Abstract
Human nuclear Dbf2-related kinases (NDRs) are up-regulated in certain cancer types, yet their precise function(s) and regulatory mechanism(s) still remain to be defined. Here, we show that active (phosphorylated on Thr444) and inactive human NDRs are both mainly cytoplasmic. Moreover, NDR kinases colocalize at the plasma membrane with human MOBs (hMOBs), which are recently described coactivators of human NDR in vitro. Strikingly, membrane targeting of NDR results in a constitutively active kinase due to phosphorylation on Ser281 and Thr444 that is further activated upon coexpression of hMOBs. Membrane-targeted hMOBs also robustly promoted activation of NDR. We further demonstrate that the in vivo activation of human NDR by membrane-bound hMOBs is dependent on their interaction and occurs solely at the membrane. By using a chimeric molecule of hMOB, which allows inducible membrane translocation, we found that NDR phosphorylation and activation at the membrane occur a few minutes after association of hMOB with membranous structures. We provide insight into a potential in vivo mechanism of NDR activation through rapid recruitment to the plasma membrane mediated by hMOBs.
The human genome encodes two highly related serine-threonine kinases, nuclear Dbf2-related kinase 1 (NDR1) and NDR2, which belong to a subfamily of kinases with roles in cell division and cell morphology (22). The two kinases represent a subclass of the AGC family of protein kinases and are partially related to protein kinase B (PKB) and protein kinase C (PKC). Human NDRs, as well as their yeast counterpart Dbf2, can be dramatically activated through inhibition of protein phosphatase 2A (PP2A) by okadaic acid (OA), demonstrating that NDR kinases require phosphorylation for activation (13, 16). Indeed, NDR1 has been shown to become autophosphorylated on Ser281 in a Ca2+-dependent manner (23), while Thr444 is phosphorylated by a hydrophobic motif kinase (M. R. Stegert et al., submitted for publication). Phosphorylation on both residues is essential for full activation of NDR1 in vitro and in vivo (16, 23). Recently, the phosphorylation of NDR2 on Ser282 and Thr442 was also shown to be required for full activation of enzymatic activity (20).
The founding member of the Mps1 one binder (MOB) family is the yeast protein MOB1p (12), a molecule functioning as a coactivator of yeast Dbf2p and Dbf20p (10, 11, 13). As previously published, human MOB1A (hMOB1A) binds NDR1 and NDR2 (NDRs) at their N termini, leading to kinase activation in vitro (3). In addition, hMOB1B and hMOB2 also stimulated NDR activity in vitro (4). However, while the MOB regulation of the NDR kinases seems to be conserved from yeast to humans, the function(s) of mammalian NDRs is unknown. Although the basic mechanism of activation of human NDR protein kinase in vitro has been described in detail (3, 16, 20, 23), the regulation of NDR in a cellular context still remains to be elucidated. Such information should bring insight into its cellular functions and, in particular, into its possible role in cancer.
While NDR1 was reported to be localized predominantly in the nucleus (14), NDR2 was shown very recently to be mostly excluded from the nucleus, displaying a cytoplasmic distribution (4, 5, 20, 21). This observation is intriguing given the high sequence similarity between these two kinases. The NDR1 nuclear localization signal (NLS) (residues 265 to 276) contains only a single conservative change in NDR2, and thus the reason for the differential localization still remains to be defined. hMOB1B and MOB2 localization was also reported to be altered by coexpression of NDRs (4).
Here, we reanalyzed the intracellular distribution of inactive and active (phosphorylated) NDR1 more precisely using cell biological and biochemical techniques. To evaluate the importance of certain subcellular compartments for NDR activation, we generated nucleus- and membrane-targeted versions of NDR and analyzed activity and subcellular localization following OA treatment. In order to understand the roles of hMOB1A, hMOB1B, and hMOB2 (hMOBs) for NDR activation in vivo, we studied their intracellular distribution and found colocalization with NDR on the plasma membrane. We also developed inducible membrane-targeting constructs of hMOB1A and showed that they were able to promote translocation of NDR to membranes and phosphorylation.
MATERIALS AND METHODS
Cell culture, chemicals, drug treatments, and transfections.
COS-7, U2-OS, HEK 293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Exponentially growing COS-7 cells were plated at a consistent confluence (3 × 105 cells/6-cm dish) and transfected the next day using Fugene 6 (Roche) as described by the manufacturer. U2-OS cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In some experiments, cells were treated for 60 min with 1 μM okadaic acid (Alexis Corp.). In some experiments, cells were serum starved for 2 h prior to transfection. The transfection mixture was removed after 4 h, and cells were serum starved overnight before stimulation with 100 ng/ml 12-O-tetradecanoylphorbol 13-acetate (TPA) (Amersham Biosciences). Leptomycin B (LMB) was purchased from Sigma.
Construction of plasmids.
All human NDR1, NDR2, hMOB1A, hMOB1B, and hMOB2 cDNAs were subcloned into pcDNA3 and pcDNA3 derivatives by using BamHI and XhoI restriction sites. pcDNA3 derivatives contained a hemagglutinin (HA) or myc epitope alone, the myristoylation/palmitylation motif of the Lck tyrosine kinase (MGCVCSSN) combined with an HA or myc epitope (mp-HA or mp-myc), or the NLS of simian virus 40 (SV40) (MLYPKKKRKGVEDQYK) combined with an HA or a myc epitope (NLS-HA or NLS-myc). All mutants of NDR1 were generated by a two-step PCR-based mutagenesis procedure using pcDNA3-HA-NDR1 as template. Individual PCR products were digested with BamHI and XhoI and cloned into pcDNA3 derivatives. To obtain pGFP-NDR1 mutants, the corresponding fragments were subcloned into pEGFP-C1 (Clontech) by using BamHI and XbaI restriction sites. To construct pcDNA3-myc-C1-hMOB1A, the C1 domain of bovine PKCα (amino acids 26 to 162) was amplified by PCR and subcloned into pcDNA3-HA by using EcoRI and XhoI restriction sites. Subsequently, the hMOB1A cDNA was inserted using BamHI and EcoRI restriction sites. To generate a construct expressing pGFP-GFP, green fluorescent protein (GFP) was amplified by PCR using pEGFP-C1 as template and first subcloned using KpnI and BamHI into pcDNA3. Then, a second GFP cDNA was inserted using EcoRI and XbaI. The NLS sequences of SV40 and NDR1 were introduced between the two GFP cDNAs by using BamHI and XhoI. SV40NLS-GFP was generated by introducing the SV40 NLS into pEGFP-N1 by using KpnI and BamHI. pNDR1-GFP was obtained by PCR cloning of NDR1 into pEGFP-N1 (Clontech), using XhoI and BamHI. All constructs were confirmed by sequence analysis. Details of the generation of constructs and sequences of primers are available upon request.
Generation and affinity purification of antibodies.
Generation and purification of phospho-specific antibodies raised against phosphorylated Ser281 and Thr444 of NDR1 have been described recently (23). Anti-NDR CT antibody has been described previously (14). Anti-NDR NT peptide antibody was raised against the synthetic peptide DEEKRLRRSAHARKETEFLRLKRTRLGL, corresponding to amino acids 59 to 86 of NDR1, by coupling to keyhole limpet hemocyanin by using glutaraldehyde. Rabbit injections and blood collections were done by Strategic Biosolutions. Antipeptide antibody was purified by coupling 60 mg of the peptide to 4 ml of Affi-Gel 10 (Bio-Rad Laboratories) according to the manufacturer's protocol. After extensive washing, the bound antibodies were eluted with 0.2 M glycine (pH 2.2) and dialyzed into phosphate-buffered saline (PBS).
Mouse monoclonal antibody against human NDR1 was purchased from Transduction Laboratories. A rat monoclonal anti-α-tubulin (YL1/2)-producing hybridoma cell line was obtained from the American Type Culture Collection. Anti-HA 12CA5 and anti-myc 9E10 antibodies were used as hybridoma supernatants. Anti-HA antibodies were from Santa Cruz (Y-11) and Roche (3F10). Anti-lamin A/C and anti-Flag M2 were from Santa Cruz Biotechnology Inc. and Sigma, respectively.
Immunoblotting.
To detect NDR or hMOB species, samples were resolved by 8% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with TBST (50 mM Tris, 150 mM NaCl, 0.5% Tween 20, at pH 7.5) containing 5% skim milk powder and then probed overnight with antibody. Bound antibodies were detected by horseradish peroxidase-linked secondary antibodies and processed with ECL (Amersham) according to the manufacturer's instructions. Immunodetection of NDR phosphorylated on Ser281/Ser282 or Thr444/Thr442 was as described previously (23). To test the specificity of anti-T444-P antibodies, duplicates of identical protein samples were immunoblotted onto one membrane, which was cut into two after blocking. Subsequently, one piece was incubated with purified anti-T444-P in the presence of 10 μg/ml dephospho-peptide (KDWVFINYTYKRFEG), while the other was incubated with anti-T444-P supplemented with 10 μg/ml phospho-peptide (KDWVFINYTpYKRFEG). For all subsequent steps, membranes were processed together in the same tube.
Immunoprecipitation.
Cells were harvested, pelleted by centrifugation at 1,000 × g for 3 min, and washed with ice-cold PBS before lysis in immunoprecipitation buffer (IP buffer) (20 mM Tris, 150 mM NaCl, 10% glycerol, 1% NP-40, 5 mM EDTA, 0.5 mM EGTA, 20 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 1 mM benzamidine, 4 μM leupeptin, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM microcystin, and 1 mM dithiothreitol [DTT] at pH 8.0). Lysates were centrifuged for 10 min at 16,000 × g at 4°C before preclearing with protein A-Sepharose, followed by immunoprecipitation with 12CA5 antibody prebound to protein A-Sepharose. Beads were washed twice with IP buffer, once with IP buffer containing 1 M NaCl, and finally once with IP buffer before samples were analyzed by SDS-PAGE.
To analyze association of NDR with hMOB species by coimmunoprecipitation, cells coexpressing HA-NDR and myc-hMOB forms were subjected to immunoprecipitation using anti-HA 12CA5 antibody as described above before analysis by SDS-PAGE and immunoblotting.
HA-NDR kinase assay.
Cells were processed for immunoprecipitation as described above and after the last wash with IP buffer were washed twice with 20 mM Tris, pH 7.5, supplemented with protease inhibitors. Beads were resuspended in 30 μl buffer containing 20 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM benzamidine, 4 μM leupeptin, 1 μM microcystin, 1 mM DTT, 1 μM cyclic AMP-dependent protein kinase inhibitor peptide, 100 μM [γ-32P]ATP (∼1,000 cpm/pmol), and 1 mM NDR substrate peptide (KKRNRRLSVA). After 60 min of incubation at 30°C, reactions were stopped with 50 mM EDTA, and 20 μl of the supernatant was spotted onto squares of P-81 phosphocellulose paper (Whatman) and washed four times for 10 min each in 1% phosphoric acid and once in acetone before counting in a liquid scintillation counter was performed. Experiments were performed in duplicate, and illustrated activities represent the averages from three independent experiments.
Fractionation of cells.
To separate cytosolic and membrane-associated proteins, cells were subjected to S100/P100 fractionation as follows. Cells were collected in PBS and incubated for 20 min at 4°C in S100/P100 buffer (20 mM Tris, 150 mM NaCl, 2.5 mM EDTA, 1 mM EGTA, 1 mM benzamidine, 4 μM leupeptin, 0.5 mM PMSF, 1 μM microcystin, and 1 mM DTT at pH 7.5). After homogenization using a 26-gauge needle (Becton Dickinson), nuclei were removed by centrifugation for 2 min at 1,000 × g at 4°C. The supernatant was then centrifuged at 100,000 × g for 60 min at 4°C. Equal amounts of supernatant (S100; cytoplasmic fraction) and pellet (P100; membrane fraction) were analyzed by SDS-PAGE and immunoblotting. Alternatively, equal amounts of cytoplasmic and membrane fractions were subjected to immunoprecipitation and subsequent kinase assays as described above.
To separate cells into nuclear, cytosolic, and membrane fractions, cells were rinsed with PBS, scraped into PBS, and pelleted in a tabletop centrifuge. Cell pellets were swollen for 30 min at 4°C in RSB buffer (10 mM HEPES, 10 mM NaCl, 1.5 mM MgCl2, 1 mM benzamidine, 4 μM leupeptin, 0.5 mM PMSF, 1 μM microcystin, and 1 mM DTT at pH 6.2), before homogenization by 20 strokes in a Dounce homogenizer. SDS loading buffer was added to an aliquot of lysed cells without further processing (total fractionation input). Nuclei were collected for 2 min at 400 × g at 4°C. The pellet was washed twice with RSB buffer before lysis in EBC buffer (50 mM Tris, 250 mM NaCl, 1% Triton X-100, 1 mM benzamidine, 4 μM leupeptin, 0.5 mM PMSF, 1 μM microcystin, and 1 mM DTT at pH 8.0). The supernatant was further fractionated by centrifugation for 90 min at 150,000 × g at 4°C. The S150 supernatant was collected (cytosolic fraction), while the membrane pellet was lysed in EBC buffer, and equal aliquots of each fraction (representing protein from the same number of cells) were analyzed by immunoblotting.
Immunofluorescence microscopy.
Cells were processed for immunofluorescence as described previously (9). Briefly, cells were grown on coverslips, washed with PBS, and fixed in 3% paraformaldehyde-2% sucrose in PBS at pH 7.4 for 10 to 15 min at 37°C or fixed with methanol (−20°C) before being permeabilized using 0.2% Triton X-100 in PBS for 2 min at room temperature. Coverslips were rinsed three times with PBS and incubated for 1 hour with primary antibody diluted in 1% bovine serum albumin-1% goat serum in PBS. After three washes with PBS for 5 min each, the appropriate secondary antibody was used. Secondary antibodies included donkey anti-rat-aminomethylcoumarin, donkey anti-mouse-fluorescein isothiocyanate (FITC) and Texas Red, and donkey anti-rabbit-FITC and Texas Red (Jackson Immunoresearch Inc.). DNA was counterstained with 1 μM TO-PRO-3 iodide (Molecular Probes Inc.). Coverslips were then inverted into 5 μl Vectashield medium (Vector Laboratories). Images were obtained with a Fluoview FV500 confocal laser scanning microscope (Olympus). Photographic images were processed using Photoshop 6.0 (Adobe Systems Inc).
RESULTS
Human NDR1 is primarily a cytoplasmic kinase.
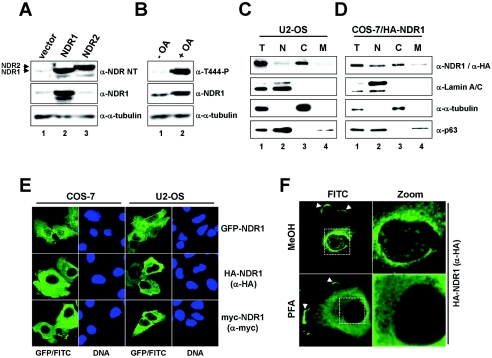
To establish the subcellular localization of native human NDR1 kinase in tissue culture cells, we generated a rabbit polyclonal antibody raised against part of the S100B-MOB1 association (SMA) domain of NDR1 (anti-NDR NT). This antibody detected overexpressed human NDR1 and NDR2 equally well by Western blotting (Fig. 1A) but also detected other unspecific molecules of higher molecular weight (see Fig. S1A in the supplemental material). Both native NDR forms were recognized from HeLa cells (data not shown), but only NDR1 was detected in U2-OS cells as a major form (Fig. 1A, lane 1). The commercially available anti-NDR1 monoclonal antibody reacted only with NDR1, as expected (Fig. 1A; see Fig. S1A in the supplemental material). The anti-T444-P antibody (raised against phosphorylated Thr444 of NDR1) recognized a single band comigrating with native NDR1 after OA treatment of U2-OS cells (Fig. 1B, lane 2; see Fig. S1B, lane 2, in the supplemental material), which was specifically competed by the corresponding phosphopeptide but by not the dephosphopeptide (see Fig. S1B in the supplemental material). Moreover, by analyzing cells overexpressing HA-NDR1, we could show that overexpressed NDR1 was detected at least 10 times better than native NDR species by anti-T444-P in our experimental settings (see Fig. S1C in the supplemental material). In summary, these data suggest that U2-OS cells express predominantly NDR1, which can be stimulated by treatment with the PP2A inhibitor OA.
FIG. 1.
Human NDR1 is a mainly cytoplasmic kinase. A. U2-OS cells expressing empty vector (lane 1), untagged human NDR1 (lane 2), or untagged human NDR2 (lane 3) were processed for immunoblotting using anti-NDR NT (top panel), anti-NDR1 (middle panel), and anti-α-tubulin (bottom panel) antibodies. Arrowheads indicate the positions of NDR1 and NDR2. B. U2-OS cells were incubated in the absence (lane 1) or presence (lane 2) of OA before processing for Western blotting using anti-Thr444-P (top panel), anti-NDR1 (middle panel), and anti-α-tubulin (bottom panel) antibodies. C. U2-OS cells were subjected to biochemical cell fractionation (T, total; N, nuclear; C, cytoplasmic; M, membrane) and processed for immunoblotting with anti-NDR1 (top panel), anti-lamin A/C (nuclear marker; top middle panel), anti-α-tubulin (cytoplasmic marker; bottom middle panel), and anti-p63/Climp (perinuclear/membrane marker; bottom panel) antibodies. D. COS-7 cells expressing HA-tagged NDR1 were subjected to biochemical cell fractionation (T, total; N, nuclear; C, cytoplasmic; M, membrane) and processed for immunoblotting with anti-HA (top panel), anti-lamin A/C (top middle panel), anti-α-tubulin (bottom middle panel), and anti-p63/Climp (bottom panel) antibodies. E. COS-7 and U2-OS cells expressing either GFP-NDR1 (top panels), HA-NDR1 (middle panels), or myc-NDR1 (bottom panels) were analyzed by indirect immunofluorescence microscopy using anti-HA Y11 (middle panels), anti-myc (bottom panels), or no (top panels) antibody. GFP/FITC signals are shown in green. Nuclei are in blue. F. COS-7 cells expressing HA-NDR1 were fixed with either methanol (MeOH) (top panels) or paraformaldehyde (PFA) (bottom panels) before permeabilization and antibody staining using anti-HA Y11 antibody. Arrowheads indicate NDR1 species found in plasma membrane structures. Enlargements of the regions indicated by white rectangles are shown on the right (Zoom).
Next, we sought to use anti-NDR NT and anti-NDR1 antibodies in indirect immunofluorescence, but despite application of various fixation and permeabilization methods, we found no specific immunofluorescence signal with either antibody. Thus, U2-OS cells were biochemically separated into nuclear, cytoplasmic, and membranous fractions (Fig. 1C). Strikingly, endogenous NDR1 protein was detected mostly in the cytoplasm (Fig. 1C, lane 3). A similar result was obtained when COS-7 cells transiently expressing HA-NDR1 were fractionated (Fig. 1D, lane 3). To confirm these observations, GFP-, HA-, and myc-tagged NDR1 proteins were expressed in COS-7, U2-OS, HEK 293, and HeLa cells before cells were processed for indirect immunofluorescence (Fig. 1E and data not shown). Significantly, regardless of N-terminal tag and cell type, a significant proportion of NDR1 was found in the cytoplasm of a vast majority of cells (more than 80%). NDR1 tagged C terminally with GFP also localized to the cytoplasm (data not shown). The antibody staining specificity was demonstrated by taking pictures illustrating transfected and untransfected cells in the same field (Fig. 1E). Moreover, the anti-HA antibody used (Y11) recognized solely a single band in immunoblotting, corresponding to the predicted molecular weight of HA-NDR1 (data not shown). Interestingly, a portion of NDR1 was further found in the nucleus and membrane (Fig. 1C, D, and F). However, localization of NDR1 in defined nuclear spots was observed only when cells were fixed with methanol (Fig. 1F, top). In contrast to previous results, we demonstrate here that human NDR1 is detected mostly in the cytoplasm. Consequently, we reanalyzed the anti-NDR antibody originally used to establish NDR as a predominantly nuclear kinase (see Fig. S2 in the supplemental material). While this antibody (anti-NDR CT) recognized HA-NDR1 and GFP-NDR1 by immunoblotting (see Fig. S2A in the supplemental material), no major colocalization of anti-NDR CT and GFP-NDR stainings was observed (see Fig. S2B in the supplemental material), arguing that NDR species were not detected by anti-NDR CT in immunofluorescence stainings. Given that anti-NDR CT signals were enriched in the nuclei of both transfected and untransfected cells, it is very likely that the originally reported localization of NDR in the nucleus represents a misinterpretation of stainings.
Human NDR1 requires the SMA domain for membrane association.
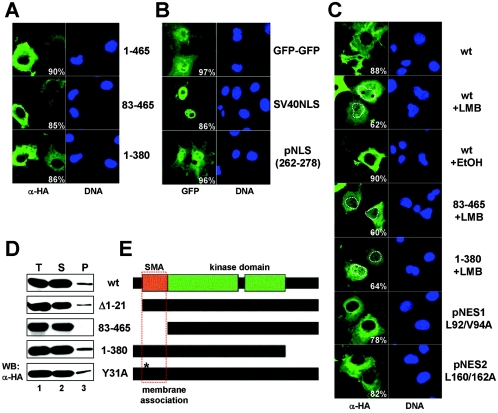
In order to define the domains in NDR1 required for its subcellular localization, we expressed N- and C-terminal truncations of NDR1 in COS-7 or U2-OS (data not shown) cells and analyzed their subcellular distribution. Neither the N terminus nor the C terminus of NDR1 was required for cytoplasmic localization (Fig. 2A). Next, the functionality of the postulated NLS (pNLS) (amino acids 262 to 278) of NDR1 was tested using a GFP-GFP construct. The GFP-GFP protein was clearly detectable in the nuclei and cytoplasm of transfected cells, while the molecule fused to the NLS of SV40 accumulated in the nucleoplasm (Fig. 2B, top and middle panels). Surprisingly, the pNLS of NDR1 did not enrich pGFP-GFP in the nucleus (Fig. 2B, bottom panels), arguing that this sequence does not function as an NLS. However, we cannot exclude the possibility that the pNLS has some very weak nuclear localization activity that was not detected in our experimental settings.
FIG. 2.
Determination of NDR1 localization domains. A. COS-7 cells expressing HA-tagged full-length (amino acids 1 to 465; top panels), N-terminally truncated (amino acids 83 to 465; middle panels), or C-terminally truncated (amino acids 1 to 380; bottom panels) NDR1 were processed for immunofluorescence using anti-HA Y11 antibody (green). Corresponding DNA stainings are shown on the right. The percentages of transfected cells displaying predominantly cytoplasmic signals are indicated. B. Cells transfected with plasmids encoding GFP-GFP alone (top panels), GFP-SV40NLS-GFP (middle panels), or GFP-NDR1(amino acids 262 to 278)-GFP (bottom panels) were subsequently analyzed by immunofluorescence. The percentages of stained cells as illustrated are indicated (>200 cells per construct were analyzed). Nuclei are shown in blue. The postulated NLS of NDR1 did not drive GFP into the nucleus, in contrast to the SV40 NLS. C. Cells expressing HA-tagged wild-type (wt), N-terminally truncated (amino acids 83 to 465), or C-terminally truncated (amino acids 1 to 380) NDR1, or selected mutants carrying point mutations at Leu92/Val94 (L92/V94A) or Leu160/Leu162 (L160/162A), were left untreated or incubated for 8 h with 10 ng/ml LMB or ethanol (EtOH), before processing for immunofluorescence using anti-HA Y11 antibody (green). The percentages of stained cells as shown are indicated (>200 cells per construct were analyzed). Nuclei are shown (right panels) and highlighted by white dashed lines in selected pictures. pNES, potential NES. D. COS-7 cells were transfected with the indicated HA-tagged NDR1 constructs and subjected to S100/P100 fractionation (T, total; S, cytoplasm; P, membrane), before immunoblotting with anti-HA 12CA5 antibody. E. Schematic representation of human NDR1 and mutant derivatives. The kinase domain (green) and the S100B/MOB association (SMA) (orange) domains are indicated. The N-terminal domain required for membrane association is marked by a red rectangle.
Further, an in silico analysis of NDR1's primary structure revealed two potential nuclear export sequence (NES) motifs (see Fig. S3 in the supplemental material), but mutating their core sequences did not alter their intracellular distribution (Fig. 2C, bottom panels). Even an HA-NDR1 L92/V94/L160/L162A mutant was localized in the cytoplasm similarly to wild-type protein (data not shown), suggesting that NDR1 does not contain a functional NES. However, to determine in more detail whether NDR1 encodes an atypical NES, we next treated cells with LMB, a well-characterized inhibitor of CRM1-dependent nuclear export, and verified the distribution of NDR1 by immunofluorescence. In the majority of cells, LMB treatment resulted in an enrichment of nuclear NDR1 species, while control cells (with solvent alone) were unaffected (Fig. 2C, top panels). However, this enrichment occurred only after treatment of cells for several hours with LMB, where a strong cytoplasmic signal always remained. Incubation with LMB for 2 hours was not sufficient to affect NDR1 localization (data not shown). We were further unable to determine a region within NDR1 required for this LMB-induced nuclear accumulation (Fig. 2C, middle panels, and data not shown). Overall, these data suggest that NDR1 accumulates rather slowly in the nucleus after LMB treatment, most likely not due to blockage of active nuclear export of NDR1 species.
Given that NDR1 was found partially in the membranous fraction, we tested whether NDR1 contains a membrane-binding domain. A series of deletion constructs analyzed biochemically for membrane association revealed that amino acids 22 to 82 of NDR1 are required for NDR1 to be recovered in the membrane fraction (Fig. 2D). Interestingly, this region perfectly matches the SMA domain of NDR1 (3). Our data suggest that NDR1 needs its N-terminal SMA domain for membrane association, but which specific MOB isoform might play a role in vivo still needs to be elucidated. An NDR1 (Y31A) mutant deficient in hMOB1A binding associated with the membrane fraction similarly to wild-type protein (Fig. 2D, bottom panel), suggesting that potentially another MOB family member is responsible for membrane recruitment in this experimental setting. Indeed, hMOB2 coimmunoprecipitated with NDR1 (Y31A) to levels comparable to those for wild-type NDR species (see Fig. S4, lanes 4 and 8, in the supplemental material), while hMOB1A/B binding to NDR1 was abolished by introducing the Y31A mutation (see Fig. S4, lanes 2, 3, 6, and 7, in the supplemental material). Thus, it is very likely that the association of NDR1 (Y31A) with membranes can be mediated by another hMOB under our experimental conditions.
NDR1 phosphorylated on Thr444 accumulates mainly in the cytoplasm.
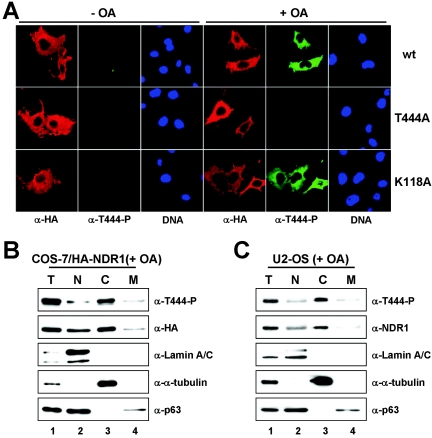
To identify where NDR1 is activated within a cell, we stimulated NDR1 phosphorylation by OA treatment and then used an antibody raised against the phosphorylated site of the NDR1 hydrophobic motif (anti-T444-P) as a marker for active NDR1 (Fig. 3). Immunofluorescence studies showed that active NDR1 accumulated in the cytoplasm similarly to the localization of total NDR1 (Fig. 3A, top panels). Anti-T444-P antibody did not detect NDR1 (T444A) carrying a Thr444-to-Ala mutation (Fig. 3A, middle panels), as expected. Similar results were obtained when NDR2 was expressed (see Fig. S5A in the supplemental material). These data were confirmed biochemically, where total and active NDR1 was also observed mainly in the cytoplasm of fractionated cells (Fig. 3B). It is important to note that both inactive and Thr444-phosphorylated native NDR1 forms were also detected mostly in the cytoplasm of fractionated U2-OS cells (Fig. 3C). Taken together, the findings from this analysis revealed that Thr444-phosphorylated NDR1 accumulated in the cytoplasm after OA treatment, further supporting the notion that NDR1 is not a predominantly nuclear kinase.
FIG. 3.
NDR1 phosphorylated on Thr444 is mostly cytoplasmic. A. COS-7 cells expressing HA-tagged wild-type (wt) (top panels), Thr444Ala (T444A) (middle panels), or kinase-dead (K118A) (bottom panels) NDR1 were either left untreated (− OA) or incubated with okadaic acid (+ OA) before processing for immunofluorescence using anti-HA 12CA5 (red) and anti-T444-P (green) antibody. Nuclei are indicated in blue. All pictures were taken using the same settings, which had been adjusted to signals of cells overexpressing NDR1. B. COS-7 cells expressing HA-tagged NDR1 were subjected to biochemical cell fractionation after OA treatment (T, total; N, nuclear; C, cytoplasmic; M, membrane) and processed for immunoblotting with anti-T444-P (top panel), anti-HA 12CA5 (top middle panel), anti-lamin A/C (middle panel), anti-α-tubulin (bottom middle panel), and anti-p63/Climp (bottom panel) antibodies. C. After OA treatment, U2-OS cells were subjected to biochemical cell fractionation (T, total; N, nuclear; C, cytoplasmic; M, membrane) and processed for immunoblotting with anti-T444-P (top panel), anti-NDR1 (top middle panel), anti-lamin A/C (middle panel), anti-α-tubulin (bottom middle panel), and anti-p63/Climp (bottom panel) antibodies.
Membrane targeting of NDR generates a constitutively active kinase.
Since a fraction of NDR1 was found to associate with membranes, we investigated whether membrane binding of NDR is part of its activation process. The myristoylation/palmitylation (mp) motif from the Lck tyrosine kinase was attached to the N terminus of NDR1 to alter its subcellular localization. Membrane-targeted NDR1 (mp-NDR1) associated mainly with the plasma membrane of transfected cells irrespective of PP2A inhibition (Fig. 4A). The intracellular localization of mp-NDR1 was further confirmed by biochemical analysis, since nearly all targeted protein was recovered in the pellet (membrane) fraction (Fig. 4B, bottom panel, lane 4). Strikingly, mp-NDR1 was already phosphorylated at the hydrophobic motif (Thr444) without OA treatment (Fig. 4B, top panel, lane 4), and elevated HA-NDR kinase activity was detected solely in the membrane fraction of mp-NDR1 (Fig. 4C, bar 4). To address this change in kinase activity in more detail, we next compared the activities of mp-NDR1 species with unstimulated and OA-treated wild-type enzyme (Fig. 4D and E) and found that mp-NDR1 was about fivefold more active than unstimulated wild-type protein (Fig. 4E, lane 3). Thr444 phosphorylation was clearly increased, whereas Ser281 phosphorylation was only slightly elevated (Fig. 4D, lane 3). Similar results were obtained when mp-NDR2 was analyzed (see Fig. S5B and C in the supplemental material). Strikingly, mp-NDR1 activity was further increased to levels comparable to those for stimulated wild-type enzyme by OA treatment (Fig. 4E, bar 4). As expected, a kinase-dead version of mp-NDR1 showed no Ser281 phosphorylation and kinase activity, but phosphorylation on Thr444 occurred even without OA treatment (Fig. 4D and E, lanes and bars 5 and 6).
FIG. 4.
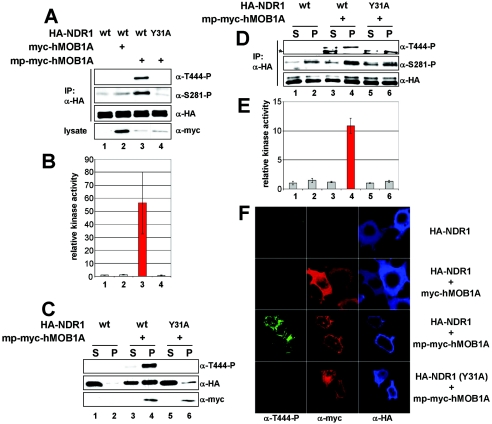
Membrane-targeted NDR1 is partially active and can be further activated by coexpression of hMOB1A without OA treatment. A. COS-7 cells transfected with membrane-targeted NDR1 (mp-HA-NDR1) were either left untreated (− OA) or incubated with okadaic acid (+ OA), before processing for immunofluorescence using anti-HA Y11 (left panels). Nuclei are in blue. B. COS-7 cells were transfected with the indicated NDR1 plasmids and subjected to S100/P100 fractionation (S, cytoplasm; P, membrane) before immunoblotting with anti-T444-P (top panel) and anti-HA 12CA5 antibody (bottom panel). C. The S100/P100 extracts described above for panel B were subjected to immunoprecipitation using anti-HA 12CA5 antibody and subsequently to kinase assays. Elevated kinase activity was found only in mp-NDR1 membrane extracts (red bar). D. Lysates of COS-7 cells expressing HA-tagged wild-type (wt), membrane-tagged wt kinase-dead (kd), or hMOB1A-binding-deficient (Y31A) NDR1 were subjected to immunoprecipitation (IP) using anti-HA 12CA5 antibody. Complexes were analyzed by Western blotting using anti-T444-P (top panel), anti-S281-P (top middle panel), and anti-HA antibody (bottom middle panel). Prior to lysis, the indicated cells were incubated with OA (+). In certain samples, myc-tagged hMOB1A was coexpressed as verified by immunoblotting using anti-myc antibody (bottom panel). An unspecific signal is indicated by an asterisk. E. In parallel, immunocomplexes were subjected to kinase assays. Membrane-targeted NDR1 activity was about fivefold elevated compared to wild type and was further increased by hMOB1A coexpression (red bars). Error bars indicate standard deviations.
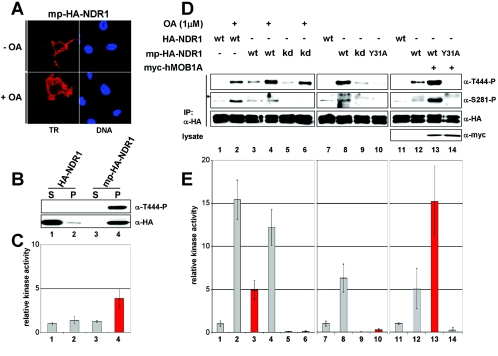
Given this absence of complete phosphorylation of mp-NDR1 on Ser281 and the recent report that hMOB1A stimulates NDR autophosphorylation on Ser281 (3), we speculated that endogenous hMOB1 does not bind sufficiently to the N terminus of mp-NDR1 to release autoinhibition of Ser281 phosphorylation. In agreement with this hypothesis, mp-NDR1 (Y31A) carrying a point mutation abolishing MOB1 binding was blocked in Ser281 phosphorylation and enzymatic activity, and phosphorylation on Thr444 was markedly reduced as well (Fig. 4D and E, lane and bar 10), despite efficient membrane targeting (data not shown). mp-NDR1 (T74A), carrying an alternative mutation also altering MOB1 association, was also affected in Ser281/Thr444 phosphorylation and enzymatic activity (data not shown). To address our hypothesis in more detail, we expressed mp-NDR1 together with hMOB1A, which led to a dramatic elevation of Ser281 and Thr444 phosphorylation and enzymatic activity relative to OA-stimulated wild-type protein (Fig. 4D and E, compare lanes and bars 12 and 13 with lanes and bars 2 and 3). These changes were dependent on the interaction of hMOB1A with NDR1, since mp-NDR1 (Y31A) showed no increase in phosphorylation or activity (Fig. 4D and E, lane and bar 14). In striking contrast, coexpression of hMOB1A with NDR1 (not membrane targeted) only slightly increased Ser281 phosphorylation, while Thr444 phosphorylation and kinase activity were not altered (see Fig. 6A and B, lanes and bars 1 and 2). Together, these data suggest that membrane targeting of NDR1 generates a constitutively active kinase which can be further activated by hMOB1A coexpression.
FIG. 6.
Membrane-targeted hMOB1A potently activates NDR1 at the plasma membrane. A. Lysates of COS-7 cells coexpressing the indicated combinations of HA-tagged wild-type (wt) or MOB1A-binding-deficient (Y31A) NDR1, myc-tagged hMOB1A, and membrane-targeted hMOB1A (mp-myc-hMOB1A) were analyzed by immunoprecipitation (IP) using anti-HA 12CA5 antibody. Complexes were assayed by Western blotting using anti-T444-P (top panel), anti-S281-P (middle top panel), and anti-HA antibody (middle bottom panel). Lysates were immunoblotted with anti-myc 9E10 antibody (bottom panel). B. In parallel, immunocomplexes were subjected to kinase assays. The robust increase of NDR1 activity by coexpression of membrane-targeted hMOB1A is marked in red. C. Cells transfected with the indicated NDR1 and hMOB1A constructs were subjected to S100/P100 fractionation (S, cytoplasm; P, membrane) before immunoblotting with anti-T444-P (top panel), anti-HA 12CA5 (middle panel), and anti-myc 9E10 (bottom panel) antibodies. D. The S100/P100 extracts described above for panel C were subjected to immunoprecipitation using anti-HA 12CA5 antibody before analysis by Western blotting using anti-T444-P (top panel), anti-S281-P (middle panel), and anti-HA (bottom panel) antibodies. An unspecific signal is indicated by an asterisk. E. In parallel, immunocomplexes were subjected to kinase assays. The increase of NDR1 activity in the membrane fraction is marked in red. F. Cells coexpressing HA-NDR1 wt/empty vector (top panels), HA-NDR1 wt/myc-hMOB1A (middle top panels), HA-NDR1 wt/mp-myc-hMOB1A (middle bottom panels), or HA-NDR1 (Y31A)/mp-myc-hMOB1A (bottom panels) were processed for immunofluorescence analysis with anti-T444-P (green), anti-myc 9E10 (red), and anti-HA 3F10 (blue) antibodies. Error bars indicate standard deviations.
hMOB1A, hMOB1B, and hMOB2 colocalize with human NDR kinases at the plasma membrane.
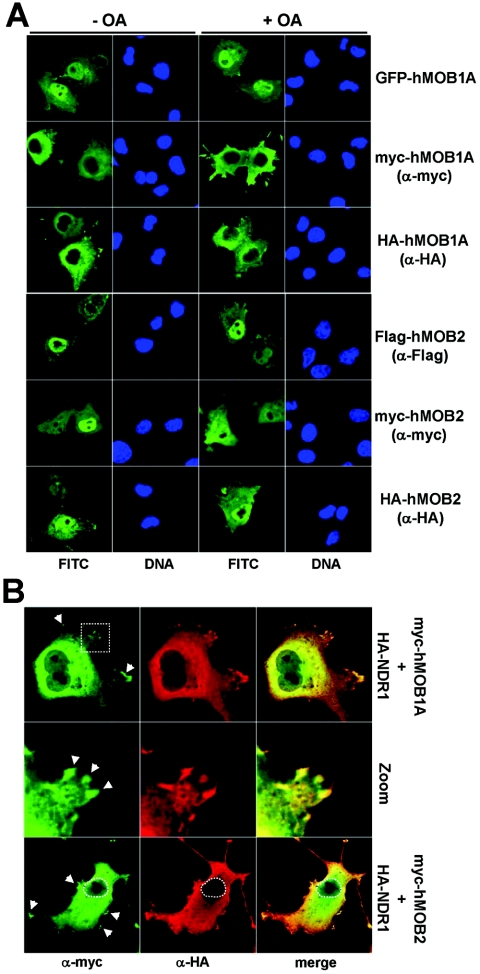
Given that hMOB1 expression increased the activity of mp-NDR1 in vivo, we analyzed the intracellular distribution of hMOBs. GFP-, myc-, and HA-tagged hMOB1A proteins were expressed in COS-7 and U2-OS cells before immunofluorescence analysis (Fig. 5A and data not shown). Significantly, the GFP-hMOB1A signal was strongest in nuclei, whereas hMOB1A carrying much smaller N-terminal tags (HA and myc) was found mostly in the cytoplasm. Similar results were obtained for hMOB1B (data not shown). This suggests that adding a GFP tag to hMOB1 apparently alters its subcellular localization and that hMOB1A and hMOB1B are mostly cytoplasmic proteins.
FIG. 5.
hMOB1A is mostly cytoplasmic, in contrast to hMOB2; however, both species colocalize with NDR1 in the cytoplasm/membrane. A. COS-7 cells expressing either GFP-hMOB1A (top panels), myc-hMOB1A (second top panels), HA-hMOB1A (middle top panels), Flag-hMOB2 (middle bottom panels), myc-hMOB2 (second bottom panels), or HA-hMOB2 (bottom panels) were either left untreated (− OA) or incubated with 1 μM okadaic acid (+ OA), before processing for immunofluorescence microscopy using no, anti-myc, anti-HA Y11, or anti-Flag M2 antibody. GFP/FITC signals are shown in green. Nuclei are indicated in blue. B. Cells cotransfected with myc-hMOB1A/HA-NDR1 (top panels) or myc-hMOB2/HA-NDR1 (bottom panels) were analyzed by immunofluorescence microscopy using anti-myc 9E10 (green) and anti-HA Y11 (red) antibodies. Merged images of hMOB and NDR1 staining are shown (merge). Enlargement of the region indicated by a white rectangle is shown in the middle panels. Arrowheads indicate colocalization sites at the plasma membrane.
In contrast, hMOB2 was detected in both nucleus and cytoplasm by using various N-terminal tags (Fig. 5A). We found no cells expressing hMOB2 solely in the cytoplasm (as we did for NDRs or hMOB1). hMOB2 was highly enriched in the nuclei of some cells, but in others it was equally present in nuclear and cytoplasmic compartments. However, all three hMOBs (hMOB1A, -1B, and -2) colocalized with NDR1 and NDR2 in the cytoplasm and at the plasma membrane (Fig. 5B and data not shown). It is noteworthy that colocalization was most apparent at certain sites of the plasma membrane (Fig. 5B, middle panels, and data not shown).
Membrane-bound hMOB strongly activates NDR kinase activity.
Since constitutively active mp-NDR1 was further stimulated by hMOB1 in vivo and hMOBs colocalized with NDR at the plasma membrane, we speculated that NDR is in part activated by hMOBs at the plasma membrane. Consequently, the membrane tag fused to NDR1 was attached to hMOB1A, membrane-targeted hMOB1A (mp-hMOB1A) was coexpressed with NDR1, and subsequently its effect on NDR1 activation was monitored (Fig. 6A). Strikingly, Ser281 and Thr444 phosphorylations were dramatically increased in wild-type protein, while NDR1 (Y31A) displayed no changes (Fig. 6A, lanes 3 and 4). This increase in phosphorylation was paralleled by a robust elevation of kinase activity (Fig. 6B, bar 3). mp-hMOB1A stimulated NDR1 activity even more than PP2A inhibition (data not shown) and also raised the phosphorylation and activity levels of NDR2 (see Fig. S5D and E in the supplemental material).
Interestingly, mp-hMOB1A recruited a significant fraction of NDR1 to the membrane, and Thr444-phosphorylated species were recovered nearly exclusively in the pellet (Fig. 6C, lane 4). In contrast, membrane levels of NDR1 (Y31A) were not increased despite efficient membrane targeting of mp-hMOB1A (Fig. 6C, lane 6). Further, phospho-S281 species and NDR kinase activity were significantly increased in membrane fractions only when wild-type NDR1 was coexpressed with mp-hMOB1A (Fig. 6D and E, lane and bar 4). NDR1 (Y31A) activity and phosphorylation were not elevated by mp-hMOB1A (Fig. 6D and E, lane and bar 6). NDR1 (T74A) was also not activated by mp-hMOB1A (data not shown), further supporting the conclusion that membrane recruitment and activation of NDR1 by mp-hMOB1A are dependent on the NDR-hMOB interaction. Importantly, mp-hMOB1B and mp-hMOB2 (mp-MOBs) also activated NDR1 at the membrane (see Fig. S6 in the supplemental material). The efficacy of activation was paralleled by the efficacy of membrane sequestration of NDR1 by mp-MOBs. mp-hMOB1B recruited more NDR1 to the membrane than mp-hMOB2 (see Fig. S6C, lanes 4 and 6, in the supplemental material), and as a result, more Thr444-P species and higher kinase activity were detected (see Fig. S6A and B, lanes 3 and 5, in the supplemental material).
These experiments did not reveal the intracellular membrane structures where NDR was activated, and we therefore examined cells expressing NDR1 and mp-hMOB1A by immunofluorescence microscopy (Fig. 6F). NDR1 was mostly cytoplasmic with or without coexpression of untargeted hMOB1A (Fig. 6F, top and top middle panels), but NDR1 was found to decorate the plasma membrane after mp-hMOB1A expression (Fig. 6F, bottom middle panels). Significantly, most forms phosphorylated on Thr444 were also observed at the plasma membrane, while NDR1 (Y31A) was not recruited to the plasma membrane, nor were Thr444-P molecules detected (Fig. 6F, bottom panels), indicating that NDR1 association with the plasma membrane is dependent on interaction with mp-hMOB1A.
Next, we expressed nontargeted and mp-hMOB1A in U2-OS cells to address the activation of native NDR1 species and found that endogenous NDR1 can also be activated once hMOB1A is targeted to the membrane (Fig. 7A). Thr444-phosphorylated NDR1 was solely detected in cells expressing mp-hMOB1A, but total NDR1 levels were also increased (Fig. 7A, lane 3). To further address the localization of excess NDR1 in mp-hMOB1A-expressing cells, cells expressing either hMOB1A or membrane-targeted hMOB1A were separated into cytoplasm and membrane and analyzed by immunoblotting (Fig. 7B), revealing that basically all excess NDR1 species accumulated at the membrane (Fig. 7B, lane 6). Strikingly, nearly all phospho-T444 protein was found enriched at the membrane as well. Whether the effect of mp-MOB1A on native NDR1 protein levels is mostly direct or indirect cannot be answered currently, but considering all the evidence provided here, it is rather unlikely that most of the effect is indirect. Since OA treatment also results in a rapid up-regulation of total NDR1 levels (Fig. 1B; see Fig. S1C in the supplemental material), it is rather tempting to speculate that activated NDR1 species are protected from rapid turnover after OA treatment or membrane accumulation. However, the protection mechanism still needs to be defined.
FIG. 7.
Membrane-targeted hMOB1A stabilizes native NDR1 proteins. A. U2-OS cells were transfected with empty vector, HA-hMOB1A, or mp-HA-hMOB1A before analysis by immunoblotting using anti-T444-P (top panel), anti-NDR1 (middle top panel), anti-HA 12CA5 (middle bottom panel), and anti-α-tubulin (bottom panel) antibodies. Arrowheads indicate NDR1 forms. B. U2-OS cells transfected with empty vector, HA-hMOB1A, or mp-HA-hMOB1A were subjected to S100/P100 fractionation (S, cytoplasm; P, membrane) before immunoblotting with anti-T444-P (top panel), anti-NDR1 (middle panel), and anti-HA 12CA5 (bottom panel) antibodies. C. U2-OS cells expressing wild-type (top panels) or membrane-targeted hMOB1A (bottom panels) were analyzed by indirect immunofluorescence with anti-T444-P (green) and anti-HA 12CA5 (red) antibodies. Nuclei are in blue.
To study the precise subcellular site of endogenous NDR1 activation at the membrane by mp-hMOB1A, U2-OS cells expressing mp-hMOB1A were subjected to immunofluorescence analysis. Similarly to overexpressed NDR1 (Fig. 6F), native Thr444-P forms localized to the plasma membrane, and in fact in selected clusters (Fig. 7C, bottom panels), suggesting activation of endogenous NDR1 at certain sites of the membrane.
Stimulated membrane association of hMOB1A leads to rapid activation of NDR1.
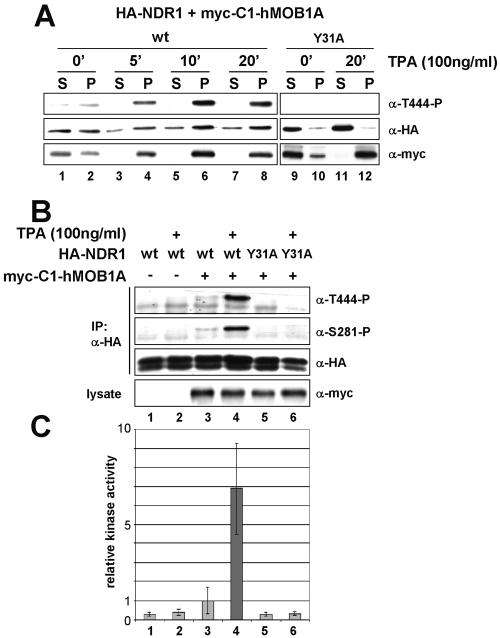
Since the effects of mp-hMOB1A accumulated over a 1-day period, any analysis of the sequence or kinetics of events leading to NDR1 activation was seriously impaired. To overcome this limitation, we created a construct that allowed rapid and inducible translocation of hMOB1A to the membrane. Earlier, a chimera of PKB and the C1 domain of PKCα was used successfully to study the kinetics of PKB activation at the membrane (2); phorbol ester (e.g., TPA) treatment of this chimera rapidly induced membrane binding. Similarly, the C1 domain fused to the N terminus of hMOB1A (C1-hMOB1A) recruited this chimeric protein exclusively to the membrane fraction after a few minutes of TPA stimulation (Fig. 8A, bottom panels). When the NDR1 (Y31A) was coexpressed, TPA-induced membrane association of C1-hMOB1A did not lead to recruitment of NDR1 species to the membrane (Fig. 8A, lane 12), suggesting that NDR1 unable to bind to C1-hMOB1A is not enriched at the membrane. Wild-type NDR1 alone was also not enriched at membranous structures after TPA treatment (data not shown). However, upon coexpression of wild-type NDR1 with C1-hMOB1A and subsequent TPA incubation, Thr444-P species accumulated rapidly in membrane fractions (Fig. 8A, lanes 4, 6, and 8). The phosphorylation reached a plateau after 20 min and remained constant for at least 60 min (data not shown). This increase in phosphorylation was accompanied by an accumulation of total NDR1 forms at the membrane, which was already obvious after 5 min (Fig. 8A, lane 4). Interestingly, a fraction of C1-hMOB1A was already present on the membrane before stimulation, causing an increase of total membranous NDR1 before TPA induction and thus the detection of some phosphorylation on Thr444 (Fig. 8A, lane 2). The increase in Thr444-P amount by TPA-induced membrane binding of C1-hMOB1A was in full agreement with NDR1 activity changes (Fig. 8C). While NDR1 coexpressed with C1-hMOB1A in nonstimulated cells was already about three- to fourfold more active than wild-type enzyme alone (due to partial membrane association of C1-hMOB1A), this activity could be further boosted sevenfold by TPA incubation (Fig. 8C, bars 3 and 4). This activation was accompanied by phosphorylation on both Ser281 and Thr444 (Fig. 8B, lanes 3 and 4). In control cells, phosphorylation of NDR1 on Ser281 and Thr444 as well as its enzymatic activity were not significantly altered by TPA treatment (Fig. 8B and C, lanes and bars 1 and 2), which excludes the possibility that the observed activation of NDR1 by membrane-bound C1-hMOB1A after TPA induction was simply due to direct activation of NDR1 by TPA. Moreover, coexpression of C1-hMOB1A and NDR1 (Y31A) resulted in neither increased enzymatic activity nor changes in phosphorylation, irrespective of TPA induction (Fig. 8B and C, lanes and bars 5 and 6). Thus, activation of NDR1 by C1-hMOB1A is a consequence of NDR1 recruitment to the plasma membrane, which is dependent on the interaction of NDR1 with hMOB1A.
FIG. 8.
Induced membrane association of hMOB1A results in rapid activation of NDR1. A. COS-7 cells expressing myc-tagged hMOB1A fused to the C1 domain of PKCα (myc-C1-hMOB1A) and wild-type (wt) HA-NDR1 or a MOB1A-binding deficient (Y31A) mutant were serum starved overnight before incubation with TPA for the indicated times. Cell lysates were subjected to S100/P100 fractionation (S, cytoplasm; P, membrane) before immunoblotting with anti-T444-P (top panel), anti-HA 12CA5 (middle panel), and anti-myc 9E10 (bottom panel) antibodies. B. Lysates of cells expressing HA-tagged wt or NDR1 (Y31A) and myc-C1-hMOB1A were analyzed by immunoprecipitation (IP) using anti-HA 12CA5 antibody. Complexes were assayed by Western blotting using anti-T444-P (top panel), anti-S281-P (top middle panel), and anti-HA (bottom middle panel) antibodies. Lysates were immunoblotted with anti-myc 9E10 antibody (bottom panel). Prior to lysis cells were incubated for 20 min with (+) or without TPA. C. In parallel, immunocomplexes were subjected to kinase assays. The significant elevation of NDR1 activity by stimulated membrane association of myc-C1-hMOB1A is highlighted in dark gray. Error bars indicate standard deviations.
Nucleus-targeted NDR accumulates in the cytoplasm after OA treatment.
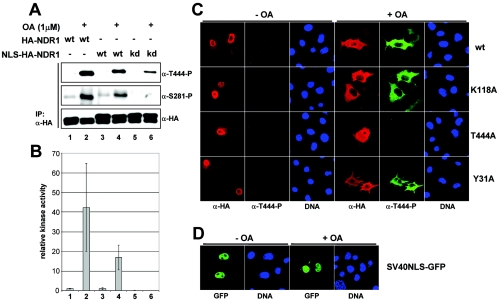
Since a fraction of NDR1 was also found to reside in the nucleus (Fig. 1 and 3), we targeted NDR1 to the nucleus to test finally whether this nuclear pool can be activated by PP2A inhibition. NDR1 tagged N terminally with the strong NLS of SV40 (NLS-NDR1) was expressed in COS-7 cells. Interestingly, NLS-NDR1 phosphorylation on Ser281 and Thr444 as well as its activity were increased by OA incubation comparably to wild-type protein (Fig. 9A and B, lanes and bars 1 to 4). As expected, when kinase-dead NLS-NDR1 was tested, Ser281 phosphorylation and kinase activity were abolished, while phosphorylation on Thr444 still occurred (Fig. 9B, bars 5 and 6). The efficient nuclear targeting of various NLS-NDR1 species was confirmed by immunofluorescence analysis (Fig. 9C, left panels). However, OA treatment led to the accumulation of Thr444-phosphorylated NLS-NDR1 in the cytoplasm (Fig. 9C, right panels). Significantly, kinase-dead (K118A) and MOB1-binding-deficient (Y31A) NLS-NDR1 proteins were observed in the cytoplasm, like their wild-type counterparts, after OA treatment (Fig. 9C), suggesting that neither kinase activity nor MOB1 binding is required for this effect. In contrast, GFP protein N-terminally tagged with the NLS of SV40 (NLS-GFP) was consistently detected in the nucleus irrespective of OA incubation (Fig. 9D), demonstrating a certain specificity for the effect of OA incubation on cytoplasmic NLS-NDR1 accumulation.
FIG. 9.
Activation of nucleus-targeted NDR1 results in cytoplasmic accumulation of NDR species. A. Lysates of COS-7 cells expressing HA-tagged wild-type (wt), SV40NLS-tagged (NLS) wt, or kinase-dead (kd) NDR1 were analyzed by immunoprecipitation (IP) using anti-HA 12CA5 antibody. Complexes were assayed by Western blotting using anti-T444-P (top panel), anti-S281-P (middle panel), and anti-HA (bottom panel) antibodies. Prior to lysis, cells were incubated with (+) or without (−) OA. B. In parallel, immunocomplexes were subjected to kinase assays. Error bars indicate standard deviations. C. COS-7 cells expressing SV40NLS-tagged wild-type (top panels), kinase-dead (K118A) (top middle panels), Thr444Ala (T444A) (bottom middle panels), or MOB1A-binding-deficient (Y31A) (bottom panels) NDR1 were either left untreated (− OA) or incubated with okadaic acid (+ OA) before processing for immunofluorescence using anti-HA 12CA5 (red) and anti-T444-P (green) antibodies. Nuclei are indicated in blue. D. COS-7 cells transfected with nucleus-targeted GFP (SV40NLS-GFP) were either left untreated (− OA) or incubated with okadaic acid (+ OA) before fixation. Nuclei are indicated in blue.
Further, coexpression of hMOBs with NLS-NDR1 changed neither NDR's phosphorylation nor its activity status (data not shown) despite recruitment of hMOBs to the nucleus by NLS-NDR1 (see Fig. S7A in the supplemental material; data not shown), while mp-NDR1 was readily stimulated by hMOB expression (Fig. 4D and E). Moreover, by showing that NLS-NDR1 recruited hMOB1A to the nucleus in an interaction-dependent manner (see Fig. S7A in the supplemental material), we could rule out the possibility that activation of nucleus-targeted NDR1 by OA only occurred in the cytoplasmic/membranous fraction due to lack of hMOB1 recruitment to the nucleus.
Finally, we addressed the effect of nucleus-targeted hMOB2 on NDR activity (see Fig. S7B, C, and D in the supplemental material). Coexpression of NLS-hMOB2 with NDR1 resulted in efficient nuclear accumulation of NDR species in most cells (see Fig. S7B in the supplemental material), but neither NDR1's phosphorylation nor its activity status was altered significantly by such a nuclear concentration (see Fig. S7C and D in the supplemental material), whereas membrane-targeted hMOBs readily could activate NDR (Fig. 6; see Fig. S6 in the supplemental material). These final data (Fig. 9; see Fig. S7 in the supplemental material) rule out the possibility that membrane activation of NDR at the membrane occurs solely as a consequence of a subcellular concentration of NDR and hMOB species but rather argue for a specific activation of NDR species at the membrane.
DISCUSSION
Even though the precise function(s) of mammalian NDR kinase is not sufficiently well defined, the available evidence suggests that under certain conditions both human NDR forms can be involved in tumor progression (1, 15, 18). NDR1 was found to be up-regulated in progressive ductal carcinoma in situ, as well as in some melanoma cell lines (1, 15). NDR1 may potentially influence the tumor metastatic potential of melanomas, since S100B can hyperactivate NDR1 (15) and S100B is overexpressed in more than 80% of metastatic melanomas (8). Furthermore, NDR2 levels are elevated in a highly metastatic non-small-cell lung cancer cell line (18). Despite these important findings, the regulation of NDR activity has not been implicated in any signal transduction pathway in vivo. hMOBs have been shown to stimulate NDR in vitro (3, 4), but overexpression of hMOB1A was not sufficient to activate NDR in vivo (3).
By readdressing the intracellular distribution of NDR1 and NDR2, we determined that both inactive and active NDR kinases were mostly cytoplasmic in our experimental settings using different cell types and epitope tags to detect NDR species. Overexpressed, as well as native, NDR1 species were detected mainly in the cytoplasm. Even nuclear targeting of NDR could not prevent its cytoplasmic accumulation after OA treatment. Interestingly, hMOB1A and hMOB1B (hMOB1) were also enriched in the cytoplasmic compartment, while hMOB2 was rather equally distributed between nucleus and cytoplasm. Nevertheless, all three hMOBs colocalized with NDR species in the cytoplasm and at the plasma membrane, suggesting that activation of NDR by hMOBs most likely occurs outside of the nucleus. The subcellular localization of NDR and hMOBs also proved to be interdependent. Targeting of NDR1 to the nucleus resulted in nuclear sequestration of hMOB1, while membrane binding of hMOBs recruited NDR forms to membranes. These findings are in contrast to previous reports identifying NDR1 as a mainly nuclear kinase (4, 5, 14). The reason for the difference is most likely due to a misinterpretation of immunofluorescence data (see Fig. S2 in the supplemental material). Nevertheless, it currently cannot be excluded that under certain conditions NDR species could be found enriched in the nucleus. However, one study (4) used only GFP-tagged hMOBs (which are shown here to affect the intracellular distribution of GFP-hMOB1B artificially), did not further test the subcellular localization of NDR by adding nuclear or membrane tags, and did not address the functionality of the postulated NLS of NDR1 or NDR2. Our current experiments tested the functionality of the NDR1 NLS (Fig. 2) and failed to provide convincing data for a strong NLS. Moreover, the N and C termini of NDR1 were not required for its cytoplasmic localization. In addition, kinase activity of NDR1 appears to be dispensable for accumulation in the cytoplasm (Fig. 3).
Residues 22 to 82, encompassing the SMA domain of NDR1, were shown to be essential for membrane association of NDR. Permanent membrane targeting of NDRs resulted in a kinase with constitutive enzymatic activity that could be further enhanced by elevation of hMOB1A levels. Membrane-bound NDR was not efficiently phosphorylated on Ser281/Ser282 in vivo. Even PP2A inhibition did not change the Ser281 phosphorylation significantly, while an increase in hMOB1A levels proved sufficient to generate Ser281-P amounts similar to those with stimulated wild-type enzyme. Although Ser281 phosphorylation was linked to NDR kinase activity, phosphorylation on Thr444 was readily seen when NDR was membrane targeted, irrespective of its own enzymatic activity or OA stimulation. This phosphorylation could be further elevated by OA treatment or hMOB1A coexpression, suggesting that phosphorylation on Thr444 is not achieved by autophosphorylation but occurs by action of an upstream kinase that can be activated by PP2A inhibition and appears also to be facilitated by hMOBs. Overall, these data are in full agreement with previous observations (3, 16, 20, 23), further supporting a model in which binding of hMOB1 to the N-terminal domain of NDR induces its Ser281 autophosphorylation activity, while phosphorylation on Thr444 is carried out by a hydrophobic motif kinase (Stegert et al., submitted for publication).
Despite giving more insight into the sequence and subcellular localization of events, these data did not reveal how fast NDR is activated at the membrane or whether MOB binding occurs prior to or after membrane association. How NDR is potentially recruited to the membrane was also not clear. We demonstrated that coexpression of membrane-targeted hMOBs is sufficient for potent stimulation of NDR activity and that activation is dependent on the efficiency of membrane recruitment of NDR species. This suggests that constitutive membrane binding of hMOBs is enough for activation of NDRs, most likely due to recruitment of NDR into spatial proximity to its upstream activators. However, due to the use of a constitutive membrane tag, these experiments were impaired in their analysis of the sequence and kinetics of events leading to NDR activation in vivo. This limitation was overcome by using a chimeric protein of hMOB1A fused to the C1 domain of PKCα. Upon phorbol ester treatment, this chimera was induced to bind to membranes, allowing analysis of the effect of rapid translocation of hMOB1A on NDR activity. NDR was found to be activated within 5 min after stimulation of membrane association of hMOB1A and remained active for at least 60 min at the membrane. Moreover, phosphorylation on Thr444 was dramatically increased and was detected solely in membrane fractions, indicating that the upstream kinase responsible for this phosphorylation is most likely constitutively active at the membrane. In Fig. 10, we propose a model for activation of NDR by hMOBs in vivo. NDR is corecruited to the membrane by hMOB, allowing its proximity to the upstream kinase of NDR and subsequent Thr444 phosphorylation. In addition, hMOB binding to the N terminus of NDR stimulates autophosphorylation on Ser281, resulting in a fully active kinase on the membrane. This active kinase is not released from the membrane (at least for 60 min under our experimental conditions), allowing local activity of NDR at certain sites close to the plasma membrane. However, the question of which hMOB form could be the main regulator of NDR is rather puzzling at the moment. Coimmunoprecipitation data (see Fig. S4 in the supplemental material) would support hMOB2, while membrane corecruitment data would rather support hMOB1A/B (Fig. 6; see Fig. S6 in the supplemental material). Of note, these differences cannot be explained by different affinities of hMOB2 and mp-hMOB2 for NDR, since mp-hMOB2 coimmunoprecipitated with NDR with much higher efficacy than mp-hMOB1A/B (data not shown). Still, in our overexpression settings, all three isoforms could activate NDR only once targeted to the membrane (Fig. 6; see Fig. S6 in the supplemental material). Overall, addressing hMOB expression patterns and putting such data into the context of NDR activation will be essential in understanding the NDR/hMOB relationship.
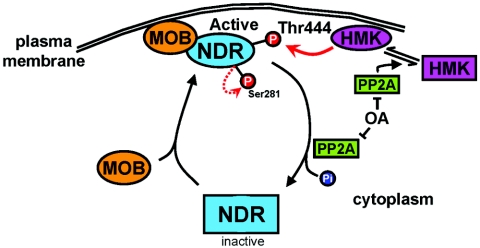
FIG. 10.
Model of potential NDR activation at the membrane. NDR is recruited by hMOB to the plasma membrane, thereby bringing NDR into the proximity of its upstream kinase (HMK [hydrophobic motif kinase]). Subsequently, NDR is phosphorylated on Thr444 by HMK, while Ser281 is phosphorylated by NDR itself, leading to full activation of NDR at the plasma membrane. The active kinase is inactivated by PP2A. The HMK is apparently also regulated by PP2A (Stegert et al., submitted for publication).
Of course, this study raises some crucial questions with respect to NDR activation on the plasma membrane, such as which physiological stimuli lead to membrane recruitment of hMOBs and which substrates are targeted at the membrane. In this context, it is important to note that, based on structural analysis of hMOB1, NDR is very likely to interact with a conserved surface exhibiting a strong negative charge on hMOB1, while a rather positively charged area located on the other side of hMOB1 might be responsible for interacting with other factors (17, 19). Potentially, this positive surface on hMOB1 could play a role in recruitment and attachment to the membrane.
The determination of a biological role of membrane-bound NDRs will be essential to fully elucidate its function. While the yeast, Drosophila, and Caenorhabditis elegans homologues of NDRs have been shown to be involved in cell cycle progression and cell morphology (22), the functions of mammalian NDR1 and NDR2 have as yet not been established. In particular, worm and fly NDR kinases have been shown previously to play central roles in neuronal processes (6, 7). However, a recent report suggests that the murine NDR2 form also plays an important role in neuronal growth and differentiation (21). Thus, it will be essential to test how much localized activation of NDR on the plasma membrane contributes to the neuronal functions described by Stork et al. (21). Nevertheless, our work provides important information relevant to the role of NDR in cancer biology. Experiments addressing the potential of NDR to transform cells, its subcellular localization in NDR-up-regulated tissues, and changes in cell morphology and migratory behavior are important in this respect. Given that NDR1 and NDR2 have been implicated in different cancer types, any molecular insights gained will provide a deeper understanding of the function(s) of NDR and its potential as a therapeutic target for treatment of human cancers.
Supplementary Material
Acknowledgments
We thank all members of our laboratory for helpful discussions. We also thank M. Pietrzak (Friedrich Miescher Institute) for DNA sequencing, F. Fischer (Friedrich Miescher Institute) for synthesis of peptides, and J. Rohrer (University of Zurich) for anti-CLIMP63 antibody. Special thanks go to P. A. Silver (Dana-Farber Cancer Institute) for providing Flag-tagged hMOB1B and hMOB2 cDNAs. We are grateful to J. Lisztwan for careful reading of the manuscript.
This work was supported by Swiss Cancer League grant KLS-01342-02-2003 (to B.A.H.), and A. Hergovich was supported by a fellowship of the Roche Research Foundation.
The Friedrich Miescher Institute is part of the Novartis Research Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adeyinka, A., E. Emberley, Y. Niu, L. Snell, L. C. Murphy, H. Sowter, C. C. Wykoff, A. L. Harris, and P. H. Watson. 2002. Analysis of gene expression in ductal carcinoma in situ of the breast. Clin. Cancer Res. 8:3788-3795. [PubMed] [Google Scholar]
- 2.Andjelkovic, M., S. M. Maira, P. Cron, P. J. Parker, and B. A. Hemmings. 1999. Domain swapping used to investigate the mechanism of protein kinase B regulation by 3-phosphoinositide-dependent protein kinase 1 and Ser473 kinase. Mol. Cell. Biol. 19:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bichsel, S. J., R. Tamaskovic, M. R. Stegert, and B. A. Hemmings. 2004. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279:35228-35235. [DOI] [PubMed] [Google Scholar]
- 4.Devroe, E., H. Erdjument-Bromage, P. Tempst, and P. A. Silver. 2004. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 279:24444-24451. [DOI] [PubMed] [Google Scholar]
- 5.Devroe, E., P. A. Silver, and A. Engelman. 2005. HIV-1 incorporates and proteolytically processes human NDR1 and NDR2 serine-threonine kinases. Virology 331:181-189. [DOI] [PubMed] [Google Scholar]
- 6.Emoto, K., Y. He, B. Ye, W. B. Grueber, P. N. Adler, L. Y. Jan, and Y. N. Jan. 2004. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119:245-256. [DOI] [PubMed] [Google Scholar]
- 7.Gallegos, M. E., and C. I. Bargmann. 2004. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron 44:239-249. [DOI] [PubMed] [Google Scholar]
- 8.Hauschild, A., G. Engel, W. Brenner, R. Glaser, H. Monig, E. Henze, and E. Christophers. 1999. S100B protein detection in serum is a significant prognostic factor in metastatic melanoma. Oncology 56:338-344. [DOI] [PubMed] [Google Scholar]
- 9.Hergovich, A., J. Lisztwan, R. Barry, P. Ballschmieter, and W. Krek. 2003. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5:64-70. [DOI] [PubMed] [Google Scholar]
- 10.Komarnitsky, S. I., Y. C. Chiang, F. C. Luca, J. Chen, J. H. Toyn, M. Winey, L. H. Johnston, and C. L. Denis. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18:2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. E., L. M. Frenz, N. J. Wells, A. L. Johnson, and L. H. Johnston. 2001. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11:784-788. [DOI] [PubMed] [Google Scholar]
- 12.Luca, F. C., and M. Winey. 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mah, A. S., J. Jang, and R. J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98:7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millward, T., P. Cron, and B. A. Hemmings. 1995. Molecular cloning and characterization of a conserved nuclear serine (threonine) protein kinase. Proc. Natl. Acad. Sci. USA 92:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millward, T. A., C. W. Heizmann, B. W. Schafer, and B. A. Hemmings. 1998. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 17:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millward, T. A., D. Hess, and B. A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847-33850. [DOI] [PubMed] [Google Scholar]
- 17.Ponchon, L., C. Dumas, A. V. Kajava, D. Fesquet, and A. Padilla. 2004. NMR solution structure of Mob1, a mitotic exit network protein, and its interaction with an NDR kinase peptide. J. Mol. Biol. 337:167-182. [DOI] [PubMed] [Google Scholar]
- 18.Ross, D. T., U. Scherf, M. B. Eisen, C. M. Perou, C. Rees, P. Spellman, V. Iyer, S. S. Jeffrey, M. Van de Rijn, M. Waltham, A. Pergamenschikov, J. C. Lee, D. Lashkari, D. Shalon, T. G. Myers, J. N. Weinstein, D. Botstein, and P. O. Brown. 2000. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24:227-235. [DOI] [PubMed] [Google Scholar]
- 19.Stavridi, E. S., K. G. Harris, Y. Huyen, J. Bothos, P. M. Verwoerd, S. E. Stayrook, N. P. Pavletich, P. D. Jeffrey, and F. C. Luca. 2003. Crystal structure of a human Mob1 protein: toward understanding Mob-regulated cell cycle pathways. Structure (Cambridge) 11:1163-1170. [DOI] [PubMed] [Google Scholar]
- 20.Stegert, M. R., R. Tamaskovic, S. J. Bichsel, A. Hergovich, and B. A. Hemmings. 2004. Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 279:23806-23812. [DOI] [PubMed] [Google Scholar]
- 21.Stork, O., A. Zhdanov, A. Kudersky, T. Yoshikawa, K. Obata, and H. C. Pape. 2004. Neuronal functions of the novel serine/threonine kinase Ndr2. J. Biol. Chem. 279:45773-45781. [DOI] [PubMed] [Google Scholar]
- 22.Tamaskovic, R., S. J. Bichsel, and B. A. Hemmings. 2003. NDR family of AGC kinases-essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73-80. [DOI] [PubMed] [Google Scholar]
- 23.Tamaskovic, R., S. J. Bichsel, H. Rogniaux, M. R. Stegert, and B. A. Hemmings. 2003. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J. Biol. Chem. 278:6710-6718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.