher. 2002. Frequent RASSF1A tumour suppressor gene promoter methylation in Wilms' tumour and colorectal cancer. Oncogene 21:7277-7282. [DOI] [PubMed] [Google Scholar]
- 41.Wang, J., J. J. Lee, L. Wang, D. D. Liu, C. Lu, Y. H. Fan, W. K. Hong, and L. Mao. 2004. Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin. Cancer Res. 10:6119-6125. [DOI] [PubMed] [Google Scholar]
- 42.Wistuba, I. I., C. Behrens, A. K. Virmani, G. Mele, S. Milchgrub, L. Girard, J. W. Fondon, H. R. Garner, B. McKay, F. Latif, M. I. Lerman, S. Lam, A. F. Gazdar, and J. D. Minna. 2000. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 60:1949-1960. [PubMed] [Google Scholar]
- 43.Yang, Q., P. Zage, D. Kagan, Y. Tian, R. Seshadri, H. R. Salwen, S. Liu, A. Chlenski, and S. L. Cohn. 2004. Association of epigenetic inactivation of RASSF1A with poor outcome in human neuroblastoma. Clin. Cancer Res. 10:8493-8500. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, S., W. Yeo, M. W. Tang, N. Wong, P. B. Lai, and P. J. Johnson. 2003. Intensive hypermethylation of the CpG island of Ras association domain family 1A in hepatitis B virus-associated hepatocellular carcinomas. Clin. Cancer Res. 9:3376-3382. [PubMed] [Google Scholar]
- 45.Zumbrunn, J., K. Kinoshita, A. A. Hyman, and I. S. Näthke. 2001. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK-3β phosphorylation. Curr. Biol. 11:44-49. [DOI] [PubMed] [Google Scholar]