Abstract
In an effort to identify novel genes involved in recombination repair, we isolated fission yeast Schizosaccharomyces pombe mutants sensitive to methyl methanesulfonate (MMS) and a synthetic lethal with rad2. A gene that complements such mutations was isolated from the S. pombe genomic library, and subsequent analysis identified it as the fbh1 gene encoding the F-box DNA helicase, which is conserved in mammals but not conserved in Saccharomyces cerevisiae. An fbh1 deletion mutant is moderately sensitive to UV, MMS, and γ rays. The rhp51 (RAD51 ortholog) mutation is epistatic to fbh1. fbh1 is essential for viability in stationary-phase cells and in the absence of either Srs2 or Rqh1 DNA helicase. In each case, lethality is suppressed by deletion of the recombination gene rhp57. These results suggested that fbh1 acts downstream of rhp51 and rhp57. Following UV irradiation or entry into the stationary phase, nuclear chromosomal domains of the fbh1Δ mutant shrank, and accumulation of some recombination intermediates was suggested by pulsed-field gel electrophoresis. Focus formation of Fbh1 protein was induced by treatment that damages DNA. Thus, the F-box DNA helicase appears to process toxic recombination intermediates, the formation of which is dependent on the function of Rhp51.
Homologous recombination not only shuffles genetic information upon sexual reproduction but also repairs damaged DNA by use of the homologous information. Furthermore, it can regenerate replication forks when they become stalled or collapsed.
Molecular mechanisms of homologous recombination in eukaryotes have been most extensively studied in the budding yeast Saccharomyces cerevisiae (reviewed in references3, 27, 42, 46, and 48). In this yeast, the MRX (Mre11 Rad50 Xrs2) complex is required for the processing of double-strand break ends to generate 3′-protruding ends. The resulting single-strand regions are coated by single-strand-binding protein RPA (replication protein A). Rad52 stimulates loading of Rad51 on RPA-coated single-strand DNA to form Rad51 nucleoprotein filament. A complex of Rad55 and Rad57, which are Rad51 paralogs, is also implicated in the assembly and stabilization of Rad51 nucleoprotein filament. Rad51 nucleoprotein filament searches homologous sequences and catalyzes the exchange of strands to form a heteroduplex joint called a D loop. Rad54 facilitates D-loop formation by remodeling chromatin structures. The annealed 3′ ends are then used as primers for repair DNA synthesis. The resulting junction molecules are resolved either by dissociation of the crossed strands or by cutting of the junction point. The Rad52 group proteins (Rad50, Rad51, Rad52, Rad54, Rad55, Rad57, Mre11, and Xrs2) are conserved throughout eukaryotes, indicating a conservation of the molecular mechanisms pertaining to homologous recombination.
In addition to the aforementioned recombination factors, DNA helicases Srs2 and Sgs1 have been implicated to be involved in the regulation of homologous recombination (reviewed in reference 6). Srs2 dissociates the Rad51 protein from nucleoprotein filament to suppress toxic recombination intermediates (26, 53). Although Srs2 is conserved in fungi, no apparent Srs2 ortholog has been found in higher eukaryotes. Sgs1 is homologous to Escherichia coli RecQ, Schizosaccharomyces pombe Rqh1, and mammalian WRN, BLM, and RTS helicases. These RecQ family helicases have been implicated to play roles in recombination at various stages such as recombination initiation, reversal or prevention of fork regression, and resolution of recombination intermediates (reviewed in reference 4). In S. cerevisiae, the srs2 sgs1 double mutant is severely impaired in growth (28). This growth defect can be overcome by rad51 mutation (15), indicating that recombination initiated by the Rad51 protein is toxic in the srs2 sgs1 background.
Replication forks become stalled when they encounters obstacles such as chemically modified bases, pyrimidine dimers that are generated by UV irradiation, proteins tightly associated with DNA, or certain DNA tertiary structures (reviewed in reference 11). The stalled replication forks are overridden by translesion polymerases, regressed to bypass the lesion by template switching or reinitiated in a manner dependent on homologous recombination (reviewed in references 6, 11, and 31-33). Replication forks collapse when they encounter a DNA nick or gap, and the nascent fork is regenerated at the site by homologous recombination (24).
In the fission yeast Schizosaccharomyces pombe, all of the above-mentioned RAD52 group genes, in addition to srs2 and the recQ homolog rqh1, are conserved and have been implicated to be involved in recombination repair (9, 14, 30, 39, 40, 49, 51).
In an effort to identify further factors involved in recombination repair, we isolated fission yeast S. pombe mutants hypersensitive to methanesulfonate (MMS) and synthetic lethal with a rad2 mutation (50). rad2 encodes flap endonuclease which removes the 5′ terminus of the Okazaki fragment (41). Mutants in flap endonuclease require recombination for survival in various organisms (24, 47, 54). In these mutants, Okazaki fragments often remain unjoined. If replication forks encounter such nonrejoined sites, they are thought to generate double-strand break (DSB) ends, and homologous recombination is required to repair the DSB ends to maintain survival (24). Thus, synthetic lethal mutants with rad2 can be expected to be defective in the regeneration of collapsed replication forks by recombination. Following the screening of such mutants, we identified rhp57 (50), rad32 (unpublished), nbs1 (51), rad60 (36), and rad62 (35). In this study, we describe the isolation and characterization of mutants of the fbh1 gene encoding the F-box DNA helicase. The Fbh1 protein was independently identified through purification of a novel S. pombe DNA helicase by Park and colleagues (43). They showed that human Fbh1 forms an SCF ubiquitin ligase complex (22, 23). However, the role of Fbh1 in vivo remains unknown. Here, we characterized the function of the fbh1 gene in S. pombe. The results showed that Fbh1 functions in recombination repair on the Rhp51 (S. pombe Rad51 ortholog) pathway downstream of Rhp51 and plays a role in processing recombination intermediates.
MATERIALS AND METHODS
S. pombe media, methods, and strains.
S. pombe cells were grown in YES or EMM medium (34), and standard genetic and molecular procedures were employed as described previously (34). The S. pombe strains used in this study are listed in Table 1. Sensitivity of S. pombe cells to γ rays and UV irradiation was analyzed as previously described (36). To determine MMS sensitivity, cells were incubated with MMS in YES medium, appropriately diluted following the specified incubation time, and then spread on YES plates. The number of colonies was scored following incubation for 3 to 5 days at 30°C.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| B54 | smt-0 rhp51::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | Y. Tsutsui |
| B63 | smt-0 rhp57::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | Y. Tsutsui |
| TE767 | h−rqh1::ura4 ura4-D18 | T. Enoch, reference (40) |
| MP110 | h−leu1-32 ura4-D18 | This study |
| MP111 | h+leu1-32 ura4-D18 | This study |
| MPM53 | h+leu1-32 ura4-D18 fbh1-1 | This study |
| MPM73 | h+leu1-32 ura4-D18 fbh1-2 | This study |
| MPF1 | h+leu1-32 ura4-D18 fbh1::LEU2 | This study |
| MPF2 | h−leu1-32 ura4-D18 fbh1::LEU2 | This study |
| MPF3 | h+leu1-32 ura4-D18 fbh1::LEU2 | This study |
| MPF21 | h+fbh1::LEU2 rhp57::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF22 | h+fbh1::LEU2 rhp57::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF23 | h+fbh1::LEU2 rhp57::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF24 | h+fbh1::LEU2 rhp51::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF25 | smt-0 fbh1::LEU2 rhp51::his3+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF41 | h−srs2::Kanrura4-D18 leu1-32 | This study |
| MPF42 | h−srs2::Kanrura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF43 | h−rqh1::ura4+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF44 | h+rqh1::ura4+ura4-D18 leu1-32 his3-D1 arg3-D1 | This study |
| MPF51 | h+leu1-32 ura4-D18 fbh1int::pREP42-EGFPN-fbh1+ | This study |
| MPF52 | h+leu1-32 ura4-D18 rhp51::his3+fbh1int::pREP42-EGFPN-fbh1+ | This study |
Cloning of the fbh1 gene that complements the fbh1-1 and fbh1-2 mutations.
fbh1-1 and fbh1-2 cells were transformed with the S. pombe genomic library (5) constructed using vector pUR19 and spread on EMM plates containing leucine (200 μg/ml) and MMS (0.004%). Transformants were examined for plasmid-dependent MMS resistance. Plasmids that complemented the MMS sensitivity of the fbh1-1 or fbh1-2 cells were isolated, transformed into E. coli DH5α, and subsequently recovered from the transformants.
Disruption of the fbh1 gene.
The chromosomal fbh1 gene was disrupted by a method employing two PCR steps as previously described (25). The region upstream of the fbh1 coding region was amplified using the primers MVF2 (5′-ACACAAAAAGTAATAGAGTC-3′) and MVD-5 (5′-GTCGTGACTGGGAAAACCCTGGCGTTACCCATAACTAAGAATTTGCTGAC-3′). The region downstream of the fbh1 coding region was amplified using the primers MVD-3 (5′-TCCTGTGTGAAATTGTTATCCGCTCACAATTAGAAACTATTTGATTTGTT-3′) and MVR3 (5′-TGAAATCATCTTTATGATG-3′). The resulting two fragments, in addition to the primers MVF2 and MVR3, were used to amplify the LEU2 sequence of pJJ282 (20) to generate the fragment for disruption. This fragment was used to transform the haploid S. pombe strain MP111, and a transformant with the appropriate disruption was verified by PCR.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis was carried out as previously described (36), except that a 0.5% Megabase agarose (Bio-Rad, Hercules, Calif.) gel and 1× TAE buffer (40 mM Tris-acetate and 1 mM EDTA) were used and run for 60 h at a 120° angle.
Indirect immunofluorescent staining of Rhp51.
S. pombe cells were fixed and stained for Rhp51 as previously described (8), except that cells were fixed using 3.7% formaldehyde for 30 min. The primary antibody consisted of a rabbit polyclonal antibody raised against recombinant Rhp51 protein expressed in E. coli and diluted 500-fold. The secondary antibody was goat anti-rabbit immunoglobulin G conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.) and diluted 1,000-fold.
Expression of the EGFP-Fbh1 fusion protein in S. pombe cells.
fbh1 cDNA was cloned into the plasmid pREP42 EGFP N (13) to express enhanced green fluorescent protein (EGFP) fusion protein under control of the medium-strength nmt1 promoter. The resulting plasmid was linearized at the unique NheI site within the fbh1 cDNA and introduced into the fbh1 locus of the S. pombe genome. The EGFP-Fbh1 fusion protein was expressed by culturing the cells in EMM medium with the appropriate supplements in the absence of thiamine. Cells were fixed with 70% ethanol and observed by fluorescence microscopy.
Measurement of cell length of cells expressing EGFP-Fbh1.
Cells expressing EGFP-Fbh1 were treated with 0.1% MMS in EMM medium containing leucine (200 μg/ml) for 1 h, washed twice with EMM2, and then fixed with 70% ethanol. Cells were then observed by fluorescence microscopy, and consecutive focal planes of 1-μm distances were photographed. The cell length was measured directly from the photographs, and cells that carried foci in any of the focal planes were scored as positive for foci.
RESULTS
Cloning of the fbh1 gene that complements S. pombe DNA repair-deficient mutations.
The S. pombe genomic library was screened for complementation of the MMS-sensitive phenotype of the two mutants previously isolated (50). Identical plasmid clones carrying exactly the same fragment of an S. pombe genomic region were obtained independently from the two mutants, and they carried a single open reading frame, SPBC336.01 (fbh1). Therefore, the two mutants will be referred to as fbh1-1 and fbh1-2, respectively. fbh1 encodes a polypeptide comprised of 878 amino acids containing at its N terminus the F-box motif which is known to interact with the Skp1 protein to form an SCF ubiquitin ligase (E3) complex (12) and seven helicase motifs of the superfamily 1 helicases (16) (Fig. 1A).
FIG. 1.
Identification of the F-box DNA helicase as a factor implicated in recombination repair. (A) Schematic presentation of the distribution of the F-box motif and seven helicase motifs in fission yeast Fbh1 and human Fbh1 proteins, including the fbh1-1 and fbh1-2 mutation sites. (B to D) Epistasis between fbh1 and rhp51. Survival curves of wild-type (MP111), fbh1Δ (MPF3), rhp51Δ (B54), and fbh1Δ rhp51Δ (MPF25) cells exposed to UV irradiation (B), MMS (0.02%) (C), and γ rays (D) are shown.
The fbh1 region of the fbh1-1 mutant was recovered utilizing the eviction method (54), and the region obtained carried a single G-to-A nucleotide change, in effect altering the GAA codon for glutamic acid 725 to an AAA codon for lysine. This glutamic acid 725 residue corresponds to a conserved residue within the helicase motif V (Fig. 1A), indicating that helicase activity is important for fbh1 function in vivo. The fbh1 region of the fbh1-2 mutant was amplified by PCR, and the nucleotide sequence was determined. It possessed a single T-to-C nucleotide change, in effect altering the TGG codon for tryptophan 839 near the C terminus to a CGG codon for arginine (Fig. 1A), suggesting that the C-terminal region is important for fbh1 function in vivo.
The fbh1Δ mutant is sensitive to MMS and a synthetic lethal with rad2Δ.
The fbh1 gene of the S. pombe haploid strain MP111 was disrupted by replacing the entire coding region of the fbh1 gene with the LEU2 marker. The resulting fbh1Δ strain showed an MMS-sensitive phenotype. The fbh1Δ strain was crossed with a wild-type strain and subjected to tetrad analysis. The segregants showed 2+:2− segregation for leucine prototroph and MMS-sensitive phenotypes, where both phenotypes always cosegregated, indicating that the fbh1Δ cells are viable and MMS sensitive. The fbh1Δ strain was crossed with a rad2Δ strain, and the resulting strain was subjected to tetrad analysis. Among the 10 tetrads dissected, no Leu+ Ura+ viable segregants were obtained, indicating that the fbh1Δ rad2Δ double mutant is lethal. Generation time and plating efficiency of fbh1 cells were 3.7 h and 39%, respectively, while in wild-type cells, they were 2.4 h and 96%, respectively, on YES medium at 30°C. Spontaneous recombination frequency between direct repeats was not affected by the fbh1 mutation (data not shown).
fbh1 works on the rhp51 pathway for recombination repair.
fbh1Δ cells were more sensitive to UV irradiation, MMS, and γ rays than wild-type cells (Fig. 1B to D). Homologous recombination represents a major pathway for the repair of radiation-induced DSBs in S. pombe. rhp51, the S. pombe ortholog of S. cerevisiae RAD51 (19, 37), plays a central role in this process (39). Therefore, the relationship between fbh1 and rhp51 was examined. As shown in Fig. 1B to D, the rhp51Δ mutant was more sensitive to UV irradiation, MMS, and γ rays than the fbh1Δ mutant. The fbh1Δ rhp51Δ double mutant and the rhp51Δ single mutant showed similar sensitivity to UV irradiation, MMS, and γ rays (Fig. 1B to D). These results indicate that rhp51 is epistatic to fbh1 with respect to DNA repair and that fbh1 works in the rhp51 pathway related to recombination repair.
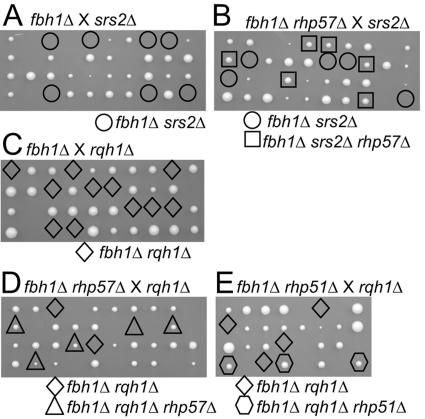
Synthetic growth defects of the fbh1Δ srs2Δ and fbh1Δ rqh1Δ double mutants are suppressed by loss of the recombination gene rhp57.
Fbh1 helicase belongs to superfamily 1 of helicases that includes Srs2. Therefore, we examined the functional relationship between fbh1 and srs2. The fbh1Δ strain was crossed with the srs2Δ strain, and the resulting spores were subjected to tetrad analysis. Of the 10 tetrads dissected, no fbh1Δ srs2Δ segregants gave viable colonies, indicating that the fbh1Δ srs2Δ double mutants are lethal (Fig. 2A). rhp57 is the S. pombe ortholog of S. cerevisiae RAD57, whose product forms a heterodimer with Rad55 and is implicated in the loading of Rad51 to RPA-coated single-strand DNA (46). Rhp57 and Rhp55 play a role in the subpathway of Rhp51 since rhp57 and rhp55 mutants are less sensitive to DNA-damaging agents than the rhp51 mutant (1, 21, 50). The fbh1Δ rhp57Δ double mutant was crossed with the srs2Δ mutant, and the resulting spores were subjected to tetrad analysis. Of the 10 tetrads dissected, six fbh1Δ srs2Δ rhp57Δ triple mutants and no fbh1Δ srs2Δ double mutants gave viable colonies (Fig. 2B). The size of the fbh1Δ srs2Δ rhp57Δ triple mutant colonies was comparable to that of rhp57Δ cells, indicating that the rhp57Δ mutation suppressed the lethality of the fbh1Δ srs2Δ double mutant. We were unable to determine whether rhp51Δ restored srs2Δ fbh1Δ inviability, since crosses between fbh1Δ rhp51Δ and srs2Δ strains produced only a few viable segregants (13 segregants out of 10 tetrads) by tetrad analysis. Low viability of the rhp51 cells could to some extent contribute to this spore inviability.
FIG. 2.
Lethality of fbh1Δ srs2Δ and fbh1Δ rqh1Δ mutants that is suppressed by loss of recombination genes. Spores of crosses between fbh1Δ (MPF3) and srs2Δ (MPF41) (A), fbh1Δ rhp57Δ (MPF21) and srs2Δ (MPF42) (B), fbh1Δ (MPF3) and rqh1Δ (MPF43) (C), fbh1Δ rhp57Δ (MPF22) and rqh1Δ (MPF43) (D), or fbh1Δ rhp51Δ (MPF25) and rqh1Δ (MPF44) (E) were subjected to tetrad analysis. Genotypes of inviable segregants were predicted by assuming Mendelian inheritance.
In S. cerevisiae, the srs2Δ sgs1Δ double mutants are lethal or severely defective in growth, and the growth defect is suppressed by loss of the RAD51 gene (15, 28). A similar relationship has been shown in S. pombe, where the srs2Δ rqh1Δ double mutants are severely impaired for growth and the growth defect is suppressed by loss of the rhp57 or rhp51 gene (14, 30). The relationship between fbh1Δ and rqh1Δ was analyzed by crossing an fbh1Δ and an rqh1Δ strain. Of the 10 tetrads dissected, no fbh1Δ rqh1Δ segregants gave viable colonies, indicating that the fbh1Δ rqh1Δ double mutants are lethal (Fig. 2C). The fbh1Δ rhp57Δ double mutant was crossed with the rqh1Δ mutant, and the resulting spores were subjected to tetrad analysis. Of the 10 tetrads dissected, four fbh1Δ rqh1Δ rhp57Δ triple mutants and no fbh1Δ rqh1Δ double mutants gave viable colonies (Fig. 2D). The size of the fbh1Δ rqh1Δ rhp57Δ triple mutant colonies was comparable to that of rhp57Δ cells, indicating that the rhp57Δ mutation suppressed the lethality of the fbh1Δ rqh1Δ double mutant. Colonies from the tetrads of the cross between fbh1Δ rhp51Δ and rqh1Δ gave rise to three fbh1Δ rqh1Δ rhp51Δ triple mutants and no fbh1Δ rqh1Δ double mutants (Fig. 2E), indicating that the rhp51 mutation suppressed the lethality of the fbh1Δ rqh1Δ double mutant. These results indicate that homologous recombination is responsible for cell death in the fbh1Δ srs2Δ and fbh1Δ rqh1Δ double mutants and suggest that the three helicase genes fbh1, rqh1, and srs2 act downstream of rhp51 and rhp57.
Nuclear chromosomal domain shrinks in the fbh1Δ mutant and extends in the rhp51Δ and rhp51Δ fbh1Δ mutants following UV irradiation.
The nuclear chromosomal domains of fbh1Δ mutant cells were examined by staining with 4′,6′-diamidino-2-phenylindole (DAPI). Six hours after UV irradiation, the nuclear chromosomal domains of approximately half of the fbh1Δ cells appeared like a compact sphere and were smaller than the hemispherical nuclear chromosomal domains of wild-type cells (Fig. 3A). Additionally, staining of the nucleolus with ethidium bromide highlighted the difference in nuclear morphology more clearly. In the fbh1Δ mutant, the nuclear chromosomal domains and nucleolus (stained less brightly) formed separate spheres, while the nuclear chromosomal domains and nucleolus of wild-type cells constituted the same spheres (Fig. 3B). In contrast to the fbh1Δ mutant, the nuclear chromosomal domains of the rhp51Δ mutant and the fbh1Δ rhp51Δ double mutant were more extended and amorphous in contrast to the hemispherical shape of wild-type cells when observed 6 h after UV irradiation (Fig. 3A). Thus, rhp51Δ is epistatic to fbh1Δ with respect to the morphology of the nuclear chromosomal domain following UV irradiation. This also suggests that fbh1 functions downstream of rhp51.
FIG. 3.
Nuclear shrinkage and chromosomal aberration of the fbh1 mutant following UV irradiation. (A) Cells of wild-type (MP111), fbh1Δ (MPF3), rhp51Δ (B54), and fbh1Δ rhp51Δ (MPF25) strains were UV irradiated (200 J/m2), cultured for 6 h at 30°C, fixed with glutaraldehyde (2.5%), stained with DAPI (1 μg/ml), and then photographed using a fluorescence microscope. Representative cells with shrunken or normal nuclei are shown. The scale bar indicates 10 μm. (B) fbh1Δ (MPF3) cells or wild-type (MP111) cells UV irradiated and fixed as described above (A) were stained with DAPI (1 μg/ml) and ethidium bromide (10 μg/ml) and then photographed using a fluorescence microscope. The chromosomal region stained with DAPI and nucleolar region (less bright) stained with ethidium bromide are shown by arrows and arrowheads, respectively. The scale bar indicates 10 μm. (C) Wild-type (MPF111), fbh1Δ (MPF3), rhp51Δ (B54), and fbh1Δ rhp51Δ (MPF25) cells were UV irradiated (200 J/m2). Chromosomes of cells before (−) and 2, 4, or 6 h after UV irradiation were analyzed by pulsed-field gel electrophoresis. Chromosomes from 108 cells were loaded onto each lane. The three chromosomes of S. pombe are indicated by I, II, and III, respectively. (D and E) Wild-type (MP111) or fbh1Δ (MPF3) cells were UV irradiated (200 J/m2) and then cultivated in YES medium at 30°C. Cell numbers were scored at the indicated time after irradiation (D). Cells were fixed in 70% ethanol and processed for fluorescence-activated cell sorter analysis (E) as described previously (45).
Recombination intermediates remain unresolved in the fbh1Δ mutant after UV irradiation.
The difference in nuclear morphology described above suggests a difference in chromosomal structure among the wild-type, fbh1Δ, and rhp51Δ cells grown after UV irradiation. Therefore, we analyzed chromosomes in these mutants by pulsed-field gel electrophoresis.
The intensity of the three chromosomal bands decreased dramatically 2 h after UV irradiation of wild-type cells, whereas the band intensity remained largely unaltered for at least up to 2 h after UV irradiation of rhp51Δ mutants (Fig. 3C). This difference probably reflects the formation of certain recombination intermediates dependent on the function of Rhp51, which migrated poorly into the gel and therefore remained at the position of the loading well. These intermediates that had accumulated in the fbh1Δ cells are not likely to be replication intermediates, since the major population of exponentially growing S. pombe cells are at G2 phase (29), and UV irradiation prevents entry of G2 cells into mitosis (2). Consistent with this, cell number and DNA contents only slightly increased during 2 h after UV irradiation in either wild-type or the fbh1Δ strains (Fig. 3D and E). The intensity of the chromosomal bands was recovered 4 h following UV irradiation of wild-type cells to the level of the unirradiated control (Fig. 3C), indicating that the recombination intermediates had been resolved. In contrast, the signals of the three chromosomes was not recovered up to 4 h after UV irradiation in the fbh1Δ mutant (Fig. 3C), indicating that recombination intermediates remained unresolved in the fbh1Δ mutant. In the fbh1Δ rhp51Δ mutant, the chromosomal signal pattern was not altered upon incubation after UV irradiation at least up to 2 h and similar to that of the rhp51Δ mutant (Fig. 3C), indicating that rhp51Δ is epistatic to fbh1Δ in this regard. These results suggested that the fbh1Δ mutant is defective in the processing of recombination intermediates, the formation of which is dependent on Rhp51 function.
The fbh1Δ mutant dies following entry into the stationary phase, and this lethality is suppressed by the rhp57Δ mutation.
During maintenance of the fbh1Δ mutant, we found that the fbh1Δ mutant has a defect in growth recovery when the culture in stationary phase was diluted with fresh medium. The growth kinetics of the fbh1Δ mutant were therefore examined from log phase to the stationary phase. The fbh1Δ mutant ceased growth at a lower cell density than the wild-type strain, i.e., at ca. 4 × 107 cells/ml for fbh1Δ and at ca. 1.5 × 108 cells/ml for the wild type (Fig. 4A). When the fbh1Δ mutant reached this maximum cell density (time, 16 h [Fig. 4A]), the proportion of viable cells dropped to ca. 10% (time, 16 h [Fig. 4B]) and continued dropping following further incubation. This suggested that the fbh1Δ mutant dies at a late growth phase and is thus unable to reach the cell density of wild-type cells at the stationary phase. The viability of the rhp51Δ and fbh1Δ rhp51Δ mutants also decreased upon entry into the stationary phase (Fig. 4B), even though the time course of their cell death was slower than that of the fbh1Δ mutant. This indicates that recombination is required for survival of the cells entering into the stationary phase. In contrast to fbh1Δ cells, fbh1Δ rhp57Δ double mutant cells did not die upon entry into the stationary phase (Fig. 4B), indicating that the rhp57Δ mutation suppressed the lethal phenotype of the fbh1Δ mutant entering the stationary phase. Similar to UV-irradiated fbh1Δ cells, fbh1Δ cells entering the stationary phase had shrunken nuclear chromosomal domains, and this shrinkage was also suppressed by the rhp57Δ mutation (Fig. 4C). Pulsed-field gel electrophoresis showed a dramatic decrease in the band intensities of the three chromosomes as fbh1Δ cells entered the stationary phase (Fig. 4D), indicating persistence of the DNA structure representing replication or recombination intermediates. Further incubation resulted in the appearance of a faster-migrating smear signal (22 and 30 h [Fig. 4D]), suggesting that some of the chromosomes eventually became fragmented, probably due to a failure in proper processing of the damaged DNA.
FIG. 4.
Cell death, chromosomal aberration, and nuclear shrinkage in the fbh1 mutant upon entry into the stationary phase. (A, B) Cells of wild-type (MP111), fbh1Δ (MPF3), rhp57Δ (B63), fbh1Δ rhp57Δ (MPF21), rhp51Δ (B54), and fbh1Δ rhp51Δ (MPF25) strains were grown in YES medium at 30°C from exponential phase to the stationary phase. Cells were sampled at the indicated time points and analyzed for cell number per milliliter (A) and number of CFU per milliliter. Viability of the cells was determined by dividing the number of CFU by the cell number at each time point (B). (C) Wild-type (MP111), fbh1Δ (MPF3), rhp57Δ (B63), and fbh1Δ rhp57Δ (MPF23) strains growing in mid-log phase (around 5 × 106 cells /ml) were cultured for a further 24 h in YES medium at 30°C to saturation, fixed with methanol, stained with DAPI, and then photographed using a fluorescence microscope. The scale bar indicates 10 μm. (D) Wild-type (MP111) or fbh1Δ (MPF3) cells at a density of 1 × 107 ∼ 2 × 107 cells/ml were grown in YES medium at 30°C. Cells were sampled at the indicated time points, and their chromosomes were analyzed by pulsed-field gel electrophoresis. Chromosomes of 108 cells were loaded onto each lane. The three chromosomes of S. pombe are indicated by I, II, and III, respectively.
Spontaneous formation of Rhp51 foci in the fbh1Δ mutant.
Rhp51 forms nuclear foci following DNA damage, and the foci are thought to represent recombination intermediates containing nucleoprotein filaments (8). In an effort to explore the localization of Rhp51 in the fbh1Δ mutant, Rhp51 foci were stained by an indirect immunofluorescent method employing anti-Rhp51 polyclonal antibodies. Wild-type and fbh1Δ mutant cells were stained either prior to or following treatment with MMS (0.025%) for 1 h (Fig. 5). Rhp51 foci were observed in 35% (44 out of 125) of the fbh1Δ mutant cells and in only 6% (7 out of 108) of wild-type cells prior to MMS treatment. This suggests that spontaneously arising Rhp51 nucleoprotein filaments persist longer in fbh1Δ cells than in wild-type cells. The proportion of cells carrying Rhp51 foci reached to 68% (89 out of 130) in wild-type and 66% (89 out of 134) in fbh1Δ cells following MMS treatment, indicating that fbh1Δ cells are proficient in Rhp51 focal induction in response to treatments that damage DNA.
FIG. 5.
Rhp51 focus formation in the fbh1Δ mutant. Wild-type (MP111) or fbh1Δ (MPF3) cells without (−) or with (+) MMS (0.025%) treatment for 1 h were processed for indirect immunofluorescence staining by employing anti-Rhp51 antibody and then photographed using a fluorescence microscope. The scale bar indicates 10 μm.
Fbh1 focus formation in response to DNA damage.
In an effort to explore the localization of the Fbh1 protein in cells, we expressed an EGFP-Fbh1 fusion protein in S. pombe cells and determined its localization by fluorescence microscopy. The EGFP-Fbh1 fusion protein was expressed under the control of the medium-strength version of the modified thiamine-repressible nmt1 promoter (7). When the expression was repressed by culturing the cells in YES medium containing thiamine, the EGFP signal could not be detected following either DNA-damaging treatments or no treatment. When the expression was derepressed in synthetic medium (EMM2 with appropriate supplements) in the absence of thiamine, a faint EGFP signal was observed throughout the cell body, with the nucleus being slightly brighter than the cytoplasm under normal growth conditions (Fig. 6A). Following a 1-h incubation after irradiation with γ rays at 500 Gy, most of the cells formed nuclear foci (Fig. 6A and B), suggesting that Fbh1 forms nuclear foci in response to DNA strand breakage. Similarly, treatment with MMS (0.1%) for 1 h gave rise to nuclear foci which were restricted essentially to relatively short cells and septated cells (Fig. 6A and B). Given that S. pombe cells septate around the S phase (29), this result suggests that Fbh1 foci are formed only in cells that are in S phase during MMS treatment. These results are consistent with the notion that Fbh1 plays a role in the recombination repair of strand breaks and stalled or collapsed replication forks. In the absence of treatments that damage DNA, EGFP-Fbh1 foci were observed in only 2% (8 out of 347) of wild-type cells, while spontaneous EGFP-Fbh1 foci were observed in 41% (93 out of 225) of the rhp51Δ mutant cells (Fig. 6C). These results suggest that both Fbh1 and Rhp51 are required to repair spontaneous DNA damage and that the focus formation of them does not require the function of the other.
FIG. 6.
EGFP-Fbh1 foci induced by DNA-damaging agents or appearing spontaneously in the rhp51Δ mutant. (A) Cells expressing the EGFP-Fbh1 fusion protein (MPF51) without treatment, irradiated with γ rays (500 Gy), and cultured for 1 h at 30°C or treated with MMS (0.1%) for 1 h at 30°C were photographed using a fluorescence microscope. The scale bar indicates 10 μm. (B) Cells expressing the EGFP-Fbh1 fusion protein (MPF51) irradiated with γ rays (500 Gy) and cultured for 1 h at 30°C or treated with MMS (0.1%) for 1 h at 30°C were photographed using a fluorescence microscope, and the distribution of cell lengths was determined. Septated cells were classified separately. Open and solid bars indicate cells with or without Fbh1 foci, respectively. (C) rhp51Δ cells expressing the EGFP-Fbh1 fusion protein (MPF52) without or with MMS (0.1%) treatment for 1 h were photographed using a fluorescence microscope. The scale bar indicates 10 μm.
DISCUSSION
In this study, we presented several pieces of evidence indicating that Fbh1 functions downstream of rhp51 and rhp57 and is involved in the processing of certain forms of recombination intermediates. First, the fbh1Δ srs2Δ and fbh1Δ rqh1Δ double mutants are lethal, and this lethality is suppressed by the rhp57Δ mutation. Second, the nuclear chromosomal domain shrinks in the fbh1Δ mutant while it extends in the rhp51Δ and rhp51Δ fbh1Δ mutants following UV irradiation. Third, pulsed-field gel electrophoretic analysis indicates that fbh1Δ cells are defective in the processing of certain forms of recombination intermediates formed following UV irradiation and that rhp51Δ and rhp51Δ fbh1Δ cells are defective in the formation. Fourth, the fbh1Δ mutant dies at late log phase before entry into the stationary phase, and this lethality is suppressed by the rhp57Δ mutation. Both the Srs2 helicase and Rqh1/Sgs1 RecQ-like helicases have been implicated to be involved in the processing of recombination intermediates (4, 6). In fact, the S. cerevisiae srs2Δ sgs1Δ double mutant is lethal (28), and this lethality is suppressed by the loss of recombination initiation (15). Thus, the absence of Srs2 or Rqh1 may cause an accumulation of spontaneously arising toxic recombination intermediates that require fbh1 for their resolution. Deleting rhp51 or rhp57 would prevent the formation of these toxic intermediates and thereby enable DNA damage to be repaired by an alternative pathway. Fbh1 is a member of the same DNA helicase family as Srs2. This family also includes PcrA, Rep, and UvrD helicases in bacteria. The Bacillus subtilis pcrA mutant is lethal, and this lethality is suppressed by recF, recO, and recR mutations (44). Similarly, the E. coli rep uvrD double mutant is lethal, and this lethality is also suppressed by recF, recO, and recR mutations (44). This suppression is analogous to the case of the fbh1Δ srs2Δ double mutant, where suppression of cell death is achieved by the rhp57 mutation. Thus, assuming functional resemblance among helicases in this family, Fbh1 and Srs2 might share a redundant function that is analogous to the function of Rep, UvrD, or PcrA. Srs2 dissociates Rad51 nucleoprotein filaments (26, 53), and UvrD dissociates RecA nucleoprotein filaments (52). Structural resemblance between these helicases suggests that Fbh1 may dissociate Rhp51 from DNA. Pulsed-field gel electrophoretic analysis indicated a defect in fbh1Δ cells for the processing of recombination intermediates. Fbh1 could dissociate Rhp51 from double-strand DNA following the strand exchange reaction, thereby resolving the D-loop structure, instead of dissociating Rhp51 from single-strand DNA. Such a role has been proposed for the Srs2 protein in the synthesis-dependent strand-annealing reaction based on the observation that Srs2 suppresses crossovers (18). As either the fbh1 or srs2 single mutant shows sensitivity to DNA-damaging agents, there would be a difference between reactions carried out by Fbh1 and Srs2. Further investigations will be required to address this possibility.
The fbh1Δ cells die when they are entering the stationary phase. As the fbh1Δ mutant ceased growth at a lower cell density than the wild-type strain, the cell death seems to have occurred during last several rounds of cell cycles, possibly during S phases. Dramatic physiological changes are assumed to occur at the late log phase. For example, glucose becomes exhausted, and utilization of nonfermentable carbon sources depends on respiration, probably leading to an increase in reactive oxygen species that damage DNA (10). Alternatively, cells may change nucleosomal structures to alter patterns of gene expression, thereby adapting to the stationary phase. Some of the nucleosomal structural changes that modify gene expression patterns may also inhibit replication fork progression. Therefore, it is possible that replication fork progression is more often inhibited during the late log phase prior to the cessation of cell proliferation and that Fbh1 is required to deal with this fork inhibition through its recombination function. Consistent with this notion, stable DNA replication that depends on recombination (24) has been detected in rapidly growing E. coli cells at the time of entry into the stationary phase (17). Deletion of rhp57 suppresses the loss of cell viability in fbh1Δ cells entering the stationary phase. As the loading of Rhp51 onto DNA and stabilization of Rhp51 nucleofilament would be compromised in the absence of Rhp57, the accumulation of toxic recombination intermediates would be reduced, negating the need for fbh1.
The fbh1 mutant is less sensitive to DNA-damaging agents than the rhp51 mutant, but this is not the case for viability upon entry into stationary phase. This lethality upon entry into stationary phase is suppressed by the rhp57 mutation. As Rhp55-57 is thought to constitute only a part of the Rhp51 pathway (1, 50), it can be explained by assuming that only certain types of recombination intermediates are made by the combination of Rhp51 and Rhp57, and these might be acted on by Fbh1. Replication forks inhibited at late log phase could be mainly acted upon by a combination of Rhp51 and Rhp57, and the absence of Fbh1 could result in failure to resolve recombination intermediates and hence could result in accumulation of toxic intermediates. In the absence of Rhp57, the inhibited forks could be resumed by recombination that does not employ Rhp57, and hence, the absence of Fbh1 would not cause accumulation of toxic recombination intermediates. rhp51 is epistatic to fbh1 in survival in DNA-damaging agents, but this is not the case for survival upon entry into stationary phase, and rhp51 and fbh1 showed mutual suppression. To explain this, we should consider Rhp51-independent recombination repair, and we leave this issue for further study.
Formation of EGFP-Fbh1 foci in γ-ray-treated cells and MMS-treated S-phase cells is consistent with the hypothesis that Fbh1 functions in the recombination repair of strand breaks and stalled or collapsed replication forks. The EGFP-Fbh1 fusion construct complemented the MMS sensitivity of the fbh1Δ cells in the presence of thiamine (data not shown), indicating that the EGFP-Fbh1 fusion protein is functional, and the repressed expression level is sufficient for this complementation. Under this repressed condition, we could not detect the EGFP signal. When cells were cultured in medium in the absence of thiamine to derepress expression, Fbh1 foci were observed in a small percentage of cells. Proportions of the cells with Fbh1 foci increased greatly after DNA-damaging treatments, suggesting that Fbh1 is recruited to the damaged sites to repair them. Under this derepressed condition, expression of the EGFP-Fbh1 fusion protein sensitizes the cells to MMS (data not shown). It could be that overexpression of EGFP-Fbh1 leads to the accumulation of more molecules per single focus, facilitating detection of the fluorescence signal, and the accumulation of too many Fbh1 molecules would be deleterious to normal DNA repair.
The Rhp51 foci were observed in 6% of exponentially growing wild-type cells. The rhp51Δ mutant cells grew more slowly than wild-type cells (38). Therefore, Rhp51-dependent recombination is required for repairing DNA damage that spontaneously arises during normal growth. Spontaneous Rhp51 foci were observed in 35% of the fbh1Δ cells, and this proportion was much higher than that in wild-type cells. In the absence of Fbh1, the Rhp51 nucleoprotein filament might be formed spontaneously, and further processing of the DNA lesion is defective, leading to the accumulation of recombination intermediates containing Rhp51 nucleofilament. Spontaneous Fbh1 foci were observed in only 2% of wild-type cells, while they were observed in 41% of the rhp51Δ cells. In the absence of Rhp51, Fbh1 is localized at the damaged DNA site, even though the DNA lesion is not processed further. Therefore, these results suggest that Rhp51 and Fbh1 are required for the processing of common DNA damage inflicted by endogenous agents.
SCF complexes act as ubiquitin ligases (E3), and the F-box component determines substrate specificity by directly interacting with the substrate at a protein recognition motif outside of the F-box motif (12). Introduction of mutations in the F-box motif sensitized S. pombe cells to MMS to a level similar to that of the fbh1Δ mutant (unpublished results), indicating that the F box is essential for Fbh1 function. The identity of the substrate of the SCF complex containing Fbh1 remains unknown. One candidate for the substrate is Rhp51, since Rhp51 is degraded when wild-type S. pombe cells enter the stationary phase, and this degradation is abolished in fbh1Δ cells (unpublished results). However, we could not detect ubiquitinated Rhp51 protein under various conditions, nor could we observe Rhp51 degradation in log-phase cultures. Further analysis is required to precisely delineate the role of the Fbh1-containing SCF complex.
Just as in yeast, RecQ family helicases in humans are required for maintaining genome stability. Indeed, mutations in three RecQ helicases, Blm, Wrn and Rts, result in the cancer predisposition syndromes Bloom, Werner, and Rothmund-Thomson syndromes, respectively (4). We suspect that the human ortholog of Fbh1 (FBH1) will also prove to be important for maintaining genome stability. Intriguingly, no ortholog of Srs2 has been identified in humans. It is possible that FBH1 fulfills the role in humans as yeast Srs2.
An fbh1 mutation was independently isolated as a suppressor of the rad22Δ mutant by Whitby's group. The phenotypes of the fbh1 mutants were in agreement with ours (41a).
Acknowledgments
We thank Matthew Whitby for communicating results prior to publication, Tamar Enoch and Yasuhiro Tsutsui for strains, Yeon-Soo Seo for helpful discussions, and Toshiji Ikeda for γ-ray irradiation.
This work was supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan to H.S., a grant from the Human Frontier Science Program Organization to A.M.C. and H. S., and grants from Cancer Research UK and the Human Frontier Science Program Organization to T.T.
REFERENCES
- 1.Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa, and H. Iwasaki. 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100:15770-15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aylon, Y., and M. Kupiec. 2004. New insights into the mechanism of homologous recombination in yeast. Mutat. Res. 566:231-248. [DOI] [PubMed] [Google Scholar]
- 4.Bachrati, C. Z., and I. D. Hickson. 2003. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 374:577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbet, N., W. J. Muriel, and A. M. Carr. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59-66. [DOI] [PubMed] [Google Scholar]
- 6.Barbour, L., and W. Xiao. 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532:137-155. [DOI] [PubMed] [Google Scholar]
- 7.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 8.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell, and N. Rhind. 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23:6564-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa, V., and P. Moradas-Ferreira. 2001. Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22:217-246. [DOI] [PubMed] [Google Scholar]
- 11.Cox, M. M. 2001. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35:53-82. [DOI] [PubMed] [Google Scholar]
- 12.Craig, K. L., and M. Tyers. 1999. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72:299-328. [DOI] [PubMed] [Google Scholar]
- 13.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 14.Doe, C. L., and M. C. Whitby. 2004. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 32:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 16.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 17.Hong, X., G. W. Cadwell, and T. Kogoma. 1996. Activation of stable DNA replication in rapidly growing Escherichia coli at the time of entry to stationary phase. Mol. Microbiol. 21:953-961. [DOI] [PubMed] [Google Scholar]
- 18.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang, Y. K., Y. H. Jin, E. M. Kim, F. Fabre, S. H. Hong, and S. D. Park. 1994. Cloning and sequence analysis of rhp51+, a Schizosaccharomyces pombe homolog of the Saccharomyces cerevisiae RAD51 gene. Gene 142:207-211. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 21.Khasanov, F. K., G. V. Savchenko, E. V. Bashkirova, V. G. Korolev, W. D. Heyer, and V. I. Bashkirov. 1999. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics 152:1557-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., J. H. Kim, S. H. Lee, D. H. Kim, H. Y. Kang, S. H. Bae, Z. Q. Pan, and Y. S. Seo. 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277:24530-24537. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. H., J. Kim, D. H. Kim, G. H. Ryu, S. H. Bae, and Y. S. Seo. 2004. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 32:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. K., R. E. Johnson, S. L. Yu, L. Prakash, and S. Prakash. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339-2342. [DOI] [PubMed] [Google Scholar]
- 29.MacNeill, S. A., and P. Nurse. 1997. Cell cycle control in fission yeast, p. 697-763. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 30.Maftahi, M., J. C. Hope, L. Delgado-Cruzata, C. S. Han, and G. A. Freyer. 2002. The severe slow growth of Δsrs2 Δrqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 30:4781-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 32.McGowan, C. H. 2003. Running into problems: how cells cope with replicating damaged DNA. Mutat. Res. 532:75-84. [DOI] [PubMed] [Google Scholar]
- 33.Michel, B., G. Grompone, M. J. Flores, and V. Bidnenko. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 101:12783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 35.Morikawa, H., T. Morishita, S. Kawane, H. Iwasaki, A. M. Carr, and H. Shinagawa. 2004. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 24:9401-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muris, D. F., K. Vreeken, A. M. Carr, J. M. Murray, C. Smit, P. H. Lohman, and A. Pastink. 1996. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109:73-81. [DOI] [PubMed] [Google Scholar]
- 39.Muris, D. F., K. Vreeken, H. Schmidt, K. Ostermann, B. Clever, P. H. Lohman, and A. Pastink. 1997. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 40.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray, J. M., M. Tavassoli, R. al-Harithy, K. S. Sheldrick, A. R. Lehmann, A. M. Carr, and F. Z. Watts. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14:4878-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Osman, F., J. Dixon, A. R. Barr, and M. C. Whitby. 2005. The F-box helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 25:8084-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, J. S., E. Choi, S. H. Lee, C. Lee, and Y. S. Seo. 1997. A DNA helicase from Schizosaccharomyces pombe stimulated by single-stranded DNA-binding protein at low ATP concentration. J. Biol. Chem. 272:18910-18919. [DOI] [PubMed] [Google Scholar]
- 44.Petit, M. A., and D. Ehrlich. 2002. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 21:3137-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97:509-516. [DOI] [PubMed] [Google Scholar]
- 46.Sung, P., K. M. Trujillo, and S. Van Komen. 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451:257-275. [DOI] [PubMed] [Google Scholar]
- 47.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsutsui, Y., F. K. Khasanov, H. Shinagawa, H. Iwasaki, and V. I. Bashkirov. 2001. Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics 159:91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a func-tional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueno, M., T. Nakazaki, Y. Akamatsu, K. Watanabe, K. Tomita, H. D. Lindsay, H. Shinagawa, and H. Iwasaki. 2003. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol. Cell. Biol. 23:6553-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veaute, X., S. Delmas, M. Selva, J. Jeusset, E. Le Cam, I. Matic, F. Fabre, and M. A. Petit. 2004. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 54.Winston, F., F. Chumley, and G. R. Fink. 1983. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 101:211-228. [DOI] [PubMed] [Google Scholar]