Abstract
The Golgi-localized, Gamma-ear-containing, Arf-binding (GGA) proteins are monomeric clathrin adaptors that mediate the sorting of transmembrane cargo at the trans-Golgi network and endosomes. Here we report that one of these proteins, GGA3, becomes transiently phosphorylated upon activation of the epidermal growth factor (EGF) receptor. This phosphorylation takes place on a previously unrecognized site in the “hinge” segment of the protein, S368, and is strictly dependent on the constitutive phosphorylation of another site, S372. The EGF-induced phosphorylation of S368 does not require internalization of the EGF receptor or association of GGA3 with membranes. This phosphorylation can be blocked by inhibitors of both the mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways that function downstream of the activated EGF receptor. Phosphorylation of GGA3 on S368 causes an increase in the hydrodynamic radius of the protein, indicating a transition to a more asymmetric shape. Mutation of S368 and S372 to a phosphomimic aspartate residue decreases the association of GGA3 with membranes. These observations indicate that EGF signaling elicits phosphorylation events that regulate the association of GGA3 with organellar membranes.
The Golgi-localized, Gamma-ear-containing, Arf-binding (GGA) proteins constitute a family of monomeric clathrin adaptors that associate with the cytosolic face of the trans-Golgi network (TGN) and endosomes (reviewed in references 6 and 17). Three GGAs (GGA1, GGA2, and GGA3) exist in humans and there are one to three in other eukaryotes. All GGAs comprise (i) a VHS domain that, in the mammalian GGAs, binds DXXLL-type sorting signals (where D is aspartate, X is any amino acid, and L is leucine) present in the cytosolic tails of various transmembrane proteins that cycle between the TGN and endosomes, (ii) a GAT domain that interacts with class I Arf GTP-binding proteins (i.e., Arf1 and Arf3 in humans; herein referred to as Arf), as well as Rabaptin-5, ubiquitin, and the tumor susceptibility gene 101 product (TSG101), (iii) a largely unstructured hinge segment that binds clathrin, and (iv) a GAE domain that binds a cohort of accessory proteins having ΨG(P/D/E)(Ψ/L/M) motifs (where Ψ is an aromatic residue, G is glycine, P is proline, E is glutamate, and M is methionine) (6, 17) (see scheme in Fig. 4A). These multiple interactions endow the GGAs with the ability to function as sorting adaptors, in much the same way as the heterotetrameric adaptor protein (AP) complexes AP-1, AP-2, and AP-3 (40).
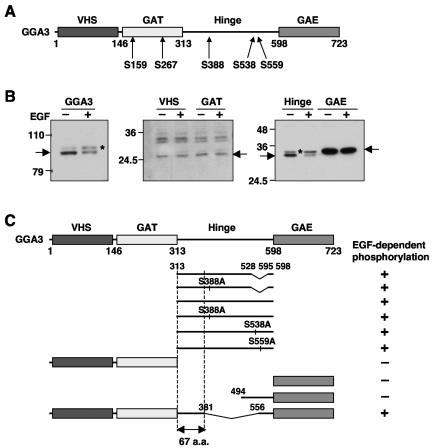
FIG. 4.
EGF induces phosphorylation of the GGA3 hinge region. (A) Schematic representation of GGA3 and location of previously reported phosphorylation sites in unstimulated cells (14, 18, 28). The different domains of GGA3 and residue numbers are indicated. (B) M1 fibroblasts transiently expressing Myc-tagged full-length GGA3, or the corresponding Myc-tagged VHS, GAT, hinge, or GAE domains were serum-starved for 5 h and incubated in the absence (−EGF) or presence (+EGF) of 10 nM EGF. Cell extracts were analyzed by SDS-PAGE and immunoblotting with antibody to the Myc epitope. Arrows indicate the positions of the different Myc-tagged proteins. Numbers on the left indicate the positions of molecular mass markers (in kilodaltons), and asterisks indicate the position of the phosphorylated species. (C) The Myc-tagged mutant forms of GGA3 represented in this panel were expressed in M1 cells an analyzed for phosphorylation as in panel B. The occurrence of an EGF-induced mobility shift is indicated by the + symbol on the right of each construct. The top two hinge constructs represent a splice variant missing residues 528 to 595. The bottom GGA3 construct has a deletion of the 381 to 556 segment. From these analyses we identified a 67-amino-acid sequence that is required for EGF-induced phosphorylation.
The GGAs are recruited from the cytosol to the TGN and endosomes by virtue of interactions with the GTP-bound form of Arf. They then promote the polymerization of clathrin onto membranes, perhaps in cooperation with other adaptors such as AP-1 and enthoprotin/epsinR. At the TGN, these clathrin “coats” enable the specific packaging of transmembrane proteins that contain DXXLL signals (such as the mannose 6-phosphate receptors [MPRs]) into clathrin-coated carriers bound for endosomes (35, 15, 38). In addition, the GGAs have been proposed to participate in the sorting of ubiquitinated transmembrane proteins from the TGN to endosomes and from early endosomes to the multivesicular body-lysosomal targeting pathway (36, 42, 44).
The spatial and temporal coordination of such a complex array of interactions likely requires tight regulatory controls. In this context, phosphorylation has been shown to regulate some aspects of GGA function. For instance, phosphorylation of serine residues adjacent to or within the DXXLL signals of the MPRs (24) and β-secretase (19, 43) increases the affinity of these signals for the VHS domain of the GGAs. The GGAs themselves are phosphorylated (14, 18, 47, 28) at multiple serine and threonine residues, including S268, T270, S355, and S480 of GGA1, and S126, S234, S355, S505, and S526 in the “short” splice form of GGA3 (they correspond to S159, S267, S388, S538, and S559 in the “long” splice form of GGA3 used in our study; see Fig. 4A) (14, 28). No tyrosine phosphorylation was detected on either GGA1 or GGA3 (28).
S355 is located just amino-terminal to an internal DXXLL sequence within the hinge segment of GGA1 and GGA3 (short splice form), and phosphorylation of this serine residue promotes an intramolecular interaction of the DXXLL sequence with the VHS domain of the same GGA molecule (14). As a consequence, these GGAs undergo large conformational changes that render them unable to bind DXXLL signals from transmembrane cargo (14, 15, 18). This has been proposed to cause dissociation of the GGAs from the TGN and the handing off of transmembrane cargo to AP-1 (15). In addition, amino acid substitutions that mimic the phosphorylation of S268 and T270 in the GAT domain of GGA1 shift the distribution of the GGAs from the TGN to peripheral puncta (28). This observation has been interpreted to mean that phosphorylation of GGA1 on S268 and T270 stabilizes its association with TGN-derived vesicles, the opposite of what has been proposed to occur upon phosphorylation of S355. Thus, the phosphorylation of different residues on the same GGA appears to exert distinct effects on the association of the GGAs with organellar membranes.
None of these phosphorylation events, however, has been shown to be modulated by external or internal stimuli. Possible regulators are signaling receptors, which are known to phosphorylate various coat proteins and to modulate trafficking (29). For example, activation of the epidermal growth factor (EGF) receptor stimulates phosphorylation of the clathrin heavy chain (48), the β2-subunit of the AP-2 adaptor complex (23), and two monomeric clathrin adaptors, Eps15 (8) and Hrs (46, 2). In all of these cases, phosphorylation occurs on tyrosine residues and is catalyzed by either the intrinsic EGF receptor tyrosine kinase or a downstream kinase such as Src (48, 23, 46, 2, 8). The EGF receptor can also lead to phosphorylation of serine and threonine residues via activation of downstream kinase cascades (49), although to date no such phosphorylation has been shown to occur for coat proteins.
Here we report that activation of the EGF receptor leads to phosphorylation of GGA3 on a previously unrecognized site in the hinge segment, S368 (all residues are herein numbered for the long splice form of GGA3). This modification is strictly dependent on the constitutive phosphorylation of another novel site, S372. The EGF-induced phosphorylation causes a conformational change in GGA3, evidenced by an increase in its hydrodynamic radius, and decreased association with membranes. These observations indicate that EGF signaling elicits phosphorylation events that regulate the association of GGA3 with organellar membranes.
MATERIALS AND METHODS
Cells and other materials.
Human HeLa and M1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Biosource, Rockville, MD) containing 4.5 g/liter glucose, 50 units/ml penicillin, 50 μg/ml streptomycin, 2 mM l-glutamine, and 10% vol/vol fetal bovine serum. Transient transfection of both cell lines was performed using FuGENE6 according to the manufacturer's instructions (Roche Molecular Biochemicals, Indianapolis, IN). HeLa DynK44A cells (gift of S. Schmid; Scripps Research Institute, La Jolla, CA) ( 11) were maintained in DMEM containing 10% vol/vol fetal bovine serum, 200 ng/ml puromycin, and 1 μg/ml tetracycline. For induction of DynK44A protein expression, cells were transferred to the same medium without tetracycline for 3 days. M1 cells expressing enhanced green fluorescent protein (EGFP)-GGA3 (referred to as GFP-GGA3) or photoactivatable GFP fused to GGA3 (PAGFP-GGA3) were created by transfection of pEGFP-GGA3 or pPAGFP-GGA3, respectively, followed by selection in 600 μg/ml G418 (the sources of the parental plasmids are indicated below). The resulting polyclonal stable transfectants were used for fluorescence recovery after photobleaching (FRAP) (34) or photoactivation (30) experiments. EGF, PD98059, wortmannin, LY294002, brefeldin A, and AG1478 were from Calbiochem (San Diego, CA), platelet-derived growth factor (PDGF) AA, basic fibroblast growth factor and transforming growth factor β1 were from Peprotek (Rocky Hill, NJ), phosphatase inhibitor cocktails 1 and 2 were from Sigma (St. Louis, MO), and protease inhibitor cocktail was from Roche. Yeast two-hybrid assays were performed as described previously (35).
Plasmids.
To construct mammalian expression vectors for human GGA3 tagged with a triple Myc epitope, a 1.6-kb EcoRI-XbaI fragment of pGBT9-GGA3 (27) and a 0.6-kb XbaI-SalI fragment of pGEX-GGA3-GAE (494 to 723) (27) were simultaneously cloned into the pCIneo-3Myc vector at EcoRI and SalI sites. The hinge region of the resultant plasmid, pCI3Myc-GGA3-WT-FL, was amplified by PCR and cloned into pCIneo-3Myc to obtain pCI3Myc-GGA3-hinge. The EcoRI-SalI fragment from pGBT9-GGA3-VHS, GAT (27) or EcoRI-NotI fragment from pGEX-GAE (13) was subcloned into pCIneo-3Myc vector to generate pCI3Myc-GGA3-VHS, -GAT, and -GAE, respectively.
To construct pEGFP-GGA3 or pPAGFP-GGA3, the 2.1-kb EcoRI-SalI fragment from pCI-3Myc-GGA3 was subcloned into the pEGFP(A206K) or pPAGFP(A206K) vector, respectively (provided by J. Lippincott-Schwartz, National Institute of Child Health and Human Development, National Institutes of Health). pEGFP-GGA2 was described previously (27). Site-directed mutagenesis was carried out with the QuikChange kit (Stratagene, La Jolla, CA) using pCI3Myc-GGA3-hinge as template. The mutagenized regions flanked by SmaI-XbaI were transferred into the corresponding region of pCI3Myc-GGA3-WT-FL to make full-length mutants. Full-length GGA3 fragments were subcloned at the EcoRI and XhoI sites of pGADT7. The plasmids pGBT9-CI-MPR-tail (35), -ubiquitin (36), and -Arf1 constructs (37) were described previously.
Antibodies.
Monoclonal antibodies to GGA3, δ-adaptin, ERK1/2, phospho-ERK1/2, Akt, and phospho-Akt (pS473) were purchased from BD Biosciences Pharmingen (San Diego, CA). Monoclonal antibodies to the Myc and hemagglutinin (HA) epitopes were purchased from Invitrogen (Carlsbad, CA) and Covance (Princeton, NJ), respectively. A rabbit polyclonal antibody to CI-MPR was raised against the C-terminal peptide (NH2-CSFHDDSDEDLLHI-COOH) of human CI-MPR (Pacific Immunology Corp., Ramona, CA). A rabbit polyclonal antibody to GGA1 was a gift of Margaret S. Robinson (University of Cambridge, United Kingdom) and a mouse monoclonal antibody to GGA2 was a gift of Doug Brooks (Women's and Children's Hospital, North Adelaide, Australia).
Stimulation of cultured cells by agonists.
HeLa or M1 cells grown on 24-well plates were incubated in serum-free medium (DMEM containing 1% wt/vol bovine serum albumin) for 18 or 5 h, respectively (referred to as “serum starvation”). Whenever indicated, serum-starved cells were treated with the same medium containing protein kinase inhibitors for 30 min at 37°C. This was followed by stimulation with agonists such as EGF, PDGF, basic fibroblast growth factor, or transforming growth factor β1 for different times at 37°C and resuspension of the cell layer in 100 μl 1x Laemmli buffer. Samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
32P labeling.
M1 cells were cultured in serum-free medium for 18 h and then labeled with 0.5 mCi/ml 32P-labeled orthophosphoric acid in phosphate-free DMEM supplemented with 0.1% wt/vol bovine serum albumin for 2 h at 37°C, followed by incubation with or without 10 nM EGF for 10 min. After labeling the cells were washed with ice-cold phosphate-buffered saline three times and resuspended in 0.5 ml of lysis buffer A (1% wt/vol Triton X-100, 0.2% wt/vol SDS, 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.4), protein phosphatase inhibitor cocktails I and II (Sigma) and 1x protease inhibitor cocktail (Roche) and incubated with protein A-Sepharose at 4°C for 30 min. This was followed by centrifugation at 15,000 × g for 15 min at 4°C to obtain detergent extracts. The extracts were incubated with 2 μl of anti-GGA1 serum, or 2 μg of anti-GGA2, anti-GGA3, or anti-GFP monoclonal antibodies, and protein A-Sepharose for 18 h at 4°C with constant tumbling. The beads were washed five times with ice-cold lysis buffer and bound proteins were eluted with 1x Laemmli buffer, then the samples were subjected to SDS-PAGE on 7% acrylamide gels. The gels were dried and scanned on a PhosphorImager (Typhoon 9200, Molecular Dynamics, Sunnyvale, CA). Immunoprecipitation of each protein was confirmed by immunoblotting.
Subcellular fractionation.
HeLa cells grown on a 10-cm culture dish were collected and washed with phosphate-buffered saline containing phosphatase and protease inhibitor cocktails. Cells were homogenized in the same buffer by passage through a 25-gauge needle 20 times, then centrifuged at 800 × g for 5 min to obtain a postnuclear supernatant fraction. The postnuclear supernatant fraction was subsequently subjected to ultracentrifugation for 1 h at 125,000 × g to generate cytosolic and membrane fractions.
Hydrodynamic analyses and immunoprecipitation.
Gel filtration and sucrose velocity gradient centrifugation were performed as described in reference 12. For immunoprecipitation of GGA3, cells were lysed with immunoprecipitation-lysis B buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% wt/vol Triton X-100, 0.1% wt/vol SDS, and 1x protease inhibitor cocktail) on ice for 30 min, and microcentrifuged at 20,800 × g for 15 min at 4°C. The resultant total cell lysate from approximately 5 × 106 cells was preincubated with protein A-Sepharose at 4°C for 1 h and centrifuged at 20,800 × g for 15 min. Three micrograms of anti-GGA3 antibody was next added to the precleared extract and this mixture was incubated at 4°C for 18 h. Antibody-GGA3 complexes were captured with protein A-Sepharose, which were then washed five times with immunoprecipitation-lysis buffer. The beads were separated in two halves, each of which was incubated for 30 min on ice in the presence or absence of 50 U/ml alkaline phosphatase (New England Biolabs, Beverly, MA). The immune complexes were eluted with 1x Laemmli sample buffer at 90°C for 5 min and subsequently subjected to SDS-PAGE and immunoblotting. Hydrodynamic parameters of each protein were calculated as described in reference 26.
Two-dimensional gel electrophoresis.
Untransfected M1 cells or M1 cells expressing wild-type or mutant Myc-tagged GGA3 were serum-starved for 5 h and incubated for 10 min in the presence or absence of 10 nM EGF. The cells were lysed in lysis buffer for two-dimensional electrophoresis (40 mM Tris base, 8 M urea and 4% wt/vol CHAPS) and subjected to isoelectric focusing using Immobiline DryStrip gel (pH 4 to 7) according to the manufacturer's protocol (Amersham Biosciences, San Francisco, CA). Proteins were further separated in the second dimension by SDS-PAGE and subjected to immunoblotting using antibody to the Myc epitope. For alignment of different blots, 1 μg of bovine serum albumin mixed in the protein sample and visualized with Ponceau S, as well as endogenous GGA3 detected by immunoblotting, were used as landmarks.
Live cell imaging.
Cells grown on glass-bottomed culture dishes (MatTek, Ashland, MA) were imaged in buffered medium using a Zeiss 510 confocal microscope equipped with a stage heated to 37°C (Carl Zeiss, Thornwood, NY). Lasers at 488 nm and 413 nm were used for excitation of GFP and activation of PAGFP, respectively. For quantitative analysis of fluorescence intensities, nonsaturated images were captured with a Plan-NEOFLUAR 25x/0.80 objective and a fully open pinhole. Selective photobleaching was performed using 488-nm laser at full power and the recovery was monitored by time-lapse imaging at 3-s intervals with low-intensity illumination. Photoactivation of PAGFP was carried out using a single hit of 413 nm laser at full power and fluorescence decay was monitored at 3-s intervals. Fluorescence quantification was done as described previously (34, 21). In brief, intensities for total cellular fluorescence or TGN-associated fluorescence were measured using NIH Image software after subtraction of background fluorescence outside the cell. Cells that exhibited no saturated pixels were used. Each experiment was repeated at least six times under identical conditions, and the mean and standard deviation were calculated.
RESULTS
EGF-induced phosphorylation of GGA3.
In the course of experiments aimed at analyzing the expression of endogenous GGAs in various human cell lines, we noticed that GGA3 migrated as a doublet on SDS-PAGE. This was in contrast to GGA1 and GGA2, which migrated as single bands (data not shown). To determine whether the GGA3 doublet arose from regulated phosphorylation, HeLa epithelial and M1 fibroblast cell lines were serum-starved for 18 h and 5 h, respectively, and subsequently incubated for 10 min in the absence or presence of 10 nM EGF (referred to as resting and stimulated conditions, respectively). GGA3 was isolated by immunoprecipitation, incubated in the absence or presence of alkaline phosphatase, and analyzed by SDS-PAGE and immunoblotting with a monoclonal antibody to GGA3. We observed that incubation with EGF resulted in a shift from the lower to the upper band in both cell lines (Fig. 1A). Treatment of the immunoprecipitates with alkaline phosphatase eliminated the upper band (Fig. 1A), demonstrating that it corresponded to phosphorylated GGA3. The proportion of phosphorylated GGA3 was higher in HeLa cells than in M1 cells (49% ± 1% versus 22% ± 7% under resting conditions, and 83% ± 5% versus 51% ± 6% under conditions of stimulation) (Fig. 1B).
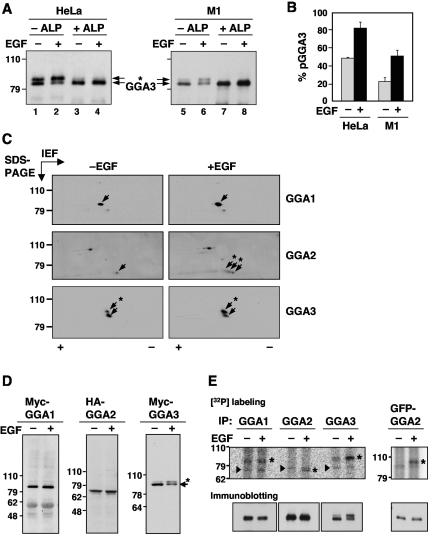
FIG. 1.
EGF-induced phosphorylation of GGA2 and GGA3. (A) Endogenous GGA3 was immunoprecipitated from resting (−EGF, lanes 1, 3, 5, and 7) or EGF-stimulated (+EGF, lanes 2, 4, 6, and 8) HeLa (lanes 1 to 4) or M1 cells (lanes 5 to 8). Immunoprecipitates were incubated in the absence (−ALP, lanes 1, 2, 5, and 6) or presence of alkaline phosphatase (+ALP, lanes 3, 4, 7, and 8), and GGA3 was analyzed by one-dimensional SDS-PAGE and immunoblotting. (B) Quantification of phosphorylated GGA3 (pGGA3) relative to total GGA3 in resting (−EGF) and EGF-stimulated (+EGF) HeLa or M1 cells analyzed as in panel A. Values are the mean ± SD of four independent determinations. (C) M1 fibroblasts were serum-starved for 5 h and incubated for 10 min in the absence (−EGF) or presence (+EGF) of 10 nM EGF, and cell extracts were subjected to two-dimensional isoelectric focusing/SDS-PAGE. Endogenous GGA proteins were detected by immunoblotting. Arrows point to the different GGA species. The symbols + and - indicate the basic and acidic ends, respectively, of the isoelectric focusing. (D) Myc-GGA1, HA-GGA2, and Myc-GGA3 expressed by transfection into resting or EGF-stimulated M1 fibroblasts were analyzed by immunoblotting with antibodies to the epitope tags as in panel A. (E) Untransfected (left panels) or GFP-GGA2-transfected (right panel) M1 fibroblasts were metabolically labeled with 0.5 mCi/ml [32P]orthophosphate for 2 h and incubated in the absence or presence of 10 nM EGF for 10 min. Each GGA was immunoprecipitated with a specific antibody (or anti-GFP antibody for GFP-GGA2) and subjected to one-dimensional SDS-PAGE. Asterisks indicate the GGA proteins, and arrowheads indicate nonspecific bands seen in all lanes. Numbers on the left indicate the positions of molecular mass markers (in kilodaltons), and asterisks in panels A, C, and D indicate the position of phosphorylated GGA3 (throughout the paper, “phosphorylated GGA3” and “pGGA3” are used to denote the more slowly migrating species that increases in intensity upon EGF treatment; the faster migrating species may have other phosphorylated residues that do not affect its electrophoretic mobility).
To determine whether GGA1 and GGA2 also underwent regulated phosphorylation despite their migration as single bands on SDS-PAGE, M1 cells were serum-starved, incubated in the absence or presence of EGF, and analyzed by two-dimensional isoelectric focusing/SDS-PAGE followed by immunoblotting with antibodies to GGA1, GGA2, and GGA3 (Fig. 1C). In agreement with the one-dimensional SDS-PAGE analyses described above, GGA3 appeared as a doublet in resting cells, with the upper, phosphorylated spot being more acidic; EGF treatment increased the intensity of this spot (Fig. 1C, lower panels). In contrast, GGA1 appeared as a single spot under both resting and stimulated conditions (Fig. 1C, upper panels). Finally, GGA2 appeared as a single spot under resting conditions (Fig. 1C, middle left panel), but stimulation with EGF resulted in the appearance of two more acidic spots that were only slightly shifted upwards in the SDS-PAGE dimension (Fig. 1C, middle right panel).
Myc-tagged GGA3 expressed by transfection into M1 cells also underwent an upward mobility shift on SDS-PAGE upon treatment of the cells with EGF (Fig. 1D, right panel), while Myc-tagged GGA1 and HA-tagged GGA2 examined under the same conditions did not show any change in mobility (Fig. 1D, left and middle panels), consistent with the behavior of the endogenous proteins described above. Finally, analysis of [32P]orthophosphate-labeled M1 cells showed that all three endogenous GGAs were phosphorylated, but only GGA2 and GGA3 exhibited increased phosphorylation upon EGF treatment (Fig. 1E). Because the EGF-induced phosphorylation of GGA3 could be easily assayed by a mobility shift on SDS-PAGE, all subsequent analyses were performed on this protein.
Characterization of EGF-induced GGA3 phosphorylation.
To characterize the EGF-induced phosphorylation of endogenous GGA3, we examined its concentration-dependence and time-course in M1 cells. Maximum GGA3 phosphorylation was attained with concentrations of EGF ≥0.1 nM after 10 min of treatment (Fig. 2A, upper left panel). Phosphorylation was transient, peaking at ∼10 min after addition of 10 nM EGF (Fig. 2A, upper right panel). Both the concentration dependence and time course of GGA3 phosphorylation by EGF were comparable to those observed for the phosphorylation of the mitogen-activated protein kinases ERK1 and ERK2 (Fig. 2A, middle panels). An inhibitor of EGF receptor tyrosine kinase, AG1478, completely blocked the EGF-induced phosphorylation of GGA3 (Fig. 2B), indicating that activation of the EGF receptor tyrosine kinase is required for GGA3 phosphorylation.
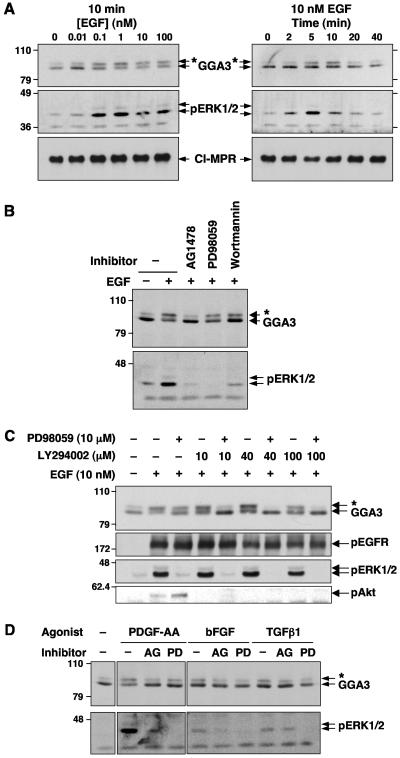
FIG. 2.
Characterization of growth factor-dependent phosphorylation of GGA3. (A) Concentration dependence and time course of EGF-induced GGA3 phosphorylation. Serum-starved M1 cells were stimulated for 10 min with the indicated concentrations of EGF (left panels) or for the indicated times with 10 nM EGF (right panels). (B) Serum-starved M1 cells were treated with 1 μM AG1478, 10 μM PD98059, or 100 nM wortmannin for 30 min prior to stimulation with 10 nM EGF. (C) Serum-starved M1 cells were treated with 10 μM PD98059 and/or the indicated concentrations of LY294002 for 30 min prior to stimulation with 10 nM EGF. (D) Serum-starved M1 cells were incubated in the presence of 0.5% dimethyl sulfoxide (− inhibitor), 1 μM AG1478 (AG), or 10 μM PD98059 (PD) for 30 min prior to stimulation with 10 ng/ml (0.35 nM) PDGF-AA, 10 ng/ml (0.58 nM) basic fibroblast growth factor (bFGF), or 1 ng/ml (0.04 nM) transforming growth factor β1 (TGFβ1) for 10 min. In all panels, GGA3, phosphorylated ERK1/2 (pERK1/2), cation-independent mannose-6-phosphate receptor (CI-MPR), phosphorylated EGF receptor (pEGFR), and phosphorylated Akt (pAkt) were analyzed by SDS-PAGE and immunoblotting, as indicated. Numbers on the left indicate the positions of molecular mass markers (in kilodaltons), and asterisks indicate the position of phosphorylated GGA3.
The kinetics of GGA3 phosphorylation (Fig. 2A), however, were slower than those of proximal targets of the EGF receptor tyrosine kinase (4), but similar to those of more distal targets such as the mitogen-activated protein kinases (Fig. 2B in reference 4), which undergo phosphorylation by the dual-specificity kinase MEK1. Importantly, the EGF-induced phosphorylation of GGA3 could be partially prevented by either the MEK1 inhibitor PD98059 (Fig. 2B and C) or the phosphatidylinositol 3-kinase inhibitors wortmannin (Fig. 2B) and LY294002 (Fig. 2C), which block kinase cascades downstream of the EGF receptor (49). Finally, combinations of PD98059 (10 μM) and LY294002 (40 to 100 μM) completely inhibited the phosphorylation induced by EGF (Fig. 2C), indicating that both the mitogen-activated protein kinase and phosphatidylinositol 3-kinase arms of EGF signaling contribute to the regulation of GGA3 phosphorylation. These results thus suggested that EGF-induced GGA3 phosphorylation is catalyzed by a protein kinase that lies downstream of the EGF receptor.
GGA3 was also phosphorylated by treatment of M1 cells with PDGF-AA (Fig. 2D), another growth factor that binds to a specific receptor tyrosine kinase (the α-PDGF receptor) and activates some of the same signaling pathways as EGF. This phosphorylation was also completely inhibited by AG1478 and partially inhibited by PD98059 (Fig. 2D). In contrast, both basic fibroblast growth factor and transforming growth factor β1, which bind to different types of receptor tyrosine kinases, caused minimal phosphorylation of GGA3 and weakly activated ERK1/2 in M1 cells (Fig. 2D). Thus, for all of these growth factors the phosphorylation status of GGA3 paralleled that of ERK1/2, again suggesting a connection between GGA3 phosphorylation and activation of downstream kinases.
The phosphorylation of some substrates downstream of the EGF receptor (e.g., Hrs) (46) requires internalization of the receptor. To determine whether this was also the case for GGA3, we used a stable HeLa cell line that can be induced to express a dominant-negative mutant form of dynamin 1 (DynK44A) (10). As previously reported (10), induction of this mutant blocked uptake of fluorescently labeled transferrin and EGF as determined by microscopy (data not shown). However, we did not observe any changes in the ability of EGF to induce phosphorylation of GGA3 (the basal phosphorylation of GGA3 in this particular cell line was higher than that in regular HeLa cells) (Fig. 3A). These observations indicate that the EGF receptor does not need to be internalized in order to induce phosphorylation of GGA3.
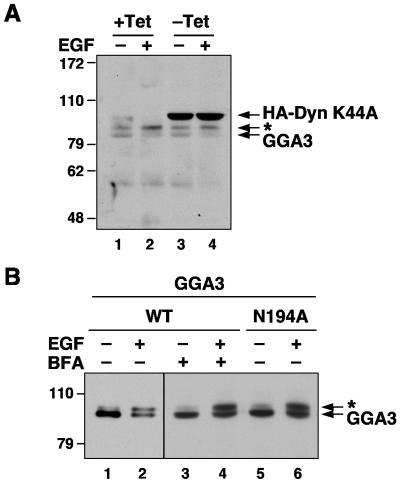
FIG. 3.
Internalization of EGF receptor and membrane association of GGA3 are not required for EGF-induced GGA3 phosphorylation. (A) Expression of HA-dynamin 1 K44A in stably transfected HeLa cells was induced by removal of tetracycline from the culture medium (−Tet) for 72 h. After serum starvation for 12 h, cells were incubated in the absence (−EGF) or the presence of 10 nM EGF (+EGF) for 10 min, and analyzed by SDS-PAGE and simultaneous immunoblotting with antibodies to GGA3 and the HA epitope. (B) M1 fibroblasts transiently expressing Myc-tagged, wild-type GGA3 (lanes 1 to 4; WT) or N194A mutant GGA3 (lane 5 and 6; N194A) were serum-starved for 5 h and incubated in the absence (−EGF) or presence of 10 nM EGF (+EGF) for 10 min. Prior to stimulation, some cells were treated with 5 μg/ml brefeldin A for 30 min (+BFA; lanes 3 and 4). Samples were analyzed by SDS-PAGE and immunoblotting with antibody to the Myc epitope. Numbers on the left indicate the positions of molecular mass markers (in kilodaltons), and asterisks indicate the position of phosphorylated GGA3 (endogenous in panel A; Myc-tagged in panel B).
We also examined whether association of GGA3 with membranes was required for its EGF-induced phosphorylation. This was done by treating M1 cells with brefeldin A, a fungal metabolite that dissociates GGAs from membranes through inactivation of Arf (5, 13, 22). We observed that treatment with brefeldin A had no effect on the EGF-induced phosphorylation of GGA3 (Fig. 3B). Moreover, a GGA3 mutant having a substitution of N194 to alanine, which prevents binding to Arf and recruitment to membranes (37), underwent EGF-induced phosphorylation to the same extent as normal GGA3 (Fig. 3B). Therefore, membrane association is not required for GGA3 phosphorylation in response to EGF. Taken together, these experiments suggested that binding of EGF to its receptor at the plasma membrane leads to activation of a downstream protein kinase that phosphorylates GGA3 irrespective of association with membranes.
Identification of EGF-induced phosphorylation sites in GGA3.
Under normal conditions of culture, GGA3 is phosphorylated only on serine residues (14, 28) (Fig. 4A). Moreover, preventing the activation of serine/threonine kinases blocks the EGF-induced phosphorylation of GGA3 (Fig. 2B and C). To determine whether the EGF-dependent phosphorylation of GGA3 indeed involved serine residues, we performed a mutational analysis of GGA3. We expressed the Myc-tagged, isolated VHS, GAT, hinge, and GAE domains of GGA3 in M1 cells and examined the effect of treatment with EGF. We observed that only the hinge construct migrated as a doublet under basal conditions and underwent an upward mobility shift upon treatment with EGF (Fig. 4B, right panel, asterisk).
The hinge region contains three of the previously reported phosphorylation sites on GGA3, S388, S538, and S559, in the “long” splice form (Fig. 4A) (28). Mutation of each of these serine residues to alanine did not affect the EGF-induced phosphorylation of the hinge (data not shown; summarized in Fig. 4C). Truncation analyses indicated that the 67-amino-acid amino-terminal region of the hinge (residues 314 to 380), comprising 18 serine and threonine residues but no tyrosine residues, contained the determinants of the shift (Fig. 4C and 5A). This region comprises a consensus site for phosphorylation by the adaptor-associated kinase AAK1, which phosphorylates the μ2 subunit of the AP-2 clathrin adaptor (9, 39). However, mutation of S363 within this site did not prevent phosphorylation of the hinge (Fig. 5B).
FIG. 5.
Identification of GGA3 residues responsible for the EGF-induced mobility shift. (A) Schematic representation of GGA3 and sequence of a 67-amino-acid hinge segment containing clusters of serine and threonine residues and a potential AAK1 phosphorylation site (boxed residues). (B to D) Myc-tagged GGA3 hinge (WT) and the corresponding S363A (B), cluster I (334 to 340; AANALAA), cluster II (347 to 351; APPAA) and cluster III mutants (368 to 372; ARAAA) (C), and cluster III single-amino-acid-substitution mutants (D) were expressed in M1 cells and examined for EGF-dependent phosphorylation as described in the legend to Fig. 4 and in Materials and Methods. (E) Alignment of GGA hinge sequences from different species. Conserved serine residues corresponding to the EGF-regulated phosphorylation sites in human GGA3 are indicated by shaded boxes. Internal DXXLL-type sequences are underlined. Amino acid numbers are indicated.
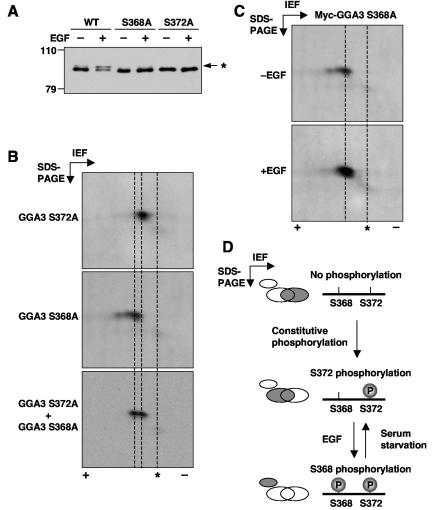
We also mutated three clusters of serine and threonine residues to alanine residues in the context of the hinge construct (Fig. 5A); only mutation of cluster III prevented phosphorylation of the hinge (Fig. 5C). Further mutagenesis revealed that substitution of either S368 or S372 to alanine completely blocked the EGF-dependent phosphorylation of both the hinge (Fig. 5D) and full-length GGA3 (Fig. 6A) constructs. These experiments thus demonstrated that both S368 and S372 are required for the EGF-induced mobility shift. We also noticed that mutation of S368 or S372 eliminated the small amount of basally phosphorylated GGA3 (upper band in Fig. 6A), indicating that basal and EGF-induced phosphorylation involve these same two residues.
FIG. 6.
Both S368 and S372 are required for the EGF-induced mobility shift of GGA3. (A) Myc-tagged, full-length GGA3 with mutations in S368A or S372A were expressed in M1 cells and analyzed for EGF-induced phosphorylation as described in the legend to Fig. 4. The asterisk indicates the position of phosphorylated GGA3. (B) Myc-tagged, full-length GGA3 with mutations in S368A (top panel) or S372A (middle panel) was expressed in resting M1 cells and analyzed by two-dimensional isoelectric focusing (IEF)/SDS-PAGE and immunoblotting with antibody to GGA3. To compare the migration of the two mutant species, both samples were also mixed and loaded together on the gels (lower panel). (C) M1 cells expressing Myc-tagged, full-length GGA3 S368A mutant were incubated in the absence (−EGF) or presence (+EGF) of 10 nM EGF and cell lysates were analyzed by SDS-PAGE/IEF and immunoblotting with antibody to GGA3. In panels B and C, asterisks indicate the position of endogenous, unphosphorylated GGA3. (E) Model for the dual-phosphorylation mechanism of GGA3 (see text for details).
Dual-phosphorylation mechanism for the EGF-induced mobility shift of GGA3.
The fact that two serine residues are required to observe the mobility shift of GGA3 could mean that EGF treatment leads to phosphorylation of both residues. However, we never observed any intermediates in which only one of these residues was phosphorylated, even when only one of the two serine residues was substituted (e.g., Fig. 5D and 6A). This suggested the alternative hypothesis that the EGF-induced phosphorylation of one of these residues required previous phosphorylation of the other.
To test this hypothesis, we analyzed by two-dimensional isoelectric focusing/SDS-PAGE the migration of GGA3-S372A and GGA3-S368A in the absence of EGF. We observed that under these conditions, GGA3-S372A was more basic than GGA3-S368A (Fig. 6B), indicating that substitution of S372 prevented a constitutive phosphorylation that occurs in the absence of added EGF. Addition of EGF did not change the mobility of GGA3-S368A on two-dimensional electrophoresis (Fig. 6C), even though it caused an upward/acidic shift of wild-type GGA3 (Fig. 1C). These results suggested that S372 is constitutively phosphorylated and that this phosphorylation is a prerequisite for subsequent phosphorylation at S368 in response to EGF stimulation, the latter being directly responsible for the mobility shift on SDS-PAGE (Fig. 6D).
Change in hydrodynamic properties caused by phosphorylation of GGA3.
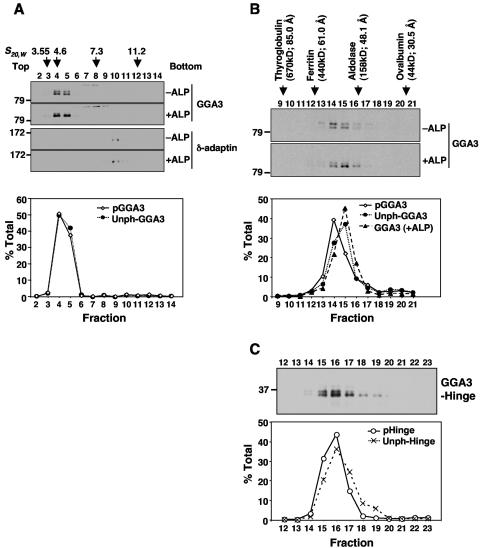
Phosphorylation often causes conformational changes in proteins or modifies their affinity for other proteins. To determine whether this was the case for GGA3, we analyzed its hydrodynamic behavior by sedimentation velocity and gel filtration. Analyses were performed with HeLa cells grown under normal conditions of culture, which exhibit comparable amounts of S368-phosphorylated and S368-unphosphorylated GGA3 (upper and lower bands, respectively, on SDS-PAGE, Fig. 1A).
Sucrose gradient centrifugation of cytosolic extracts showed equal migration of both GGA3 species with a sedimentation coefficient of ∼5 S (Fig. 7A). In contrast, gel filtration on Superdex 200 showed that S368-phosphorylated GGA3 peaked one fraction earlier than S368-unphosphorylated GGA3 (corresponding to Stokes radii of ∼54 Å versus ∼49 Å, respectively) (Fig. 7B). Pretreatment of the cytosolic extract with alkaline phosphatase caused the majority of the GGA3 to elute as a single peak corresponding to that of the lower band in the untreated sample (Fig. 7B). Immunoprecipitation of 35S-labeled GGA3 from HeLa cells did not reveal any additional associated proteins in response to EGF stimulation (data not shown). Therefore, the faster elution of the S368-phosphorylated GGA3 on gel filtration is likely due to a conformational change. Similar gel filtration analysis of a Myc-tagged GGA3 hinge construct showed that the S368-phosphorylated form eluted faster than the S368-unphosphorylated form (Fig. 2C), indicating that the conformational change is intrinsic to the hinge.
FIG. 7.
EGF-induced phosphorylation changes the hydrodynamic properties of GGA3. (A) Cytosol prepared from normally growing HeLa cells was incubated in the absence (−ALP) or presence (+ALP) of alkaline phosphatase and fractionated by ultracentrifugation on 3 to 15% sucrose gradients followed by analysis of the fractions by SDS-PAGE and immunoblotting with antibodies to GGA3 and the δ-adaptin subunit of AP-3 (control). Numbers on top indicate the sedimentation coefficients (S20,w) of marker proteins: ovalbumin (3.55), albumin (4.6), aldolase (7.3), and catalase (11.2). (B) Cytosol prepared as in panel A was analyzed by gel filtration on Superdex 200 followed by SDS-PAGE and immunoblotting of fractions with antibody to GGA3 as described in Materials and Methods. (C) Cytosol prepared from EGF-stimulated, Myc-GGA3-hinge-expressing M1 cells was subjected to gel filtration analysis as in panel B. Notice the difference in the ratios of S368-phosphorylated and unphosphorylated GGA3 in fractions 15 and 17. The positions, molecular masses, and Stokes radii of marker proteins are indicated on top. Numbers on the left of the top panels in panels A, B, and C indicate the positions of molecular mass markers in the SDS-PAGE (in kilodaltons). The lower graphs show quantifications of the different forms of GGA3 expressed as a percent of total GGA3.
Phosphorylation of GGA3 decreases its association with membranes.
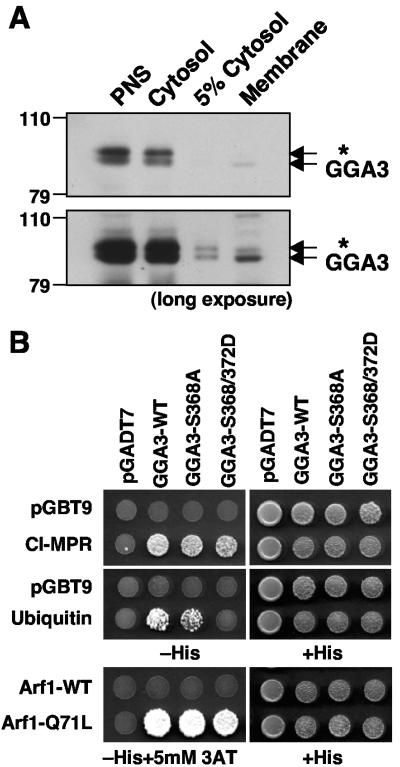
We next examined the effects of phosphorylation and the ensuing conformational change of GGA3 on its association with membranes and interactions with various binding partners (Fig. 8). Upon disruption of cells, most of the GGA3 appeared in the supernatant and only a small fraction remained associated with the particulate fraction (Fig. 8A). However, the ratio of S368-phosphorylated to S368-unphosphorylated GGA3 was much lower in the particulate fraction than in the cytosolic fraction (Fig. 8A), implying that this phosphorylation decreases the association of GGA3 with membranes.
FIG. 8.
Phosphorylation of GGA3 on S368 decreases its recruitment to membranes and binding to ubiquitin. (A) Postnuclear supernatant (PNS), cytosol, and membrane fractions from HeLa cells, prepared as described in Materials and Methods, were subjected to SDS-PAGE and immunoblotting with anti-GGA3 antibody. Short (upper panel) and long (lower panel) exposures of the same blot are shown. (B) Yeast two-hybrid analysis of the binding of GGA3 constructs to the cytosolic tail of the CI-MPR, ubiquitin, wild-type Arf1, and the constitutively activated Arf1-Q71L mutant. pGBT9 and pGADT7 represent “empty” plasmid controls. Cotransformed yeast strains were grown on plates without histidine (−His) or with histidine (+His), and in some cases in the presence of 5 mM 3-aminotriazole (3AT), as needed to suppress nonspecific growth.
GGA3 interacts with many binding partners, and a weakened interaction with any of them could underlie the decreased association of the S368-phosphorylated form with membranes. To assess whether any of the known interactions was impaired by phosphorylation, we examined the binding of various partners to GGA3 or variants of this protein in which S368 was mutated to alanine or both S368 and S372 were mutated to phosphomimic aspartate residues (Fig. 8B). We observed that binding to the GTP-locked form of Arf1 (Arf1-Q71L) and to the cytosolic tail of the cation-independent MPR (which has a DXXLL-type signal) was not affected by the mutations (Fig. 8B). However, binding to ubiquitin was decreased by mutation of S368 and S372 to aspartate (Fig. 8B). Therefore, the decreased association of S368-phosphorylated GGA3 with membranes could be due to impaired interactions with ubiquitin.
Phosphoacceptor serine 368 regulates the kinetics of association of GGA3 with the TGN.
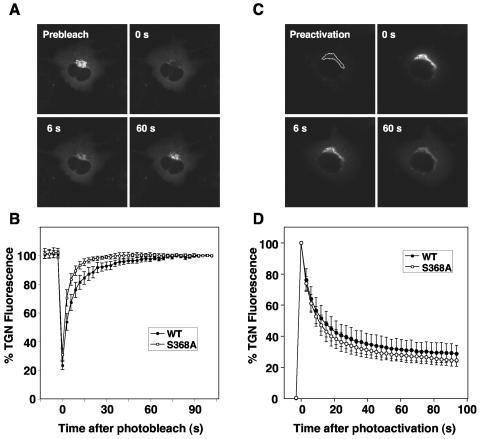
To determine whether phosphorylation affected the kinetics of GGA3 association with the TGN, we examined the rate of fluorescence recovery after photobleaching (FRAP) (34) of wild-type GFP-GGA3 and S368A mutant GFP-GGA3 in stably transfected M1 cells (Fig. 9A). We observed that wild-type GFP-GGA3 (which exists as a mixture of S368-phosphorylated and unphosphorylated protein) and the S368A mutant of GFP-GGA3 reassociated with the TGN with initial rates of 13% ± 3%/s and 21% ± 2%/s, respectively (Fig. 9B).
FIG. 9.
Dynamics of GGA3 binding to membranes in live cells analyzed by photobleaching and photoactivation. (A) The kinetics of association of GFP-GGA3 with the TGN in stably transfected M1 cells were studied by fluorescence recovery after photobleaching. The dashed line indicates the area that was photobleached with high laser intensity at time zero. Fluorescence recovery was assessed at different times after photobleaching. (B) Quantification of the FRAP data for wild-type GFP-GGA3 and the S368A mutant from a set of eight experiments similar to that described for panel A. Values are the mean ± standard deviation. (C) The kinetics of dissociation of photoactivatable GFP (PAGFP)-GGA3 from the TGN in stably transfected M1 cells were studied after photoactivation of the area delimited by the dashed line. (D) Quantification of the dissociation data for wild-type PAGFP-GGA3 and the S368A mutant from a set of six experiments similar to that described for panel C. Values are the mean ± standard deviation.
We also determined the rate of dissociation from the TGN of wild-type and S368A mutant GGA3 fused to a photoactivatable GFP (PAGFP) (30). We found that the TGN fluorescence of both proteins decreased while the cytoplasmic fluorescence increased with indistinguishable kinetics (initial rate ∼25% ± 5%/s) (Fig. 9C and D). From these experiments, we concluded that the phosphoacceptor S368 regulates the initial rate of association of GGA3 with the TGN, though not its rate of dissociation from this organelle.
DISCUSSION
The results presented here show that endogenous GGA3 exists in resting HeLa and M1 cells as two electrophoretically distinct species that differ in the phosphorylation state of a novel phosphoacceptor site in the hinge, S368. Treatment of cells with physiological (subnanomolar) concentrations of EGF induces a transient increase in the amount of S368-phosphorylated GGA3. Both basal and EGF-induced phosphorylation of S368 requires constitutive phosphorylation of a neighboring residue in the hinge, S372. Phosphorylation of S368 causes a conformational change in GGA3 and alters the association of GGA3 with membranes.
Agonist-regulated phosphorylation of GGA3.
A previous study used tandem mass spectrometry to identify several phosphoserine residues on the GAT and hinge domains of epitope-tagged GGA3 expressed in transfected cells (28). None of these residues, however, corresponded to S368, nor were they shown to be regulated by any physiological stimuli. We find that S368 is a major site of phosphorylation, accounting for 22 to 49% and 51 to 83% of endogenous GGA3 under resting and EGF-stimulated conditions, respectively, in two different cell lines. The EGF receptor has intrinsic tyrosine kinase activity, so the enhancement of GGA3 phosphorylation on a serine residue elicited by EGF points to downstream serine/threonine kinases as the probable catalysts. Indeed, both basal and EGF-induced S368 phosphorylation can be prevented with inhibitors of not only the EGF receptor tyrosine kinase, but also mitogen-activated protein kinases and phosphatidylinositol 3-kinase, which are key components of the EGF receptor-activated serine/threonine kinase cascades.
Notably, the electrophoretic mobility shift that results from S368 phosphorylation is strictly dependent on another residue, S372. This residue is constitutively and quantitatively phosphorylated, since its substitution by alanine causes a basic shift in isoelectric point of the entire population of GGA3. S368 and S372 are contained within a sequence context that fits consensus motifs for phosphorylation by glycogen synthase kinase 3 (GSK-3) (25) and casein kinase II (31), respectively. This particular arrangement of putative GSK-3 and casein kinase II sites is conserved in vertebrate GGA3 orthologs from human to chicken, and even in the only GGA protein encoded in the zebrafish genome (Fig. 5E); this suggests evolutionary pressure to maintain a regulatory element within the otherwise poorly conserved sequence of the hinge region. Glycogen synthase itself has a similar configuration of phosphoacceptor sites that are phosphorylated by a dual-kinase mechanism involving casein kinase II and GSK-3 (16). However, GSK-3 is inhibited, not activated, upon EGF receptor activation (41). Furthermore, overexpression of a constitutively active GSK-3α mutant did not result in a mobility shift of endogenous GGA3 on SDS-PAGE (data not shown). Thus, it is unlikely that GGA3 phosphorylation on S368 is effected by GSK-3.
None of the known serine/threonine kinases that lie downstream of the activated EGF receptor (e.g., Akt, S6K, mitogen-activated protein kinase, or JNK) (49) would be expected to phosphorylate S368, based on their substrate consensus sequences (http://scansite.mit.edu/). Therefore, phosphorylation of GGA3 on S368 in response to EGF is likely catalyzed by an as yet unidentified kinase that receives activating signals from both the mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways.
Conformational change of GGA3 elicited by S368 phosphorylation.
The GGAs are predicted to have a highly extended, flexible conformation resembling “beads on a string,” with the beads representing the folded VHS, GAT, and GAE domains and the string representing two connecting, largely unstructured sequences (the more carboxy-terminal being the hinge) (7). Indeed, the frictional ratio of endogenous GGA3 calculated from our gel filtration and sedimentation velocity data (Fig. 7) is ∼3, much greater than the ∼1 value typical of globular proteins. Thus, GGA3 is quite asymmetric, in large part due to the length and unstructured nature of the hinge. Such a structure is ideally suited for conformational changes.
We found that phosphorylation of S368 resulted in faster elution of GGA3 on gel filtration. Since the molecular mass of a single phosphate group is negligible in this type of analysis, and no other proteins coprecipitate quantitatively with GGA3 upon EGF treatment of the cells (data not shown), the faster elution of S368-phosphorylated GGA3 must be due to increased asymmetry. This could result from the disruption of an interdomain interaction, such as those previously reported to occur between the VHS-hinge (14) and VHS-GAT (20) domains. Alternatively, the hinge domain could have local stretches of secondary structure that unfold upon phosphorylation of S368. This possibility appears more likely, since the isolated hinge itself exhibits phosphorylation-dependent mobility shifts on SDS-PAGE (Fig. 4B) and gel filtration (Fig. 7C).
The occurrence of a conformational change in GGA1 has already been reported to occur upon phosphorylation of hinge residue S355 by casein kinase II (18). This phosphorylation and the ensuing conformational change promote interaction of the VHS domain with an internal DXXLL-type motif of GGA1, which in turn inhibits binding of GGA1 to DXXLL motifs from cargo proteins (14, 15, 18). Our findings on GGA3 differ in several respects from these previous observations on GGA1. First, phosphorylation of GGA3 S368 causes a mobility shift on SDS-PAGE that is not observed for phosphorylation of GGA1 S355. Second, and more significantly, phosphorylation of GGA3 S368 increases the Stokes radius of the protein from 49 to 54 Å (Fig. 7B), whereas phosphorylation of GGA1 S355 decreases the Stokes radius from 60 to 40Å (18). Hence, the phosphorylation-induced conformational change of GGA3 is of a smaller magnitude and opposite in sense relative to that of GGA1. Third, unlike the phosphorylation of GGA1 S355 (14, 15), phosphorylation of GGA3 S368 apparently does not affect the binding of the VHS domain of this protein to the DXXLL signal-containing tail of the cargo cation-independent MPR (Fig. 8B). Thus, it appears that these two modifications, casein kinase II-dependent phosphorylation of GGA1 and EGF-stimulated phosphorylation of GGA3, have fundamentally different effects on the conformation and signal recognition activities of the corresponding GGA proteins.
S368 regulates the association of GGA3 with membranes.
Subcellular fractionation analyses indicated that phosphorylation on S368 decreases association of GGA3 with membranes. Consistent with this notion, mutation of S368 to alanine increased the initial rate of association of fluorescently tagged GGA3 to the TGN in vivo, without affecting the initial rate of dissociation (Fig. 9). The decreased recruitment of S368-phosphorylated GGA3 to membranes could be due to impaired interaction with some of its binding partners. Interestingly, two-hybrid analyses showed that phosphomimic mutations of S368 and S372 to aspartate decreased interactions of GGA3 with ubiquitin, though not with the DXXLL signal-containing cytosolic tail of the cation-independent MPR or the GTP-locked form of Arf1 (Fig. 8B). Ubiquitin binds to two sites on the three-helix bundle subdomain of the GAT domain of GGA3 (3, 36, 44, 32). The two-hybrid data would therefore suggest that the phosphomimic mutations in the hinge region result in occlusion of the ubiquitin binding sites in the GAT domain. This occlusion would only affect the three-helix bundle subdomain, however, as the binding of GTP-Arf to the helix-loop-helix subdomain of GAT is unchanged by these mutations. Another possibility is that the S368-unphosphorylated hinge region enhances interactions with ubiquitin, an enhancement that could be abrogated by S368 phosphorylation. In any event, reduced binding to ubiquitinated cargo or the loss of a regulatory ubiquitin interaction might underlie the decreased association of S368-phosphorylated GGA3 with membranes. At this point we cannot rule out, however, that changes in interactions with other partners not tested in our study also contribute to this decrease.
The physiological consequences of the regulation of GGA3 association to membranes by EGF remain to be determined. One possibility is that EGF regulates the fate of its own receptor by modulating the activity of GGA3 in the recognition of ubiquitinated cargo in endosomes (36). An alternative possibility is that EGF controls the sorting of proteins at the TGN by either reducing the direct recognition of ubiquitinated cargo or the sorting of nonubiquitinated cargo through regulatory ubiquitin interactions. In this regard it is worth mentioning that oncogenically transformed cells exhibit altered secretion of lysosomal hydrolases, the cargo for the MPRs (45, 1). Moreover, short-term treatment of fibroblasts with PDGF diverts the lysosomal hydrolase cathepsin L from lysosomal delivery to secretion (33). It is then possible that the transforming signals elicited by PDGF and EGF impair GGA3 association with membranes and, consequently, sorting of MPRs, eventually leading to increased lysosomal enzyme secretion.
Concluding remarks.
The results presented here constitute the first demonstration that serine kinases downstream of signaling receptors such as the EGF and PDGF receptors regulate the phosphorylation and membrane association of an adaptor coat protein, GGA3. This indirect phosphorylation on serine residues may exemplify a novel mode of regulation of components of the endosomal-lysosomal trafficking machinery, in particular those that are not immediately responsible for internalization of signaling receptors but function in intracellular sorting events.
Acknowledgments
We thank X. Zhu for excellent technical assistance and Y. Wakabayashi for helpful discussions. We also thank S. Schmid, J. Lippincott-Schwartz, M. S. Robinson, and D. Brooks for kind gifts of reagents.
REFERENCES
- 1.Achkar, C., Q. M. Gong, A. Frankfater, and A. S. Bajkowski. 1990. Differences in targeting and secretion of cathepsins B and L by BALB/3T3 fibroblasts and Moloney murine sarcoma virus-transformed BALB/3T3 fibroblasts. J. Biol. Chem. 265:13650-13654. [PubMed] [Google Scholar]
- 2.Bache, K. G., C. Raiborg, A. Mehlum, I. H. Madshus, and H. Stenmark. 2002. Phosphorylation of Hrs downstream of the epidermal growth factor receptor. Eur. J. Biochem. 269:3881-3887. [DOI] [PubMed] [Google Scholar]
- 3.Bilodeau, P. S., S. C. Winistorfer, M. M. Allaman, K. Surendhran, W. R. Kearney, A. D. Robertson, and R. C. Piper. 2004. The GAT domains of clathrin-associated GGA proteins have two ubiquitin-binding motifs. J. Biol. Chem. 279:54808-54816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blagoev, B., I. Kratchmarova, S. E. Ong, M. Nielsen, L. J. Foster, and M. Mann. 2003. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21:315-318. [DOI] [PubMed] [Google Scholar]
- 5.Boman, A. L., C. J. Zhang, X. Zhu, and R. A. Kahn. 2000. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11:1241-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S. 2004. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell. Biol. 5:23-32. [DOI] [PubMed] [Google Scholar]
- 7.Collins, B. M., P. J. Watson, and D. J. Owen. 2003. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev. Cell 4:321-332. [DOI] [PubMed] [Google Scholar]
- 8.Confalonieri, S., A. E. Salcini, C. Puri, C. Tacchetti, and P. P. Di Fiore. 2000. Tyrosine phosphorylation of Eps15 is required for ligand-regulated, but not constitutive, endocytosis. J. Cell Biol. 150:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conner, S. D., and S. L. Schmid. 2002. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J. Cell Biol. 156:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damke, H., T. Baba, D. E. Warnock, and S. L. Schmid. 1994. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127:915-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damke, H., M. Gossen, S. Freundlieb, H. Bujard, and S. L. Schmid. 1995. Tightly regulated and inducible expression of dominant interfering dynamin mutant in stably transformed HeLa cells. Methods Enzymol. 257:209-220. [DOI] [PubMed] [Google Scholar]
- 12.Dell'Angelica, E. C., C. Mullins, and J. S. Bonifacino. 1999. AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem. 274:7278-7285. [DOI] [PubMed] [Google Scholar]
- 13.Dell'Angelica, E. C., R. Puertollano, C. Mullins, R. C. Aguilar, J. D. Vargas, L. M. Hartnell, and J. S. Bonifacino. 2000. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doray, B., K. Bruns, P. Ghosh, and S. A. Kornfeld. 2002. Autoinhibition of the ligand-binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc. Natl. Acad. Sci. USA 99:8072-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doray, B., P. Ghosh, J. Griffith, H. J. Geuze, and S. Kornfeld. 2002. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297:1700-1703. [DOI] [PubMed] [Google Scholar]
- 16.Fiol, C. J., A. M. Mahrenholz, Y. Wang, R. W. Roeske, and P. J. Roach. 1987. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262:14042-14048. [PubMed] [Google Scholar]
- 17.Ghosh, P., and S. Kornfeld. 2004. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell Biol. 83:257-262. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, P., and S. Kornfeld. 2003. Phosphorylation-induced conformational changes regulate GGAs 1 and 3 function at the trans-Golgi network. J. Biol. Chem. 278:14543-14549. [DOI] [PubMed] [Google Scholar]
- 19.He, X., G. Zhu, G. Koelsch, K. K. Rodgers, X. C. Zhang, and J. Tang. 2003. Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase) cytosolic domain with the VHS domain of GGA proteins. Biochemistry 42:12174-12180. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch, D. S., K. T. Stanley, L. X. Chen, K. M. Jacques, R. Puertollano, and P. A. Randazzo. 2003. Arf regulates interaction of GGA with mannose-6-phosphate receptor. Traffic 4:26-35. [DOI] [PubMed] [Google Scholar]
- 21.Hirschberg, K., C. M. Miller, J. Ellenberg, J. F. Presley, E. D. Siggia, R. D. Phair, and J. Lippincott-Schwartz. 1998. Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J. Cell Biol. 143:1485-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst, J., W. W. Lui, N. A. Bright, N. Totty, M. N. Seaman, and M. S. Robinson. 2000. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, F., X. Jiang, and A. Sorkin. 2003. Tyrosine phosphorylation of the beta2 subunit of clathrin adaptor complex AP-2 reveals the role of a di-leucine motif in the epidermal growth factor receptor trafficking. J. Biol. Chem. 278:43411-43417. [DOI] [PubMed] [Google Scholar]
- 24.Kato, Y., S. Misra, R. Puertollano, J. H. Hurley, and J. S. Bonifacino. 2002. Phosphoregulation of sorting signal VHS domain interactions by a direct electrostatic mechanism. Nat. Struct. Biol. 9:532-536. [DOI] [PubMed] [Google Scholar]
- 25.Kennelly, P. J., and E. G. Krebs. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266:15555-15558. [PubMed] [Google Scholar]
- 26.Marks, M. S. 1998. Determination of molecular size by sedimentation velocity analysis on sucrose gradients, p. 5.3.1-5.3.33. In J. S. Bonifacino, M. Dasso, J. B. Harford, J. Lippincott-Schwartz, and K. Yamada (ed.), Current protocols in cell biology. John Wiley & Sons, New York, N.Y.
- 27.Mattera, R., C. N. Arighi, R. Lodge, M. Zerial, and J. S. Bonifacino. 2003. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 22:78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKay, M. M., and R. A. Kahn. 2004. Multiple phosphorylation events regulate the subcellular localization of GGA1. Traffic 5:102-116. [DOI] [PubMed] [Google Scholar]
- 29.McPherson, P. S., B. K. Kay, and N. K. Hussain. 2001. Signaling on the endocytic pathway. Traffic 2:375-384. [DOI] [PubMed] [Google Scholar]
- 30.Patterson, G. H., and J. Lippincott-Schwartz. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297:1873-1877. [DOI] [PubMed] [Google Scholar]
- 31.Pinna, L. A., and M. Ruzzene. 1996. How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1314:191-225. [DOI] [PubMed] [Google Scholar]
- 32.Prag, G., S. Lee, R. Mattera, C. N. Arighi, B. M. Beach, J. S. Bonifacino, and J. H. Hurley. 2005. Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins. Proc. Natl. Acad. Sci. USA 102:2334-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prence, E. M., J. M. Dong, and G. G. Sahagian. 1990. Modulation of the transport of a lysosomal enzyme by PDGF. J. Cell Biol. 110:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Presley, J. F., T. H. Ward, A. Pfeiffer, E. D. Siggia, R. D. Phair, and J. Lippincott-Schwartz. 2002. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature 417:187-193. [DOI] [PubMed] [Google Scholar]
- 35.Puertollano, R., R. C. Aguilar, I. Gorshkova, R. J. Crouch, and J. S. Bonifacino. 2001. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292:1712-1716. [DOI] [PubMed] [Google Scholar]
- 36.Puertollano, R., and J. S. Bonifacino. 2004. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 6:244-251. [DOI] [PubMed] [Google Scholar]
- 37.Puertollano, R., P. Randazzo, L. M. Hartnell, J. Presley, and J. S. Bonifacino. 2001. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105:93-102. [DOI] [PubMed] [Google Scholar]
- 38.Puertollano, R., N. N. van der Wel, L. E. Green, E. Eisenberg, P. J. Peters, and J. S. Bonifacino. 2003. Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol. Biol. Cell 14:1545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricotta, D., S. D. Conner, S. L. Schmid, K. von Figura, and S. Honing. 2002. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, M. S. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14:167-174. [DOI] [PubMed] [Google Scholar]
- 41.Saito, Y., J. R. Vandenheede, and P. Cohen. 1994. The mechanism by which epidermal growth factor inhibits glycogen synthase kinase 3 in A431 cells. Biochem. J. 303:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott, P. M., P. S. Bilodeau, O. Zhdankina, S. C. Winistorfer, M. J. Hauglund, M. M. Allaman, W. R. Kearney, A. D. Robertson, A. L. Boman, and R. C. Piper. 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6:252-259. [DOI] [PubMed] [Google Scholar]
- 43.Shiba, T., S. Kametaka, M. Kawasaki, M. Shibata, S. Waguri, Y. Uchiyama, and S. Wakatsuki. 2004. Insights into the phosphoregulation of beta-secretase sorting signal by the VHS domain of GGA1. Traffic 5:437-448. [DOI] [PubMed] [Google Scholar]
- 44.Shiba, Y., Y. Katoh, T. Shiba, K. Yoshino, H. Takatsu, H. Kobayashi, H. W. Shin, S. Wakatsuki, and K. Nakayama. 2004. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 279:7105-7111. [DOI] [PubMed] [Google Scholar]
- 45.Troen, B. R., D. Ascherman, D. Atlas, and M. M. Gottesman. 1988. Cloning and expression of the gene for the major excreted protein of transformed mouse fibroblasts. A secreted lysosomal protease regulated by transformation. J. Biol. Chem. 263:254-261. [PubMed] [Google Scholar]
- 46.Urbe, S., I. G. Mills, H. Stenmark, N. Kitamura, and M. J. Clague. 2000. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol. Cell. Biol. 20:7685-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakasugi, M., S. Waguri, S. Kametaka, Y. Tomiyama, S. Kanamori, Y. Shiba, K. Nakayama, and Y. Uchiyama. 2003. Predominant expression of the short form of GGA3 in human cell lines and tissues. Biochem. Biophys. Res. Commun. 306:687-692. [DOI] [PubMed] [Google Scholar]
- 48.Wilde, A., E. C. Beattie, L. Lem, D. A. Riethof, S. H. Liu, W. C. Mobley, P. Soriano, and F. M. Brodsky. 1999. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell 96:677-687. [DOI] [PubMed] [Google Scholar]
- 49.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]