Abstract
The SR protein SRp38 is a general splicing repressor that is activated by dephosphorylation during mitosis and in response to heat shock. Here we describe experiments that provide insights into the mechanism by which SRp38 functions in splicing repression. We first show that SRp38 redistributes and colocalizes with snRNPs, but not with a typical SR protein, SC35, during mitosis and following heat shock. Supporting the functional significance of this association, a micrococcal nuclease-sensitive component, i.e., an snRNP(s), completely rescued heat shock-induced splicing repression in vitro, and purified U1 snRNP did so partially. SRp38 contains an N-terminal RNA binding domain (RBD) and a C-terminal RS domain composed of two subdomains (RS1 and RS2 domains). Unexpectedly, an RS1 deletion mutant derivative specifically inhibited the second step of splicing, while an RS2 deletion mutant retained significant dephosphorylation-dependent repression activity. Using chimeric SRp38/SC35 proteins, we show that SC35-RBD/SRp38-RS can function as a general splicing activator and that the dephosphorylated version can act as a strong splicing repressor. SRp38-RBD/SC35-RS, however, was essentially inactive in these assays. Together, our results help to define the unusual features of SRp38 that distinguish it from other SR proteins.
Splicing of mRNA precursors (pre-mRNA) is an essential step in gene expression in eukaryotic organisms. A large fraction (40 to 60%) of human genes are now suspected to be subject to alternative splicing, highlighting the importance of splicing as a regulatory mechanism (19, 21, 34, 36). Splicing of pre-mRNA occurs in the spliceosome, which is formed by the assembly onto the pre-mRNA of five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4/U6, and U5) and many non-snRNP proteins (reviewed in references 4 and 20). Many of these factors play an important role in the recognition of 5′ and 3′ splice sites in pre-mRNAs. Among non-snRNP splicing factors, SR proteins play key roles not only in constitutive splicing but also in alternative splicing, frequently by functioning in a combinatorial manner with other regulatory factors (reviewed in reference 44).
SR proteins constitute a group of splicing factors that are highly conserved throughout the metazoans (reviewed in references 14 and 33). SR proteins contain one or two N-terminal RNP-type RNA binding domains (RBD) and a C-terminal arginine- and serine-rich domain of various lengths and compositions (RS domain). The RBDs of SR proteins are capable of sequence-specific RNA binding, while the RS domains are involved in protein-protein interactions during early spliceosome assembly and are subject to phosphorylation-dependent regulation (51, 52). Most RS domains are functionally interchangeable in vivo (49), indicating that classical SR proteins are modular splicing factors with independent activation domains.
SR proteins affect splicing both generally and in a sequence-specific manner. The general splicing activation function of typical SR proteins is mainly mediated by cooperative interactions involving RS domain-containing general splicing factors such as the U1 snRNP 70K protein (U1-70K) and U2AF35 (25, 50). Sequence-specific interactions with RNA do not seem to play a significant role in this case. On the other hand, SR proteins utilize sequence-specific activity when interacting with exonic splicing enhancers in modulating splicing of specific target transcripts (reviewed in references 2 and 48). RS domain-mediated protein-protein interactions again play a significant role in activating splicing (25, 50), but recent studies suggest that the RS domain of an SR protein bound to an exonic splicing enhancer may contact the branchpoint RNA sequence to promote prespliceosome assembly (39). In addition to their activity in splicing, SR proteins have also been shown to function as adapters for nucleocytoplasmic shuttling of mRNA (18), in influencing mRNA stability (30, 54), and in the stimulation of mRNA translation (38).
Immunofluorescence and confocal studies showed that SR proteins localize in the nucleoplasm and in interchromatin granule clusters, or speckles, in interphase cells (45; reviewed in reference 28). snRNPs have been shown to localize in the nucleoplasm, Cajal bodies, and speckles (46; reviewed in references 10 and 27). SR proteins also seem to be recruited to sites of transcription, where they can participate in the splicing of nascent transcripts (8, 11, 35). During mitosis, some SR proteins localize in mitotic interchromatin granules (MIGs), structures that appear similar to the interchromatin granule clusters in interphase cells (37; reviewed in reference 28). After a short heat shock, which transiently inhibits splicing (e.g., see reference 3), the localization of SC35 speckles does not change significantly, whereas snRNPs distribute uniformly throughout the nucleoplasm (45).
Recently, we described an SR protein, SRp38, that functions as a splicing repressor when activated by dephosphorylation (41, 42). Although the domain organization of SRp38 is typical of SR proteins, SRp38 does not function as a splicing activator in standard splicing assays, and it is unclear what features of SRp38 are responsible for its unusual behavior. SRp38 is dephosphorylated and activated as a splicing repressor during mitosis and in response to heat shock (41, 42). Splicing inhibition is due, at least in part, to an interaction between dephosphorylated SRp38 (dSRp38) and U1 snRNP (and possibly other snRNPs) that interferes with the ability of the snRNP to stably recognize the 5′ splice site (41). However, a number of issues regarding SRp38 structure and function remain to be elucidated.
In this paper, we provide insights into the mechanism by which SRp38 functions as a splicing repressor. We first show that SRp38 redistributes in cells following heat shock or during mitosis, such that it colocalizes with snRNPs but not with SC35. Consistent with this, a micrococcal nuclease-sensitive component in S100 extracts very efficiently restores splicing in heat shock-inhibited nuclear extracts, and purified U1 snRNP displays partial activity. We then characterize the domains of SRp38 that are essential for splicing inhibition. Using RS domain deletion mutants and chimeric SRp38-SC35 recombinant proteins, we show that SRp38 contains two RS subdomains, one of which has a unique, second-step repression activity. However, both function together as a modular, transferable splicing repression domain and are necessary for complete dephosphorylation-activated splicing repression.
MATERIALS AND METHODS
Immunostaining and confocal microscopy.
HeLa cells were grown to 80% confluence on coverslips, with or without heat shock at 45°C for 30 min or 75 min. Coverslips with attached cells were rinsed once in phosphate-buffered saline (PBS) and incubated with 4% paraformaldehyde in PBS (pH 7.4) for 15 min at room temperature. After fixation, cells were rinsed three times for 10 min each time with PBS and permeabilized with 0.2% Triton X-100 plus 1% fetal bovine serum (FBS) in PBS for 5 min on ice. Cells were subsequently rinsed three times for 10 min each time in PBS-1% FBS. Fixed cells were incubated with the indicated concentration of the appropriate primary antibody diluted in PBS-1% FBS for 1 h at room temperature. The following antibodies were used: affinity-purified anti-SRp38 polyclonal antibody (1:10), Y12 ascites (against Sm B/B′) (1:20), anti-SC35 monoclonal antibody (1:100), and MAb2.73 (against U1-70K) (1:20). Subsequently, cells were washed three times for 10 min each time with PBS-1% FBS and incubated with secondary antibodies diluted in PBS-1% FBS for 1 h. Labeled goat anti-rabbit immunoglobulin G and goat anti-mouse immunoglobulin G antibodies (Molecular Probes) were used. Finally, cells were washed four times for 10 min each time with PBS and rinsed in water. Coverslips were mounted on 1 drop of mounting medium (Polysciences Inc.).
Micrococcal nuclease treatment.
Increasing amounts of micrococcal nuclease (USB), as indicated in the legend to Fig. 3, were added to cytoplasmic S100 extracts for 30 min at 30°C in the presence of 1 mM CaCl2 with or without 2 mM EGTA. Reactions were stopped by the addition of EGTA to 2 mM.
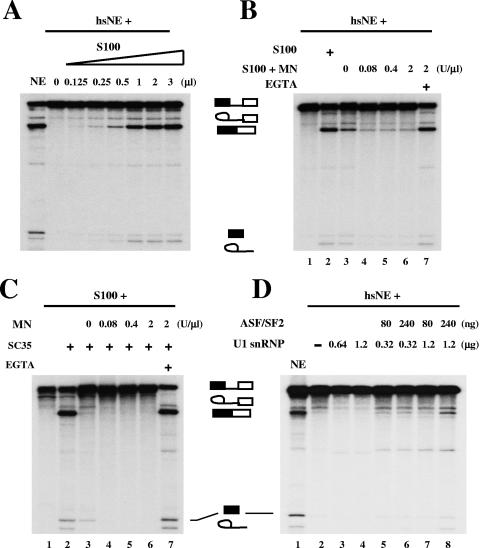
FIG. 3.
Rescue of heat shock-induced splicing inhibition by snRNPs. (A) S100 extract can rescue heat shock-induced splicing repression. Increasing amounts of S100 extract were added to splicing reactions containing β-globin pre-mRNA and hsNE. (B) MN-sensitive components in S100 extract can rescue splicing inhibition induced by heat shock. Lane 1, β-globin pre-mRNA splicing in hsNE; lane 2, splicing in hsNE complemented with 1 μl of S100 extract; lanes 3 to 6, complementation of hsNE with pretreated S100 extract with the indicated amounts (units/microliter of S100 extract) of MN; lane 7, hsNE plus MN (2 U/μl)-pretreated S100 extract in the presence of EGTA. (C) snRNPs are defective in MN-pretreated S100 extract. The S100 extracts described for panel B were tested in complementation assays with recombinant SC35 protein. Lane 1, S100 extract alone; lane 2, S100 extract complemented with SC35; lanes 3 to 6, complementation assays of MN-pretreated S100 extracts from panel B with SC35; lane 7, MN-pretreated S100 extract in the presence of EGTA and SC35. (D) U1 snRNP and ASF/SF2 partially rescue heat shock-induced splicing inhibition. Lane 2, splicing of β-globin pre-mRNA in hsNE; lanes 3 and 4, splicing in hsNE complemented with purified U1 snRNP; lanes 5 to 8, splicing in hsNE complemented with purified U1 snRNP and baculovirus-produced recombinant ASF/SF2 protein. Splicing in normal NE was used as a control (lane 1).
Recombinant proteins.
His-tagged SC35, SRp38ΔRS1, SRp38ΔRS2, SC35-RBD/SRp38-RS, and SRp38-RBD/SC35-RS proteins were prepared from recombinant baculovirus-infected Sf9 cells. His-tagged recombinant proteins were purified under denaturing conditions (6 M guanidine-HCl or 8 M urea) by Ni2+-agarose chromatography and renatured by dialysis against buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 20% glycerol, and 0.5 mM dithiothreitol) containing decreasing concentrations of urea, followed by a final dialysis step in buffer D alone. Dephosphorylation of the recombinant proteins was performed as previously described (42). The purity and concentration of proteins were determined by Coomassie blue staining of sodium dodecyl sulfate (SDS) gels. Detailed information concerning plasmid constructs is available upon request.
In vitro splicing assays.
Nuclear extracts (NE) from heat-shocked HeLa cells (hsNE), as shown in Fig. 3, were prepared as described previously (41). In vitro splicing reactions were performed as described previously (47) in either 12.5- or 25-μl reaction mixtures, which contained 12% NE, 15% hsNE, or 32% S100 extract supplemented or not with purified proteins as described in the figure legends. The final concentrations of buffer components were 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 0.12 mM EDTA, 0.3 mM dithiothreitol, 12% glycerol, 2.5 mM MgCl2, 20 mM creatine phosphate (di-Tris), 2 mM ATP, 2.4% polyvinyl alcohol, and 0.5 U RNasin (Promega). For in vitro spliceosome assembly assays, polyvinyl alcohol was replaced with glycerol, and heparin was added prior to loading of the samples, as described previously (16). The quantitation shown in Fig. 4F was performed using a phosphorimager analyzer (FUJIFILM BAS-2500), and the production rate (slope) was calculated from the raw data for the given time periods.
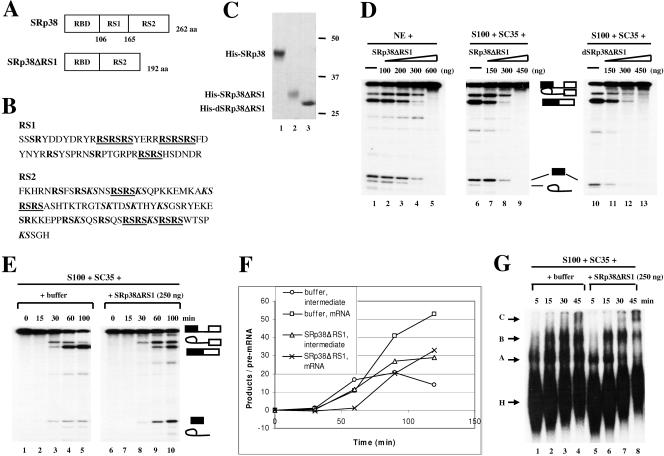
FIG. 4.
Characterization of an internal RS1 domain deletion mutant of SRp38. (A) Diagrams of SRp38 and an internal RS1 domain deletion mutant of SRp38 (SRp38ΔRS1). (B) Sequences of RS1 and RS2 subdomains of SRp38. RS dipeptides are shown in bold, and RS tetra- or hexapeptides are shown in bold and underlined. KS dipeptides are shown in bold italics. (C) Coomassie blue-stained SDS gel of the baculovirus-produced recombinant His-tagged proteins diagrammed in panel A. Dephosphorylated SRp38ΔRS1 (dSRp38ΔRS1, lane 3) was prepared from phosphorylated SRp38ΔRS1 (lane 2) by treatment with CIP and repurification. (D) SRp38ΔRS1 inhibits the second step of splicing. Increasing amounts of SRp38ΔRS1 (lanes 2 to 5 and lanes 7 to 9) or dSRp38ΔRS1 (lanes 11 to 13) were added to in vitro splicing reaction mixtures containing β-globin pre-mRNA and nuclear extract (NE) or S100 extract plus SC35. (E) Time course of β-globin pre-mRNA splicing in S100 extract complemented with 250 ng of SC35 in the presence (lanes 6 to 10) or absence (lanes 1 to 5) of 250 ng of SRp38ΔRS1. (F) Quantitative analysis of β-globin pre-mRNA spliced product (mRNA) and splicing intermediate (lariat/exon2) shown in panel E. (G) SRp38ΔRS1 does not affect spliceosome assembly. Spliceosome assembly was performed with (lanes 5 to 8) or without (lanes 1 to 4) SRp38ΔRS1. Spliceosome complexes were analyzed by 4% native polyacrylamide gel electrophoresis and autoradiography.
RESULTS
SRp38 colocalizes with snRNPs during mitosis and in response to heat shock.
Dephosphorylated SRp38 has been shown to repress splicing in mitotic extracts prepared from nocodazole-arrested HeLa cells (42) and in heat shock-treated cell extracts (41). In addition, dSRp38 interacts strongly with U1-70K in glutathione S-transferase “pull-down” assays (41). To characterize further the physiological relevance of these in vitro results, we analyzed the subcellular localization of SRp38 with other splicing factors in HeLa cells using indirect immunofluorescence analysis and confocal microscopy. HeLa cells were stained with an affinity-purified anti-SRp38 polyclonal antibody (green labeling), a Y12 monoclonal antibody (which recognizes snRNPs; red labeling), or an SC35 monoclonal antibody (red labeling). HeLa cells in interphase of the cell cycle are shown in the upper two rows of Fig. 1A and B (two asynchronous cells in panels a to d and one enlarged single cell in panels e to g), and cells in mitosis are shown in the lower two rows (Fig. 1A and B; two mitotic cells in panels h to k and one enlarged dividing cell in panels l to n). Phase-contrast images are shown in the left panels. SRp38 displayed weak, diffuse staining throughout the nucleus but localized predominantly in relatively small perinucleolar foci (typically two per nucleus, shown as arrowheads in Fig. 1A, panels b and e) (see Discussion). snRNPs stained by the Y12 antibody were found in the nucleoplasm, in speckles, and in two large dots corresponding to Cajal bodies (shown as arrowheads in Fig. 1A, panels c and f) (reviewed in reference 28). Significantly, while the SRp38 foci did not colocalize with the snRNPs in interphase (Fig. 1A, panels a to g), they redistributed and colocalized with snRNPs during mitosis (shown by arrows in Fig. 1A, panels h to n). These findings provide in vivo support for our biochemical studies showing that the inhibition of splicing during mitosis is due to an interaction of dSRp38 and U1 snRNP (and possibly other snRNPs) that interferes with the ability of the snRNP to stably recognize the 5′ splice site (41; reviewed in reference 43).
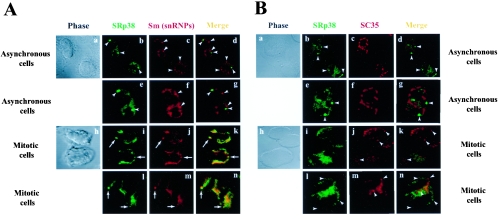
FIG. 1.
Colocalization of SRp38 with snRNPs during mitosis. (A) SRp38 colocalizes with snRNPs during mitosis but not during interphase of the cell cycle. HeLa cells were fixed in 4% paraformaldehyde and immunostained with an affinity-purified SRp38 polyclonal antibody (green) and the Y12 monoclonal antibody (red). Confocal optical sections of double-labeled cells were analyzed. In the left panels, a and h, phase-contrast images of the cells are shown; in the right panels, d, g, k, and n, merged images of the two antibody-stained fields are shown. Arrows indicate dots that colocalize, and arrowheads indicate dots that do not colocalize. (B) SRp38 does not colocalize with SC35 either in interphase or during mitosis. In the left panels, a and h, phase-contrast images of the cells are shown; in the right panels, d, g, k, and n, merged images of the two antibody-stained fields are shown. Arrowheads indicate dots that are not colocalized.
Since SRp38 has a primary structure typical of other SR proteins, we next examined the subcellular localization of SRp38 relative to the SR protein SC35. SC35 displays a well-characterized speckled pattern in nuclei of interphase cells (e.g., see Fig. 1B) and MIG granules in mitotic cells (37; reviewed in reference 28). Importantly, SRp38 dots did not colocalize with SC35 speckles, either in interphase (Fig. 1B, panels a to g) or during mitosis (Fig. 1B, panels h to n), as indicated by arrowheads in the figure. The MIG granules containing SC35 are distinct from the SRp38 dots, supporting the idea of a distinct role for SRp38 in splicing, compared with other characterized SR proteins.
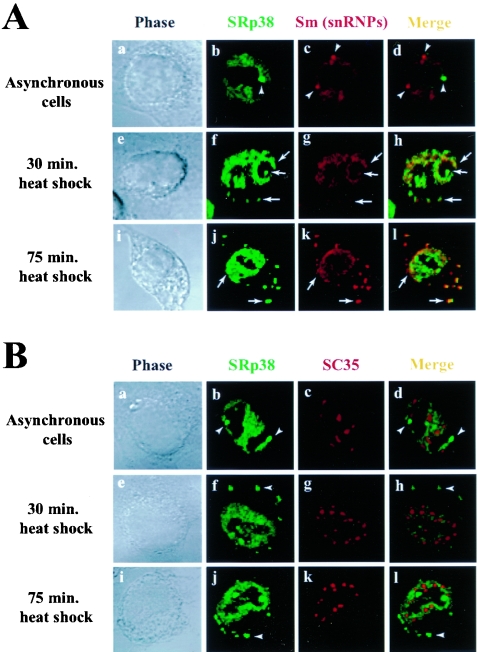
We next examined the subcellular localization of SRp38 and other splicing factors following heat shock. We incubated HeLa cells at 45°C for 30 or 75 min, stained the cells with the antibodies used above, and examined the distribution of the proteins by confocal microscopy (Fig. 2). As shown above, SRp38 did not colocalize with snRNPs in the control cells (Fig. 2A, panels a to d). However, as in M phase, SRp38 and snRNPs were observed to colocalize after 30 min of heat treatment (Fig. 2A, panels e to h). Interestingly, granules with both SRp38 and snRNPs appeared in the cytoplasm after 30 min and became larger and more numerous after 75 min (Fig. 2A, panels i to l). As a reflection of this increase in the number and size of granules, we observed a stronger colocalization of SRp38 and snRNPs. The identity of the granules is currently unknown, but they may be related to stress granules (SGs) (23) or cytoplasmic processing bodies (P bodies) (40). Confirming these results, confocal studies with an anti-U1-70K monoclonal antibody (MAb2.73) (1) indicated a colocalization of U1 snRNP with SRp38 and their accumulation in cytoplasmic granules after heat treatment (data not shown). As in mitotic cells, SRp38 did not colocalize significantly with SC35 under any of the heat shock conditions analyzed (Fig. 2B). This is consistent with our biochemical results showing that SC35 could not restore in vitro splicing inhibited by dSRp38 in mitotic extracts or in nuclear extracts from heat-shocked cells (41, 42).
FIG. 2.
Colocalization of SRp38 with snRNPs in response to heat shock. (A) SRp38 colocalizes with snRNPs upon heat shock. Asynchronously grown HeLa cells were incubated at 45°C for the indicated times (e to h, 30 min; i to l, 75 min) and were fixed in 4% paraformaldehyde. Fixed cells were immunostained with an affinity-purified SRp38 polyclonal antibody (green) and the Y12 monoclonal antibody (red). In the left panels, a, e, and i, phase-contrast images of the cells are shown; in the right panels, d, h, and l, merged images of the two antibody-stained fields are shown. Arrows indicate dots that colocalize, and arrowheads indicate dots that do not colocalize. (B) SRp38 does not colocalize with SC35 upon heat shock. In the left panels, a, e, and i, phase-contrast images of the cells are shown; in the right panels, d, h, and l, merged images of the two antibody-stained fields are shown. Arrowheads indicate dots that are not colocalized.
Together, these results indicate that SRp38 colocalizes with U1 snRNP during mitosis and in response to heat shock, cellular conditions where splicing is generally repressed by dSRp38. These data support the idea that dSRp38 represses splicing through interactions with U1 and possibly other snRNPs (41).
Heat shock-induced splicing inhibition can be rescued by snRNPs.
To better understand the mechanism(s) involved in the inhibition of splicing induced by dSRp38, we next wished to test whether heat shock-induced splicing repression could be rescued by complementation with other splicing factors. We first attempted to rescue the inhibition brought about by dSRp38 or inactive nuclear extracts from heat-shocked HeLa cells (hsNE) by the addition of recombinant SR proteins, but without success (results not shown; also see reference 42). However, the addition of small amounts of S100 extract, which by itself is inactive in splicing but can be complemented by the addition of SR proteins, efficiently restored splicing in hsNE (Fig. 3A). This implies that a factor(s) in S100 extract might be the target of splicing repression by dSRp38.
The localization data described above and our previous biochemical experiments (41) suggest that the rescuing factor might be an snRNP. To test this hypothesis, S100 extract was pretreated with different concentrations of micrococcal nuclease (MN) and then added to splicing reactions with hsNE (Fig. 3B). S100 extract pretreated with MN did not restore splicing (lanes 4 to 6), while S100 pretreated with MN in the presence of EGTA was fully active in rescuing splicing (lane 7). As a control, SC35 could not reconstitute splicing activity in the S100 extract pretreated with MN, although the S100 extract pretreated with MN plus EGTA was activated by SC35 (Fig. 3C). Together, these results suggest that a component(s) containing RNA (i.e., an snRNP) becomes limiting for splicing in extracts from heat-shocked cells.
We previously showed that dSRp38 interacts with U1 snRNP and that this interaction interferes with ASF/SF2-stabilized 5′ splice-site recognition by U1 snRNP (41). To examine the functional significance of the interaction and at the same time to test whether U1 snRNP might be able to rescue splicing in hsNE, we determined the effect of purified U1 snRNP, in the presence or absence of ASF/SF2, on splicing in hsNE (Fig. 3D). U1 snRNP (lanes 3 and 4) and ASF/SF2 (data not shown) alone were unable to rescue splicing, nor were combinations of 80 and 240 ng of ASF/SF2 with 0.32 μg U1 snRNP (lanes 5 and 6). However, the addition of 240 ng of ASF/SF2 with 1.2 μg of U1 snRNP resulted in significant levels of splicing (lane 8). These results indicate that exogenous U1 snRNP plus ASF/SF2 can partially rescue heat shock-induced splicing repression, and they support the functional significance of the previously described interactions of both ASF/SF2 (25) and dSRp38 (41) with U1 snRNP. We suspect that rescue was incomplete because the specific activity of the U1 snRNP preparation was low and/or additional factors (e.g., other snRNPs) are inhibited by dSRp38.
Characterization of an internal RS1 domain deletion mutant of SRp38.
SRp38 has a primary structure typical of SR proteins, containing an N-terminal RBD and a C-terminal RS domain. However, the RS domain is distinctive, as it is composed of two subdomains (RS1 and RS2 domains) (Fig. 4A and B). While classical SR proteins typically contain RS domains with a continuous stretch(s) of alternating arginine and serine residues (RS repeats), the RS2 domain contains fewer consecutive RS dipeptides but has a large number of KS dipeptides (Fig. 4B). To characterize the function of the RS2 domain in splicing and to help define the region of SRp38 responsible for repression activity, we purified a baculovirus-produced His-tagged RS1 domain deletion protein (SRp38ΔRS1; Fig. 4C, lane 2) and employed it in splicing assays with β-globin pre-mRNA. Like full-length SRp38 (42), the recombinant deleted protein did not activate splicing in S100 complementation assays. The addition of larger amounts of the protein inhibited in vitro splicing in NE (Fig. 4D, lanes 2 to 5) and in S100 extract complemented with SC35 (Fig. 4D, lanes 7 to 9). However, the inhibition pattern displayed was entirely different from that of the full-length protein. While full-length SRp38 resulted in a decreased accumulation of both first-step intermediates and second-step products, likely reflecting competitive inhibition of splicing activators binding to the pre-mRNA (42), SRp38ΔRS1 resulted in only a modest decrease in first-step intermediates but a drastic decrease in second-step products. To investigate this apparent second-step inhibition more closely, we employed a time course experiment (Fig. 4E). Compared with the case for the control S100 extract supplemented with SC35 alone (lanes 1 to 5), second-step splicing products diminished in the presence of 250 ng of SRp38ΔRS1, while first-step intermediates accumulated (lanes 6 to 10), consistent with second-step inhibition. Quantitation (Fig. 4F) revealed that reactions with SRp38ΔRS1 displayed a reduction in the second-step mRNA production rate (for example, a nearly twofold reduction between 30 and 60 min) and an increase in first-step intermediate accumulation (fourfold increase between 30 and 60 min) compared to reactions without SRp38ΔRS1.
Both catalytic steps of splicing take place after formation of the spliceosome. If indeed SRp38ΔRS1 preferentially inhibits the second step of splicing, then spliceosome assembly should not be affected by SRp38ΔRS1. To examine if this is the case, spliceosome assembly assays were employed (Fig. 4G). Compared with the control (S100 plus SC35; lanes 1 to 4), the formation of spliceosomal A, B, and C complexes was not affected in the presence of 250 ng of SRp38ΔRS1 (lanes 5 to 8). These results together indicate that SRp38ΔRS1 preferentially represses the second step of splicing.
We next wished to determine whether SRp38ΔRS1 was able to bring about the potent repression observed with full-length dephosphorylated SRp38. To this end, we dephosphorylated SRp38ΔRS1 by treating the protein (Fig. 4C, lane 2) with calf intestinal phosphatase (CIP) and repurified it to generate dSRp38ΔRS1 (lane 3). When the activity of dSRp38ΔRS1 was measured in S100 extract supplemented with SC35, the protein was found not only to be incapable of the strong repression detected with the full-length protein but also to be less active in the second-step repression described above (Fig. 4D). These experiments indicate that SRp38 lacking subdomain RS1 preferentially represses the second step of splicing and is incapable of strong, dephosphorylation-dependent splicing repression.
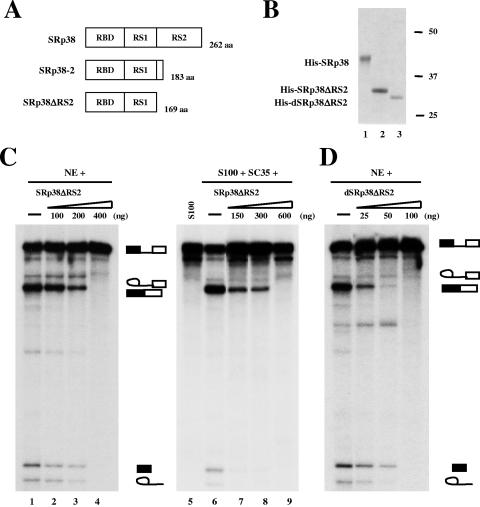
Characterization of an RS2 domain deletion mutant of SRp38.
We next wished to investigate the function of the RS1 subdomain. This region, while shorter than RS2, more closely resembles a typical RS domain containing a high density of RS repeats (Fig. 4B). We constructed a C-terminal RS2 domain deletion mutant of SRp38 (SRp38ΔRS2) and purified the protein from recombinant baculovirus-infected cells (Fig. 5A). SRp38ΔRS2 was unable to activate splicing of β-globin pre-mRNA in S100 extract (data not shown), indicating that the protein lacks the typical activating function of standard SR proteins. However, large amounts of the protein (200 to 400 ng) inhibited splicing in NE (Fig. 5C, lanes 2 to 4) and in S100 extract supplemented with SC35 (Fig. 5C, lanes 7 to 9). This behavior is similar to that observed with full-length, phosphorylated SRp38 (42).
FIG. 5.
Characterization of a C-terminal RS2 domain deletion mutant of SRp38. (A) Diagrams of SRp38, SRp38-2, and a C-terminal RS2 domain deletion mutant of SRp38 (SRp38ΔRS2). (B) Coomassie blue-stained SDS gel of the baculovirus-produced recombinant His-tagged proteins diagrammed in panel A. Dephosphorylated SRp38ΔRS2 (dSRp38ΔRS2, lane 3) was prepared from phosphorylated SRp38ΔRS2 (lane 2) by treatment with CIP and repurification. (C) SRp38ΔRS2 inhibits splicing at high concentrations in nuclear extract (NE; lanes 2 to 4) or S100 extract complemented with recombinant SC35 (lanes 7 to 9). (D) dSRp38ΔRS2 inhibits β-globin pre-mRNA splicing. Twenty-five to 100 ng of dSRp38ΔRS2 was added to splicing reactions with NE.
To analyze the effect of dephosphorylation on SRp38ΔRS2 activity, dephosphorylated SRp38ΔRS2 (dSRp38ΔRS2) protein was prepared by an identical method to that used with SRp38ΔRS1 (Fig. 5B, lane 3). When increasing amounts of dSRp38ΔRS2 were added to splicing reactions with NE (Fig. 5D), a strong splicing inhibition was observed. Significant repression was detected with 25 ng of dSRp38ΔRS2, and <100 ng was required for full repression, an amount substantially lower than that observed with either phosphorylated SRp38ΔRS2 (400 ng; Fig. 5C) or dSRp38ΔRS1 (450 ng; Fig. 4D). However, the protein was not as active as dSRp38 in repressing splicing, where 10 ng or less of protein was found to be sufficient for complete repression (42). These results indicate that the RS1 subdomain, despite its similarity to typical activating RS domains, displays significant dephosphorylation-dependent splicing inhibitory activity.
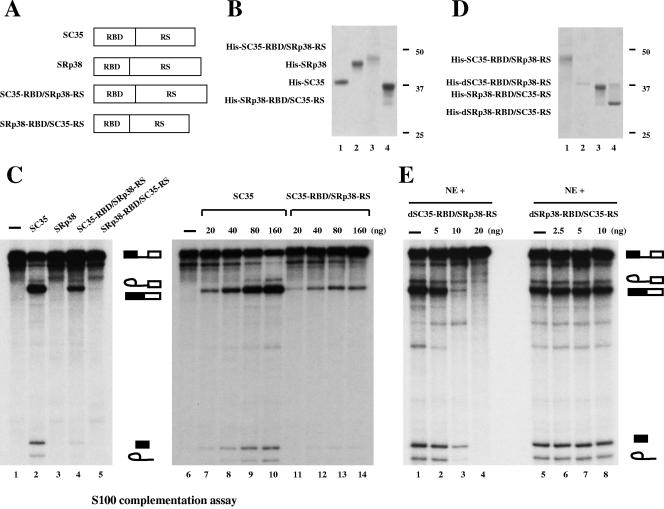
Properties of SRp38-SC35 chimeric proteins.
To investigate further the domain(s) of SRp38 responsible for the protein's potent repression activity, we constructed chimeric proteins in which the RBD and the RS domain of SRp38 were swapped with those of SC35 (Fig. 6A). Baculovirus-produced recombinant His-tagged proteins were purified (Fig. 6B), and the splicing activities of the chimeric proteins were analyzed by S100 complementation assays (Fig. 6C). Surprisingly, the fusion protein consisting of the RBD of SC35 and the RS domain of SRp38 (SC35-RBD/SRp38-RS; Fig. 6B, lane 3) activated in vitro splicing of β-globin pre-mRNA, whereas the protein with the RBD of SRp38 and the RS domain of SC35 (SRp38-RBD/SC35-RS; Fig. 6B, lane 4) did not (Fig. 6C, lanes 4 and 5, respectively). SRp38-RBD/SC35-RS was not totally inactive, as it can activate splicing of a β-globin-derived RNA that contains high-affinity SRp38 binding sites (Y. Feng, C. Shin and J. L. Manley, unpublished data). Complementation assays with SC35 or SRp38 (Fig. 6B, lanes 1 and 2, respectively) are shown as controls (Fig. 6C, lanes 2 and 3). To examine the activity of SC35-RBD/SRp38-RS more closely, increasing amounts of the protein were added to S100 extract (lanes 11 to 14), and the splicing activity of the fusion protein was compared with that of SC35 (lanes 7 to 10). Relative to SC35, SC35-RBD/SRp38-RS weakly activated splicing; the maximum splicing activity observed with the chimera was ∼20% that of SC35. Nonetheless, these results indicate that the SRp38 RS domain is capable of functioning positively in the appropriate context. In contrast, the SRp38 RBD was unable to activate splicing, even with the SC35 RS domain. Together, our results suggest that the SRp38 RBD has unusual properties that prevent it from functioning as a general splicing activator, regardless of the nature of the RS domain.
FIG. 6.
Functional domains of SRp38. (A) Diagrams of SC35, SRp38, and domain-swap mutants of these two proteins (RBD of SC35 and RS domain of SRp38 [SC35-RBD/SRp38-RS] and RBD of SRp38 and RS domain of SC35 [SRp38-RBD/SC35-RS]). (B) Coomassie blue-stained SDS gel of the baculovirus-produced recombinant His-tagged proteins diagrammed in panel A. Lane 1, SC35; lane 2, SRp38; lane 3, SC35-RBD/SRp38-RS; lane 4, SRp38-RBD/SC35-RS. (C) SC35-RBD/SRp38-RS weakly activates in vitro splicing of β-globin pre-mRNA splicing in S100 extract. One hundred nanograms of recombinant protein was added to the S100 assay mixtures in lanes 2 to 5, and 20 to 160 ng of protein was used for the assays shown in lanes 7 to 14. (D) Dephosphorylated proteins used for panel E. Dephosphorylated SC35-RBD/SRp38-RS protein (dSC35-RBD/SRp38-RS, lane 2) was prepared from phosphorylated SC35-RBD/SRp38-RS (lane 1) by treatment with CIP and repurification. Dephosphorylated SRp38-RBD/SC35-RS (dSRp38-RBD/SC35-RS, lane 4) was prepared from SRp38-RBD/SC35-RS (lane 3) in the same way as dSC35-RBD/SRp38-RS. (E) The RS domain of SRp38 functions as a modular repression domain. Five to 20 ng of dSC35-RBD/SRp38-RS (lanes 2 to 4) or 2.5 to 10 ng of dSRp38-RBD/SC35-RS (lanes 6 to 8) was added to in vitro splicing reaction mixtures containing β-globin pre-mRNA with nuclear extract (NE).
We next wished to determine whether either of the chimeric proteins was able to carry out the potent dephosphorylation-dependent repression that characterizes SRp38. To test this, dephosphorylated versions of the two fusion proteins (dSC35-RBD/SRp38-RS and dSRp38-RBD/SC35-RS; Fig. 6D, lanes 2 and 4, respectively) were prepared by CIP treatment and repurification of SC35-RBD/SRp38-RS (lane 1) and SRp38-RBD/SC35-RS (lane 3). Significantly, 5 ng of dSC35-RBD/SRp38-RS weakly inhibited splicing, 10 ng resulted in strong inhibition, and ∼20 ng gave rise to a complete inhibition of splicing (Fig. 6E, lanes 2 to 4). This profile is very similar to that of dSRp38 itself (42). The addition of dephosphorylated SC35 to the nuclear extract had no effect on splicing (42). On the other hand, the addition of similar concentrations of dSRp38-RBD/SC35-RS did not affect splicing (Fig. 6E, lanes 6 to 8). These results indicate that the SRp38 RS domain is responsible for dephosphorylation-dependent splicing repression and can function as a modular, transferable repression domain.
DISCUSSION
SRp38 is an unusual SR protein. Despite its similarity in primary sequence to other SR proteins, the principal function of SRp38 is to repress, rather than to activate, splicing. In this paper, we have described several additional properties of SRp38 that provide insight into its unusual behavior. We first showed that SRp38 localizes in cells in a distinctive pattern that does not normally coincide either with that of the typical SR protein SC35 or with snRNPs. However, SRp38 and snRNPs do colocalize during mitosis and following heat shock, and our biochemical data, presented both here and previously (41), support the functional significance of this colocalization. We also described a functional analysis of the SRp38 RBD and RS domain, which revealed that both have unusual properties that contribute to the protein's unique behavior. Below, we discuss these findings from two perspectives, first by showing how they provide mechanistic insight into SRp38 function, and second by showing how they help to explain why SRp38 functions differently from the other SR proteins.
SRp38 displays a distinctive localization pattern in normally growing interphase cells. Rather than the well-documented speckles characteristic of typical SR proteins (reviewed in reference 28), SRp38 localizes predominantly in a diffuse granular pattern throughout the nucleus, although significantly, it also accumulates in a small number (typically two per nucleus) of dots that are invariably localized adjacent to nucleoli. Several perinucleolar structures have been described previously, notably the perinucleolar compartment and Sam68 nuclear bodies (SNBs) (reviewed in reference 17). Intriguingly, both of these structures have been shown to contain splicing factors. These include the splicing repressor PTB in the case of perinucleolar compartments (13) and the regulator Sam68 in the case of SNBs (6). SNBs also contain another factor, HAP/SAF-B, that has been implicated in transcription and possibly splicing (reference 9 and references therein). Two facts in addition to their similar localization are consistent with a possible link between SNBs and SRp38. First, both SRp38 and HAP/SAF-B interact with the splicing regulator Tra2 (C. Shin and J. L. Manley, unpublished data). Second, SNBs seem to change their properties upon heat shock, such that Sam68 and HAP/SAF-B relocalize to distinct, albeit related, structures (9). Although the pattern does not resemble that which we described here for SRp38, it is intriguing that these nucleolus-associated factors show related changes following heat shock. The significance of these similarities, if any, will require further study.
The redistribution of SRp38 and snRNPs to sites in the cytoplasm as well as the nucleus following heat shock is intriguing. A possible explanation for the cytoplasmic granules is that dSRp38 sequesters snRNPs and brings them to transient sites used during recovery from heat shock. In fact, as cells recover from heat shock, these granules appear to decrease or disappear, which correlates with the rephosphorylation of SRp38 and the restoration of splicing (unpublished data). In another scenario, recent studies have shown that shuttling SR proteins can stimulate translation (38), raising the possibility that SRp38 might be involved in translational regulation following heat shock. Indeed, in response to environmental stresses, including heat shock, untranslated mRNAs accumulate in SGs, which contain TIA-1 and TIA-1-related protein, factors implicated in both splicing and translation control, poly(A) binding protein 1, and certain components of the translational preinitiation complex (22, 23). If the SRp38/snRNP-containing structures correspond to SGs, then SRp38 could function in the repression of translation after heat shock. In keeping with this, recent findings that SRp38 can inhibit neuronal differentiation and interact with 28S rRNA suggest a possible role of the protein in translational silencing in undifferentiated neural cells (32). On the other hand, cytoplasmic P bodies contain enzymes involved in mRNA degradation, such as decapping enzymes and a 5′ to 3′ mRNA exonuclease (29, 40). As with the perinucleolar bodies described above, future studies examining the identity and significance of the SRp38/snRNP cytoplasmic bodies should be informative. However, in any case, our results have shown that SRp38 localizes in a pattern distinct from those of other SR proteins and colocalizes with snRNPs during mitosis or following heat shock.
It was surprising that the SRp38 mutant containing only the RS1 subdomain retained a significant fraction of the dephosphorylation-dependent repression activity. This region is relatively small (∼60 residues) and bears the greatest similarity to typical RS domains. The “strength” of RS domains as activators reflects the number of RS dipeptides and was found to correlate best with the number of RS tetrapeptides (15). Thus, it might be expected that RS1 (two hexapeptides, one tetrapeptide, and several RS dipeptides) would behave as a modest activation domain. We suspect its failure to do so reflects an interaction with the SRp38 RBD, which may have unusual properties (see below). Consistent with this, the entire SRp38 RS domain does display a weak activation activity when fused to the SC35 RBD. We also noted that the RS2 deletion mutant very closely resembles the variant arising from alternative splicing, SRp38-2 (26, 42, 53). We were unable to make recombinant SRp38-2, especially the dephosphorylated version, in a soluble form (unpublished data), but our data thus suggest that SRp38-2 behaves similarly to SRp38, except with weaker activity. Transient transfection experiments have suggested that both proteins can influence the alternative splicing of reporter transcripts, although with somewhat different specificities (26, 53). Whether this reflects the relative activities of SRp38 and SRp38ΔRS2 is not clear.
The RS2 subdomain also displays unusual properties. Although it is significantly longer than RS1 (∼90 residues) and contains four RS tetrapeptides, it displays no activation or dephosphorylation-dependent repression activity. Instead, it produces a novel second-step repression. We have no data indicating what is responsible for this, but we speculate that it may be the unusual abundance of SK dipeptides. SK dipeptides are not typically found in RS domains, and the conversion of RS dipeptides to KS inactivated the SR protein ASF/SF2 in assays conducted in vitro (5). Additionally, an SR-related protein, SRrp86, contains a KE-rich domain that represses splicing (31) and which could resemble a phosphorylated KS-rich region. Phosphorylation indeed enhanced second-step repression, and this may reflect the phosphorylation of KS serines. The SR protein kinase Clk/Sty has been shown to be capable of phosphorylating KS-containing peptides (7). The rationale for inhibiting splicing at the second step and how it might occur are not clear. However, it is well known that transcription can be regulated, including regulation by repression, at multiple different steps, in some instances with a specific repressor protein, for example, the Rb tumor suppressor, functioning at multiple points (reviewed in reference 12). This is likely to facilitate the most efficient possible repression, and it could be that this is the case with SRp38 and splicing as well. If so, then an intriguing issue is how differential states of phosphorylation might be controlled. For example, might the phosphorylation status of RS and KS dipeptides be maintained by different kinases and/or phosphatases? During heat shock, we observed a distinct dephosphorylation intermediate (41), and it will be interesting to determine whether this reflects differential phosphorylation of RS versus KS serine residues.
The SRp38 RBD is also atypical and likely contributes to the unusual properties of SRp38. It is unable to provide a general activation function, even in the context of a positive-acting RS domain, and also prevents the SRp38 RS domain from displaying the positive activity that it can have when fused to the SC35 RBD. This is surprising, on the one hand, as the SRp38 RBD displays significant similarity to several SR protein RBDs and is in fact most similar to the SC35 RBD (46% identity). However, the SRp38 RBD displays another feature that distinguishes it from those of other SR proteins, which is that it shares similarity with a group of RBDs that constitute a newly defined U2AF homology motif (UHM) family (24). This grouping of RBDs is based on sequence similarities with RBDs from the splicing factor U2AF, specifically the third RBD of the U2AF65 subunit and the sole RBD of U2AF35. These two RBDs have unusual properties and may function primarily in protein-protein rather than protein-RNA interactions. Although SRp38 shares only limited similarity with this group of RBDs and although we have shown that the SRp38 RBD does possess a high-affinity sequence-specific RNA binding activity (42; Y. Feng and J. L. Manley, unpublished data), SRp38 is the only SR protein to display an identifiable similarity with UHM proteins (24). Although the significance of this similarity will require further study, it may contribute to the unusual properties of SRp38 that we have described here and previously.
Acknowledgments
We thank Y. Shi, S. Millhouse, Y. Feng, and S. Kaneko for materials and discussions, Viji M. Draviam for comments on the manuscript, A. Norris, A. Doty, and K. Schwartz for technical assistance, and I. Boluk for help preparing the manuscript. We also thank R. Luhrmann and B. Kastner for purified U1 snRNP, X.-D. Fu for the SC35 monoclonal antibody, J. A. Steitz and Y. Huang for the Y12 monoclonal antibody, and W. J. van Venrooij and S. O. Hoch for MAb2.73.
This work was supported by NIH grant R37 GM48259.
REFERENCES
- 1.Billings, P. B., R. W. Allen, F. C. Jensen, and S. O. Hoch. 1982. Anti-RNP monoclonal antibodies derived from a mouse strain with lupus-like autoimmunity. J. Immunol. 128:1176-1180. [PubMed] [Google Scholar]
- 2.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 3.Bond, U. 1988. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 7:3509-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Caceres, J. F., and A. R. Krainer. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12:4715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T., F. M. Boisvert, D. P. Bazett-Jones, and S. Richard. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Cell. Biol. 10:3015-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colwill, K., L. L. Feng, J. M. Yeakley, G. D. Gish, J. F. Caceres, T. Pawson, and X. D. Fu. 1996. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271:24569-24575. [DOI] [PubMed] [Google Scholar]
- 8.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA Pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 9.Denegri, M., I. Chiodi, M. Corioni, F. Cobianchi, S. Riva, and G. Biamonti. 2001. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Cell. Biol. 12:3502-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gall, J. G. 2003. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4:975-980. [DOI] [PubMed] [Google Scholar]
- 11.Gall, J. G., M. Bellini, Z. Wu, and C. Murphy. 1999. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10:4385-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaston, K., and P. S. Jayaraman. 2003. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 60:721-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghetti, A., S. Pinol-Roma, W. M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNPs. Nucleic Acids Res. 20:3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 17:6747-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, S. 2000. Review: perinucleolar structures. J. Struct. Biol. 129:233-240. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 20.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 21.Kan, Z., E. C. Rouchka, W. R. Gish, and D. J. States. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motif: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu, M., E. Kominami, K. Arahata, and T. Tsukahara. 1999. Cloning and characterization of two neural-salient serine/arginine-rich (NSSR) proteins involved in the regulation of alternative splicing in neurones. Genes Cells 4:593-606. [DOI] [PubMed] [Google Scholar]
- 27.Lamond, A. I., and J. E. Sleeman. 2003. Nuclear substructure and dynamics. Curr. Biol. 13:R825-R828. [DOI] [PubMed] [Google Scholar]
- 28.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 29.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 30.Lemaire, R., J. Prasad, T. Kashima, J. Gustafson, J. L. Manley, and R. Lafyatis. 2002. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 16:594-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J., D. C. Barnard, and J. G. Patton. 2002. A unique glutamic acid-lysine (EK) domain acts as a splicing inhibitor. J. Biol. Chem. 277:39485-39492. [DOI] [PubMed] [Google Scholar]
- 32.Liu, K. J., and R. M. Harland. 2005. Inhibition of neurogenesis by SRp38, a neuroD-regulated RNA-binding protein. Development 132:1511-1523. [DOI] [PubMed] [Google Scholar]
- 33.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 34.Mironov, A. A., J. W. Fickett, and M. S. Gelfand. 1999. Frequent alternative splicing of human genes. Genome Res. 9:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 36.Modrek, B., A. Resch, C. Grasso, and C. Lee. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29:2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasanth, K. V., P. A. Sacco-Bubulya, S. G. Prasanth, and D. L. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 14:1043-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanford, J. R., N. K. Gray, K. Beckmann, and J. F. Caceres. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, H., J. L. Kan, and M. R. Green. 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13:367-376. [DOI] [PubMed] [Google Scholar]
- 40.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, C., Y. Feng, and J. L. Manley. 2004. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427:553-558. [DOI] [PubMed] [Google Scholar]
- 42.Shin, C., and J. L. Manley. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407-417. [DOI] [PubMed] [Google Scholar]
- 43.Shin, C., and J. L. Manley. 2004. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 5:727-738. [DOI] [PubMed] [Google Scholar]
- 44.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 45.Spector, D. L., X. D. Fu, and T. Maniatis. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10:3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spector, D. L., G. Lark, and S. Hunag. 1992. Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell 3:555-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tacke, R., and J. L. Manley. 1999. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11:358-362. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., S. H. Xiao, and J. L. Manley. 1998. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12:2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, S. H., and J. L. Manley. 1997. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11:334-344. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, S. H., and J. L. Manley. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17:6359-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, L., L. J. Embree, and D. D. Hickstein. 2000. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol. Cell. Biol. 20:3345-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Z., and A. R. Krainer. 2004. Involvement of SR proteins in mRNA surveillance. Mol. Cell 16:597-607. [DOI] [PubMed] [Google Scholar]