Abstract
Induction of brown adipocytes in white fat depots by adrenergic stimulation is a complex genetic trait in mice that affects the ability of the animal to regulate body weight. An 80-fold difference in expression of the mitochondrial uncoupling gene (Ucp1) at the mRNA and protein levels between A/J and C57BL/6J (B6) mice is controlled by allelic interactions among nine quantitative trait loci (QTLs) on eight chromosomes. Overlapping patterns of these QTLs also regulate expression levels of Pgc-1α, Pparα, and type 2 deiodinase. Independent validation that PPARα is associated with Ucp1 induction was obtained by treating mice with the PPARα agonist clofibrate, but not from the analysis of PPARα knockout mice. The most upstream sites of regulation for Ucp1 that differed between A/J and B6 were the phosphorylation of p38 mitogen-activated protein kinase and CREB and then followed by downstream changes in levels of mRNA for PPARγ, PPARα, PGC-1α, and type 2 deiodinase. However, compared to Ucp1 expression, the two- to fourfold differences in the expression of these regulatory components are very modest. It is proposed that small variations in the levels of several transcriptional components of the Ucp1 enhanceosome interact synergistically to achieve large differences in Ucp1 expression.
Stimulation of the sympathetic nervous system by cold exposure or β-adrenergic agonists induces brown adipocytes in traditional white fat depots of mice, rats, dogs, and cats, as demonstrated by their multilocular morphology, abundant mitochondria, and the expression of Ucp1 (2, 14, 21, 34, 35). This inducible brown adipocyte phenotype is significant because it reverses both diet-induced and genetic obesity (12, 21) and diabetes (19). In addition, transgenic studies have demonstrated that increasing Ucp1 expression in white fat is associated with a reduction of adiposity (15, 26, 27, 44, 48). Although humans have an abundance of brown adipocytes in fat depots at birth for regulation of body temperature (30), Ucp1 expression is only detectable in adults at very low levels (18). Several studies suggest that adult human white adipocytes have a latent ability to express Ucp1 and can be converted into brown adipocytes (11, 16, 46).
Given the potential for brown fat thermogenesis to reduce obesity and diabetes, it is important to identify mechanisms that can activate brown adipocyte differentiation in response to drug treatment. Transcriptional regulation of the Ucp1 gene is controlled by regulatory elements that have been shown to be critical for both white and brown fat adipogenesis. This includes members of the peroxisome proliferator activated receptor (PPAR) and CCAAT/enhancer binding protein (C/EBP) families and cyclic AMP (cAMP) response binding protein (CREB) (36, 40). Pivotal to this regulation is a PPRE site in the distal Ucp1 enhancer that forms a complex with PPARγ2-RXR and the coactivator PGC-1α (37, 42, 47). In addition, half-site cAMP response elements (CRE) in both the proximal promoter and the distal enhancer are critical for expression. These CRE sites interact with CREB and ATF2, and mutations within them abolish Ucp1 expression in transient expression assays (4, 7, 29, 49). The role of PPARγ in adipogenesis has been extensively documented in both tissue culture and in vivo models of adipogenesis and brown adipocyte expression (3, 20, 39, 42). While C/EBPα appears not to be required for brown fat expression (33), C/EBPβ expression and C/EBPδ expression have critical roles in brown fat differentiation (45). Also located in the Ucp1 enhancer region is a site for regulation by the thyroid hormone receptor (9), which accordingly links Ucp1 regulation to the potential influence of thyroid hormones on thermogenesis (43).
The adrenergic signaling mechanism for regulation of Ucp1 expression and brown adipocyte differentiation is a G-protein receptor mechanism coupling cAMP production to protein kinase A (PKA)-dependent phosphorylation of CREB and p38 mitogen-activated protein (MAP) kinase. The latter has been implicated in ATF2 activation by phosphorylation, with subsequent regulation of both Pgc-1α and Ucp1 transcription (7). Interactions between retinoblastoma protein and FOXC2, which has been postulated to act in the signaling cascade by inducing the level of the PKA-RIα regulatory protein, has also been implicated in the regulation of Ucp1 (10, 22).
Accordingly, the number of potential sites for regulation of Ucp1 and brown adipocyte induction is large and many of the signaling molecules and transcription factors could be involved in other aspects of adipocyte biology, as well as muscle and liver structure and function. A fundamental question is how one can modulate these pathways pharmacologically to selectively induce brown fat differentiation in white fat depots without adversely affecting regulation in other organ systems. To address this question, we have pursued a genetic analysis based upon the variation in brown adipocyte differentiation observed between A/J and C57BL/6J (B6) mice. The large differences in Ucp1 mRNA and protein levels (in some experiments up to 80-fold) between these strains suggested that the quantitative genetic analysis of signaling and transcription pathways would reveal how natural genetic variation is able to achieve selective induction of brown adipocytes in white fat depots by adrenergic signaling.
Our earlier studies quantifying Ucp1 mRNA variation in retroperitoneal (RP) fat after cold exposure of progeny from genetic crosses revealed quantitative trait loci (QTLs) on chromosomes (Chr) 2, 3, 8, and 19 that interact synergistically to control the induction of Ucp1 mRNA levels in white fat (28). This study also showed how interactions of A/J- and B6-derived alleles between QTLs lead to transgressive variation. Except for Ucp1 on Chr 8, none of the QTLs were located in regions carrying genes associated with Ucp1 expression by molecular studies. When it was proposed that Pgc1α was specifically expressed in brown adipocytes and a master regulator of brown fat differentiation and Ucp1 expression (37), we took advantage of a difference in Pgc1α mRNA levels between A/J and B6 mice to map QTLs controlling Pgc1α mRNA levels and to link its expression to the regulation of Ucp1 (13). The findings of this study showed that (i) some QTLs regulated both Ucp1 and Pgc1α, whereas others were unique to each gene; (ii) there were no mutations in the Pgc1α coding region or in a 10-kb region upstream of the transcription start site to account for variable expression; (iii) the region of Chr 5 in which the Pgc-1α gene was located did not carry a QTL for regulation of Ucp1 or Pgc1α mRNA, indicating that differences in Pgc-1α expression were determined by allelic differences in genes on other Chr; and (iv) the introduction of an environmental variation, an HF diet, had profound effects on the QTLs controlling the expression of both Ucp1 and Pgc1α. These data suggested that environmental influences on body weight such as diet can modulate the genetic regulation of thermogenesis in white fat. With the genetic analysis of Pgc1α, the number of QTLs controlling genes associated with brown fat induction had grown to 7 and, except for the Ucp1 structural gene on Chr 8, none of them showed linkage to genes encoding signaling or transcription factors controlling Ucp1.
We have expanded our delineation of Ucp1 induction to both adrenergic signaling and transcription pathways associated with brown adipogenesis in white fat depots. We propose a model in which the 80-fold variation in Ucp1 expression is determined by synergistic interactions of modest two- to fivefold differences in the levels of several signaling and transcription factors that are part of a regulatory cascade. Large variations in PPARα mRNA levels together with linkage of a QTL to the Pparα gene on Chr 15 led us to validate a role for PPARα in the Ucp1 induction with further analysis of the Pparα knockout (KO) mouse and in mice treated with a PPARα agonist. The absence of this transcription factor was found to have no influence on the levels of Ucp1 mRNA despite evidence for a strong association between Pparα and Ucp1 mRNA levels in backcross progeny; however, mice treated with a PPARα agonist show increased levels of Ucp1 mRNA. The most upstream site of regulation for Ucp1 that differed between A/J and B6 was the phosphorylation of p38 MAP kinase and CREB; however, no differences in the total amount of protein or the level of phosphorylation of the regulatory subunits of PKA were detected that could account for this variation. We propose that the large differences in the induction of Ucp1 in RP fat are determined by modest variations in several components of both signaling and transcription that function in a synergistic manner to achieve both tissue specificity and a high level of redundancy.
MATERIALS AND METHODS
Animals.
The A/J and B6 strains were obtained from The Jackson Laboratory (Bar Harbor, ME). The F1 hybrid and (B6 × A/J)F1 × A/J backcross mice were set up in Pennington Biomedical Research Center. Pparα KO mice were purchased from The Jackson Laboratory, and the HcB-19/Dem recombinant congenic strain carrying a spontaneous null mutation in the gene encoding the thioredoxin-interacting protein (TXNIP) was generously provided by Peter Demant (5). All parental mice and their pups were kept at room temperature (22 ± 1°C) with a 12-h light-dark cycle. Animals exposed to cold were carefully monitored in accordance with procedures approved by the Animal Care and Use Committee.
For genetic analysis of the regulation of PPARα and type II deiodinase (Dio2) in RP fat, A/J, B6, F1 hybrid, and (B6 × A/J)F1 × A/J backcross mice from dams fed a low-fat chow diet (LF; 11.9 kcal% fat, 23.6 kcal% protein, and 64.5 kcal% carbohydrate; Picolab 5053 Rodent Diet 20) were weaned at 21 days of age onto either the LF chow diet or a high-fat diet (HF, 58.0 kcal% fat, 16.5 kcal% protein, and 25.5 kcal% carbohydrate; Research Diets D12331) fed ad libitum. Male mice at 2 months of age, fed either an HF or an LF diet, were placed in the cold (5°C) for 7 days. Mice were sacrificed by cervical dislocation, and RP fat was dissected for RNA isolation and the tail was removed for DNA isolation and genotyping. Two-month-old male B6 and A/J mice were treated with the PPARα agonist clofibrate (Sigma, St. Louis) by feeding it ad libitum at a concentration of 0.1% (wt/wt) in powdered rodent chow (Picolab 5053) during the 7 days that mice are exposed to the cold. RP fat tissue and liver was removed for the preparation of total RNA.
Phenotypic assays. (i) Quantitative RT-PCR.
Total RNA was isolated using TriReagent (Molecular Research Center, Cincinnati, OH). Ucp1 and other mRNA levels were measured by quantitative reverse transcription (RT)-PCR using either the ABI Prism 7700 or the ABI 7900 sequence detection system (Applied Biosystems, Foster City, CA) as described previously (28). Briefly, RNA was diluted to 50 ng/μl (RP fat) or 20 ng/μl (interscapular brown adipose tissue [BAT]) in formamide and then diluted in deionized, RNase-free water prior to analysis. Ten microliters of diluted RNA (5 ng of RP fat or 2 ng of interscapular BAT) was used in a 50-μl reaction mixture with single-reporter measurement. The sequences of the primers and probes are available upon request. RNA from the liver was used to generate a standard curve for PPARα. All other genes were quantified from a standard curve generated by using interscapular BAT RNA as previously described (28). Both standards and samples were run in duplicate. Each transcript was corrected by cyclophilin.
(ii) Mitochondrial DNA measurement.
Total DNA was isolated using TriReagent (Molecular Research Center Inc., Cincinnati, OH) after the isolation of RNA according to manufacturer's instructions. Mitochondrial mass was measured in DNA samples by quantitative PCR as described above using primers and probe corresponding to cytochrome c oxidase subunit I (COXI), which is encoded by mitochondrial DNA, and corrected with Ucp2, a nucleus-encoded gene. DNA from interscapular BAT was used as the standard.
(iii) Western blot analysis.
Interscapular BAT and RP tissue were homogenized in buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM sucrose, 1 mM EDTA, 1% Triton X-100, and 1% each protease, serine phosphatase, and tyrosine phosphatase inhibitor cocktail (Sigma, St. Louis, MO) and incubated on ice for 1 h, and supernatant was collected after centrifugation at 5,000 × g for 15 min at 4°C. After determining protein concentrations by a modified Lowry method (Bio-Rad, Hercules, CA), 60 μg of protein was mixed with 2× Laemmli sample buffer and subjected to 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (29a), depending on the size of target proteins. After transfer to a polyvinylidene difluoride membrane (Millipore, Bedford, MA), the blot was blocked with 5% nonfat dry milk-0.01 M phosphate-buffered saline (PBS) for 4 h at room temperature and incubated with primary antibodies diluted in 1% nonfat dry milk-PBS at 4°C overnight with gentle shaking. Rabbit anti-UCP1 (1:10,000) and anti-PGC1α (1:4,000) polyclonal antibodies were kindly provided by Thomas Gettys (Pennington Biomedical Research Center, Baton Rouge, LA). Rabbit anti-Pparα (1:1,000) polyclonal antibodies were purchased from Santa Cruz (Santa Cruz, CA). The blot was washed and incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Amersham Pharmacia Biotech, Piscataway, NJ) diluted 1:10,000 in PBS at room temperature for 2 h. Protein signals were visualized using the enhanced chemiluminescence system (ECL; Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
(iv) Immunohistochemistry.
RP tissues were fixed in 4% paraformaldehyde, paraffin embedded, cut into 3-μm-thick sections, and mounted onto slides (SuperFrost Microscopy Slides. Fisher Scientific, Pittsburgh, PA). Immunohistochemistry was performed according to Fekete et al. (18a), with some modifications and using the rabbit anti-UCP1 antibody at a 1:2,000 dilution.
(v) Type II deiodinase (Dio2) activity measurement.
Dio2 activity was measured using 1 nM 125I-labeled T4 (specific activity, 1,000 μCi/μg; Perkin-Elmer, Boston, MA) as described by Schneider et al. (41). The labeled substrate was purified using LH-20 (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) column chromatography on the day of the experiment. Nonenzymatic deiodination was corrected by subtracting the 125I− released in tissue-free incubations. Dio2 activity was reported as femtomoles of I− released per milligram per hour. Results were multiplied by 2 to correct for random labeling at the equivalent 3′ and 5′ positions.
Genotyping.
Genotyping was performed using microsatellite markers with tail DNA as described previously (28). Four hundred (B6 × A/J)F1 × A/J backcross mice each on either an LF or an HF diet and exposed to cold at 5°C for 7 days at 2 months of age were analyzed. A genome-wide scan was performed separately on mice with the 50 highest and 50 lowest levels of mRNA in both the LF and HF groups, with markers spaced at 20-cM intervals on each Chr. Fine mapping was performed on Chr showing significant associations with the expression of Pparα and Dio2 using all 400 mice in each group. Data were analyzed using single-factor analysis of variance (Microsoft Excel).
RESULTS
Regulation of transcript levels.
Genetic variation in Ucp1 between A/J and B6 mice is principally observed in RP white fat and not at all in interscapular brown fat. Using QTL analysis of mRNA levels, the variation provided an opportunity to delineate controlling steps within the complex regulatory pathways that begin with adrenergic signaling on the surface of the adipocyte and end with the transcriptional regulation of Ucp1 (17). We began by using quantitative RT-PCR to evaluate the variation of mRNA levels for Pref1, Nfe2l2, SREBP-1c, PPARγ, PPARγ2, CEBP-α, CEBP-β, CEBP-δ, PPARα, FoxC2, β3-adrenergic receptor, and type 2 T4 deiodinase (Dio2), in addition to Ucp1 and Pgc-1α, in RP fat of A/J and B6 mice after 7 days of cold exposure. The β3-adrenergic receptor mRNA was about threefold higher in B6 mice, FoxC2 was unchanged, while all other genes showed higher expression in A/J mice (data not shown). Except for PPARα and Dio2 (Fig. 1G and H, and Fig. 2), the other genes showed less than a twofold difference in induction between A/J and B6 mice and were not included in the analyses because the extent of variation was considered too small for genetic analysis.
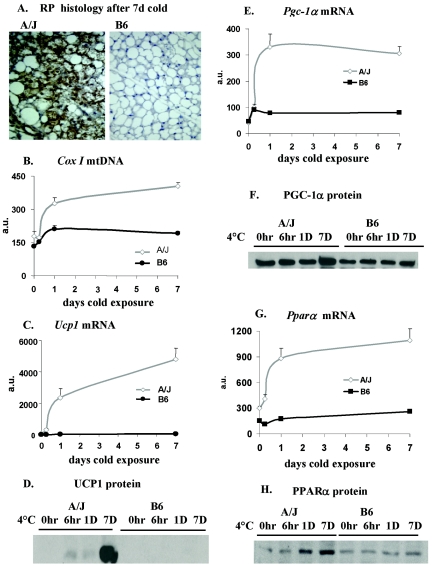
FIG. 1.
Illustration of the morphological and molecular phenotypes associated with differences in the induction of brown adipocytes in the RP fat depots of A/J and B6 mice. Immunohistologic analysis and determinations of mRNA and protein levels were carried out as described in Materials and Methods. The data in the graphs are based upon five A/J and three B6 mice analyzed in duplicate and are presented as the mean ± the standard error. D or d, days; a.u., arbitrary units.
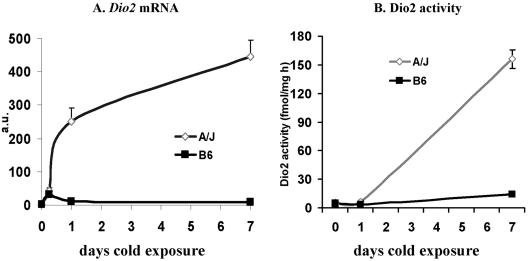
FIG. 2.
Changes in Dio2 mRNA and enzymatic activity levels in the RP fat depots of mice exposed to cold for the indicated times. The analysis is based upon three to five mice for each time point. After 1 and 7 days of cold exposure, the difference in mRNA levels between strains was significant at P < 0.01. a.u., arbitrary units.
For this experiment, the acquisition of brown fat morphology in the RP tissue of A/J mice following cold exposure and the absence of this induced phenotype in B6 mice are shown in Fig. 1A. The increase in brown adipocytes was accompanied by an increase in mitochondrial DNA content (Fig. 1B) and a large, 80-fold increase in Ucp1 mRNA (Fig. 1C) and UCP1 protein (Fig. 1D). A 3.5-fold increase in Pgc-1α mRNA occurred selectively in A/J mice (Fig. 1E); however, the increase in Pgc-1α protein was less than twofold (Fig. 1F). PPARα mRNA was induced 4.5-fold, and this was accompanied by a comparable increase in its protein (Fig. 1G and H). Finally, type 2 deiodinase mRNA and enzymatic activity were induced 47- and 15-fold, respectively, in A/J mice, with virtually no induction observed in B6 mice (Fig. 2A and B). These robust differences in RP fat deiodinase and PPARα expression between A/J and B6 mice were considered to be sufficient for QTL linkage analysis. They also lend support to the hypothesis that PPARα and deiodinase play key roles in the regulation of brown adipocyte induction in white fat.
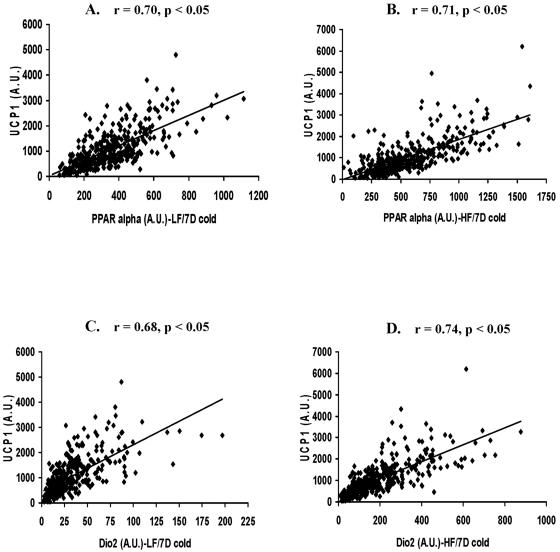
Our previous results showed that in addition to the use of allelic variation to establish functional interactions among genes, dietary manipulations, that is, an HF diet, caused perturbations in expression that can further test the role of specific components in a regulatory model (13). Whereas an HF diet caused small increases in Ucp1 and Pgc-1α (13), Pparα and Dio2 mRNA levels were strongly induced in the A/J, B6, F1 parental, and backcross mice exposed to cold (Table 1). The mRNA levels of Pparα and Dio2 were determined in two cohorts of approximately 400 (B6 × A/J)F1 × A/J backcross males, one fed an LF diet and the other on an HF diet, that were used previously in a QTL analysis of Ucp1 and Pgc-1α expression after 7 days of cold exposure (13). Among these backcross mice, PPARα mRNA levels in RP tissue varied 60-fold and levels of Dio2 varied 80-fold (Fig. 3). The strength of the associations between mRNA levels for the regulatory molecules and Ucp1 were estimated by regression analysis (Fig. 3 and Table 2). The results suggest that in the presence of an LF diet about 50% of the variation in Ucp1 mRNA levels is associated with variation in the expression of Pgc-1α, Pparα, and Dio2 and significant, though lower, correlations occurred among these regulatory molecules. The HF diet has a major effect on the strength of the correlations involving Pgc-1α, particularly with Ucp1, where the R2 value drops from 0.55 to 0.16 (Table 2). An HF diet did not affect the correlations of Ucp1 with Pparα and Dio2. To provide evidence that correlations among Ucp1, Pparα, and Pgc1α are specific, we showed no significant correlations with TXNIP, which is encoded by a candidate gene located within the Chr 3 QTL (described below).
TABLE 1.
Expression of Dio2 and PPARα after 7 days of cold exposure in RP tissue of A/J, B6, F1, and backcross animals on either an HF or an LF dieta
| Gene and strain | LF, 7 days of cold | HF, 7 days of cold |
|---|---|---|
| Dio2 | ||
| A/J | 427.23 ± 24.07 (n = 15) | 710.15 ± 128.32 (n = 8) |
| B6 | 30.25 ± 4.19 (n = 12)* | 118.30 ± 15.70 (n = 10) |
| F1 | 74.34 ± 10.40 (n = 7) | 278.11 ± 65.92 (n = 11) |
| BC | 31.58 ± 1.35 (n = 388)* | 194.34 ± 7.67 (n = 384) |
| PPARα | ||
| A/J | 813.54 ± 49.20 (n = 15) | 1120.12 ± 150.26 (n = 8) |
| B6 | 201.92 ± 18.62 (n = 12) | 366.79 ± 70.3 (n = 10) |
| F1 | 260.08 ± 22.85 (n = 7) | 526.31 ± 46.67 (n = 11) |
| BC | 343.49 ± 8.67 (n = 387) | 574.44 ± 15.30 (n = 386) |
Values (arbitrary units) labeled with the same symbol are not significantly different from each other at P < 0.05. The values shown are means and standard deviations.
FIG. 3.
Correlation analysis between Ucp1 and Pparα mRNA levels in backcross progeny exposed to cold at 5°C and fed either an LF diet (A) or an HF diet (B). A correlation analysis was also made between Ucp1 and Dio2 mRNA levels in backcross mice fed an LF diet (C) or an HF diet (D). There were 387 and 386 mice in the LF and HF groups, respectively. D, days; A.U., arbitrary units.
TABLE 2.
Regression analysis of mRNA levels in backcross progeny
| Gene comparison | Diet | R2 value |
|---|---|---|
| Ucp1 vs Pgc1α | LF | 0.55 |
| Ucp1 vs Pgc1α | HF | 0.16 |
| Ucp1 vs PPARα | LF | 0.49 |
| Ucp1 vs PPARα | HF | 0.50 |
| Ucp1 vs Dio2 | LF | 0.46 |
| Ucp1 vs Dio2 | HF | 0.55 |
| PGC-1α vs PPARα | LF | 0.425 |
| PGC-1α vs PPARα | HF | 0.311 |
| PGC-1α vs Dio2 | LF | 0.382 |
| PGC-1α vs Dio2 | HF | 0.172 |
| PPARα vs Dio2 | LF | 0.228 |
| PPARα vs Dio2 | HF | 0.262 |
| TXNIP vs Ucp1 | LF | <0.01 (R = −0.04) |
| TXNIP vs PGC-1α | LF | <0.01 |
| TXNIP vs PPARα | LF | <0.01 |
| TXNIP vs Dio2 | LF | <0.01 |
QTL analysis.
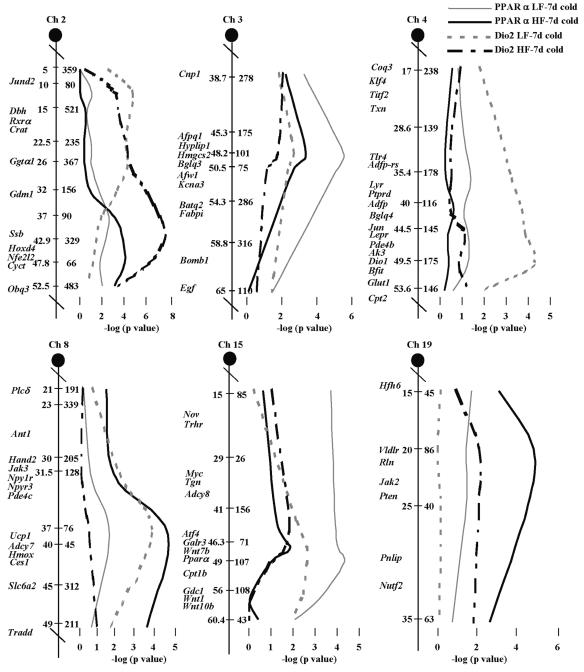
The regression analysis described above suggested that if the levels of mRNA are highly correlated among backcross progeny, it may indicate that genetic variation of a gene acting upstream of both Ucp1 and Pgc-1α will coordinately affect their expression. For example, changes in the signaling properties of p38 MAP kinase could affect both Ucp1 and Pgc-1α, since there is evidence that ATF2 is a transcription factor for both genes and is activated by phosphorylation by p38 MAP kinase (7). Accordingly, a QTL that is associated with the expression of several genes could encode an upstream regulatory gene. Genome-wide scans were performed to determine whether related genetic pathways, as determined by the presence of overlapping QTLs, regulate the expression of Pparα, Dio2, Pgc-1α, and Ucp1. We first showed that four QTLs on Chr 2, 3, 8, and 19 had a strong linkage with levels of Ucp1 mRNA (28). A subsequent linkage analysis showed that the QTL on Chr 3 also controlled the levels of Pgc-1α mRNA along with QTLs on Chr 4 and 10 (13). In this study, a QTL with a strong association with Pparα and a weak association with Dio2 is also located at D3Mit101 on Chr 3 (Table 3). Linkage and regression analyses also showed that Pgc-1α expression was associated with Pparα and/or Dio2 mRNA levels, particularly with an LF diet (Table 2), in a manner similar to its association with Ucp1 (13). Furthermore, on an LF diet, the QTL on Chr 3 had strong linkages to all mRNAs examined (Table 3 and reference 13), possibly by acting to coordinately regulate multiple pathways involved in brown adipocyte induction. Two additional QTLs on Chr 4 and 15 have been identified with strong linkage to Dio2 and Pparα. Since D15Mit107 shows the most significant association with Pparα mRNA levels and is located within 1 cM of the structural gene for Pparα, the possibility that polymorphisms in the Pparα promoter or coding regions between the A/J and B6 strains are determining variation in Pparα expression needed to be evaluated.
TABLE 3.
Chr linkage to cold-induced Dio2 and PPARα expression in backcross mice
| Chr | Marker | Position (cM) |
P value
|
|||
|---|---|---|---|---|---|---|
|
Dio2
|
PPARα
|
|||||
| LF diet | HF diet | LF diet | HF diet | |||
| 2 proximal | D2Mit80 | 10 | 5.3 × 10−5 | 5.6 × 10−4 | NSa | NS |
| 2 distal | D2Mit66 | 47.8 | 5.0 × 10−3 | 2.9 × 10−6 | 0.01 | 1.9 × 10−4 |
| 3 | D2Mit101 | 48.2 | 3.2 × 10−3 | NS | 4.9 × 10−6 | 0.01 |
| 4 | D4Mit175 | 49.5 | 5.3 × 10−5 | NS | NS | NS |
| 5 | D5Mit182 | 24 | 2.1 × 10−3 | NS | 1.2 × 10−3 | NS |
| 8 | UCP1 | 37 | 2.0 × 10−4 | NS | 0.03 | 1.1 × 10−5 |
| 15 | D15Mit107 | 49 | 2.8 × 10−3 | NS | 7.4 × 10−5 | NS |
| 19 | D19Mit86 | 20 | NS | 7.0 × 10−3 | 0.03 | 9.5 × 10−6 |
NS, no significant difference.
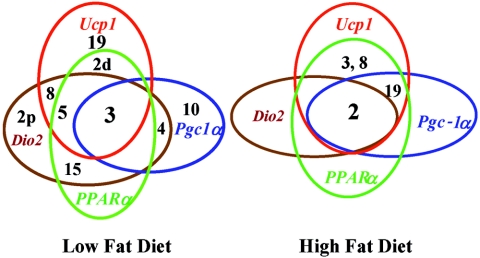
The dramatic effects of high dietary fat on the number and strength of the QTLs regulating Pparα and Dio2 are shown in Table 3 and the chromosomal linkage maps in Fig. 4. As observed in an earlier study of QTLs regulating Ucp1 and Pgc-1α in this cohort of backcross mice generated in our laboratory (13), Chr 2 and 19 have much a stronger linkage in mice fed the HF diet and Chr 3 has a reduced linkage. There is also a marked reduction in the association of phenotypes with Chr 4 and 15 with an HF diet. Interestingly, linkage of the Chr 8 QTL, using Ucp1 as a chromosomal marker, becomes significant for Pparα with an HF diet but disappears for Dio2 (Table 3). A Venn diagram summarizing the linkage relationships of QTLs associated with Ucp1 mRNA levels during brown adipocyte induction in RP white fat is shown in Fig. 5. Several observations are noteworthy. First, the number of QTLs controlling the expression of these genes is drastically reduced when mice are fed an HF diet. Second, no new QTLs are uncovered in the presence of an HF diet. Third, Chr 2 and 3 appear to be broadly involved in the regulation of brown adipocyte induction, since they influence the expression of all four genes; however, all QTLs seem to be involved in the regulation of more than one gene.
FIG. 4.
Interval maps of QTLs controlling Pparα and Dio2 mRNA levels in mice fed an LF or an HF diet based upon recombinations between microsatellite markers using 387 and 386 mice fed an LF or an HF diet. Microsatellite markers are shown on the right side of each Chr, and centimorgan (cM) distances and candidate genes are shown on the left side. The statistical significance of the association between mRNA levels and chromosomal markers is given as the negative log of the P value. The P values for peak markers are given in Table 3. Linkage maps were generated by analysis of variance of microsatellite markers with gene expression levels and positioned on the Chr with the use of the Jackson Laboratory genome database. d, days.
FIG. 5.
Venn diagrams for Chr and gene expression illustrating the overlapping function of selective Chr in regulating genes associated with the regulatory network controlling Ucp1 function. The diagrams also underscore the profound effects of diet on genes associated with regulatory networks of Ucp1 expression. Numbers refer to Chr carrying QTLs, and those within an oval are those QTLs associated with regulation of the indicated gene.
Candidate gene analysis.
Linkage maps for the QTLs regulating Pparα and Dio2 in the presence and absence of an HF diet are shown in Fig. 4. Linkage maps for Ucp1 and Pgc-1α have been previously published and are in good agreement with these maps (13, 28). While the locations of the QTLs generally cover a fairly broad region, we nonetheless selected two regions, the QTLs on Chr 3 and 15, for further analyses; Chr 3 was selected because the marker D3Mit101 was the peak marker for four mRNA phenotypes in three different crosses. Accordingly, it was considered to contain a gene that could coordinate the expression of Ucp1, Pgc-1α, Pparα, and Dio2. We also evaluated whether the QTL on Chr 15 was the structural gene for PPARα by analyzing the PPARα KO mouse for expression of Ucp1 and evaluating the effects of a PPARα agonist on expression.
The Celera database listed 152 potential genes in a 6.5-Mb region on Chr 3 flanked by D3Mit76 and D3Mit75. Seventy-five of these were found to be expressed in RP fat by RT-PCR and then analyzed further by SYBR Green1 quantitative RT-PCR with RNA isolated from RP tissue of A/J and B6 mice after cold exposure. Two transcripts were induced more than twofold higher in A/J mice relative to B6. One of these candidate genes, encoding TXNIP, was also induced by an HF diet in a manner that reduced the difference in expression between A/J and B6 mice, consistent with the suppression of the Chr 3 QTL in the presence of an HF diet. To further evaluate a role for TXNIP in Ucp1 induction, we analyzed expression in the HcB-19 mouse, which carries a spontaneously generated null allele for TXNIP (5). Compared to the control strain, predominantly derived from C3H, no reduction in Ucp1 expression was found in RP tissue after cold exposure. This result, together with the lack of correlation between TXNIP and Ucp1 expression (Table 2), suggests that TXNIP is not a candidate for the QTL for Ucp1-related gene regulation on Chr 3. The other gene with twofold greater expression in A/J mice encoded a synaptobrevin-related vesicle trafficking protein and was considered not a promising candidate gene.
The strong correlation between Ucp1 and Pparα mRNA levels shown in Fig. 3 suggested that Pparα was important for Ucp1 expression. We further evaluated whether the Pparα structural gene on Chr 15 was involved in determining Ucp1 expression in the Pparα KO mouse and by treating B6 and A/J mice with a PPARα agonist. Analysis of Pparα KO mice on a 129Sv/ImJ background (Ucp1 is induced by cold in 129Sv/ImJ mice to a similar extent as in A/J mice) failed to show a reduction in Ucp1 mRNA in the RP fat of mutant mice after cold exposure of the mutant (Table 4). In addition, PPARγ protein and Pparδ mRNA levels were unchanged between the mutant and wild-type mice, indicating that these PPARs did not compensate for lack of PPARα with increased expression. Pgc-1α and Dio2 mRNA levels were also not reduced in the Pparα KO mice. In contrast to these results, treatment of A/J and B6 mice with PPARα enhanced the expression of Ucp1. Although drug treatment of A/J mice did not lead to a statistically significant difference, there was a striking reduction in the variance in A/J mice treated with the agonist (Table 4). On the other hand, B6 mice, which have much lower levels of Ucp1 expression, showed a significant increase in Ucp1 mRNA levels.
TABLE 4.
Ucp1 expression in mice with an inactive PPARα gene or treated with a PPARα agonista
| Strain, genotype or treatment | RP tissue, Ucp1 | Liver, AOXb |
|---|---|---|
| 129, Pparα−/− | 133.8 ± 16.5* | NDc |
| 129, Pparα+/+ | 111.5 ± 23.3* | ND |
| A/J, control diet | 42.7 ± 7.3* | 3.57 ± 0.28* |
| A/J, clofibrate | 46.3 ± 1.8* | 8.83 ± 0.66† |
| B6, control diet | 0.91 ± 0.39* | 9.93 ± 1.32* |
| B6, clofibrate | 4.54 ± 1.09† | 19.12 ± 1.49† |
Statistical comparisons were made between Pparα genotypes and between A/J and B6 strains treated or not treated with PPARα agonists. Groups with the same symbol are not significantly different from each other, while those with different symbols are significantly different at P < 0.01.
AOX, acyl coenzyme A oxidase.
ND, not determined.
Protein levels and phosphorylation patterns of signaling molecules.
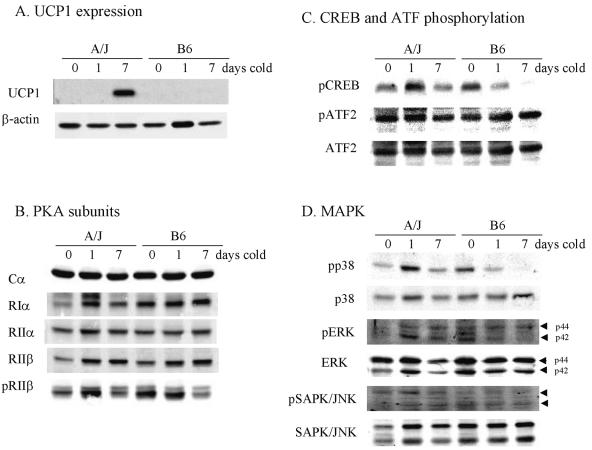
The analysis of the signaling pathway for activation of Ucp1 transcription by adrenergic stimulation and developmental mechanisms has identified several molecules in the pathway that could be targets for regulation of Ucp1 expression. Evidence suggests that a retinoblastoma pathway and an adrenergic receptor/adenylyl cyclase pathway converge on PKA, where they can modulate both the levels and phosphorylation state of PKA regulatory subunits (7, 8, 10, 15, 22). We have compared the levels of total protein and phosphoprotein for PKA subunits, pCREB, ATF2, p38 MAP kinase, ERK, and pSAPK/JNK to UCP1 in A/J and B6 mice following 1 and 7 days of cold exposure (Fig. 5). Shorter exposures to cold showed rapid phosphorylation of CREB and p38 MAP kinase in interscapular BAT but not in RP fat (38). This may reflect the rapid physiological response of fully differentiated adipocytes in interscapular BAT but a slower response to the process of differentiation in RP fat. The difference in the levels of UCP1 protein between strains was robust and very similar in magnitude to differences observed for mRNA (Fig. 6A). On the other hand, variations in the signaling molecules, either in total protein or in the phosphorylated state, were modest. Consistently higher levels of three- to fivefold were found in A/J mice for total protein and phosphoprotein for PKA subunits RIα and RIIβ (Fig. 6B) and for pCREB and p38 MAP kinase (Fig. 6C and D) following 1 day of cold exposure. Inexplicably, B6 mice showed higher signals for these targets prior to exposure to cold (0 days of cold), but these signals were reduced after 1 day of cold exposure. There were no differences in the levels of retinoblastoma protein and its phosphorylated form as measured by immunoblotting. FoxC2 mRNA levels were determined in two groups of 12 backcross progeny each that were selected for low and high levels of Ucp1 mRNA. In the group with low Ucp1 mRNA levels (18.0 ± 3.41 [standard deviation]) and high Ucp1 mRNA (178 ± 30.9), the levels of FoxC2 mRNA were not significantly different (40.9 ± 40.5 and 47.1 ± 31.4, respectively). These data do not support a role for the FoxC2 pathway as being important for regulating the genetic variation in Ucp1 induction in RP fat depots.
FIG. 6.
Protein expression and phosphorylation patterns for Ucp1 expression and cell signaling components in total tissue extracts of RP fat from A/J and B6 mice exposed to cold for the indicated times. Procedures are described in Materials and Methods.
DISCUSSION
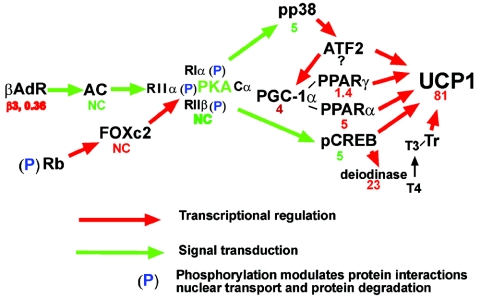
Brown adipocyte induction in white adipose tissue is a complex genetic trait that is highly sensitive to environmental factors. We have shown that B6 mice are missing the robust cold-stimulated increases in expression of Ucp1 and Pgc-1α in RP fat observed in A/J mice. Previous genetic analyses of this variable trait in backcross progeny derived from A/J and B6 mice indicate that differences in the levels of Ucp1 and Pgc-1α are determined by genes in several QTLs. In the present study, we have extended the analysis of genetic variability to additional transcription factors and signaling molecules of adipogenesis and Ucp1 expression. Differences between A/J and B6 mice in the levels of mRNA expression of several transcription factors, including the phosphorylation state of the factors and signaling molecules, were found to be very modest compared to the large differences in Ucp1 expression. The regulatory pathways depicted in Fig. 7 identify those steps for which data have been obtained, where we have compared the levels of expression between A/J and B6 mice, in some cases at the level of transcripts (Fig. 7, red) and in others at the level of protein or protein phosphorylation (Fig. 7, green). A striking finding is that the difference between strains in the level of Ucp1 gene expression is much greater than the differences found for any of the components in the signaling and transcription pathways; the large difference in the expression of Dio2 is an exception. We had anticipated that mutation(s) of one or a few regulatory molecules would cause differences in their expression comparable to that observed for Ucp1 itself. In contrast, rather modest reductions in expression for several components of the regulatory pathway from the adrenergic receptor to the Ucp1 target gene are sufficient to prevent brown adipogenesis in B6 RP tissue. These data on signaling and transcription molecules associated with Ucp1 expression and brown adipogenesis suggest a model of transcriptional synergy, as described for regulation of the beta interferon gene (25, 31), whereby cooperative interactions between low concentrations of components of the enhanceosome generate an amplified response to produce large differences in transcription. According to this model, transcription factors in B6 mice fail to reach the critical levels necessary for the formation of functional transcriptional complexes on the Ucp1 promoter. In addition to levels of transcription factor protein, the failure of a transcriptional complex to assemble will also be influenced by the phosphorylation state of some factors, for example, CREB and p38 MAP kinase.
FIG. 7.
Signaling and transcription components of Ucp1 regulation. The number below each regulatory factor refers to the ratio of expression in A/J versus B6 mice. NC is no change, red is mRNA expression, and green represents total protein levels or protein phosphorylation or activity.
The goal of the QTL analysis has been to identify genetically variant genes that regulate the capacity of an animal to induce brown adipocytes. Our evidence indicates that as many as nine QTLs control Ucp1 expression in RP fat. The expectation of analysis of quantitative variation in Ucp1 gene expression was that several of these QTLs would map to chromosomal sites that encode either these factors or other genes that regulate Ucp1. However, we have evidence that only the QTL on Chr 15, encoding PPARα, is associated with Ucp1 expression. As outlined in Fig. 7, many of the genes encoding factors associated with Ucp1 regulation are variable between A/J and B6 mice. In addition to the mapping data, much evidence supports a role for PPARα as one of the QTLs controlling Ucp1 expression in RP tissue. PPARα is an important transcriptional factor required for the regulation of genes of fatty acid oxidation (1). Pparα is expressed in brown fat, and it has been shown that PPARα activators can increase expression of the Ucp1 gene both in vivo and in transient expression assays where Pparα expression vectors stimulated transcriptional activity of reporter genes driven by Ucp1 promoter constructs (4). Cold exposure also induces total PPARα protein levels and its nuclear translocation in rodent brown fat (38). Our QTL data showed that there is a strong correlation between the expression of Pparα and that of Ucp1 in backcross mice. In addition, QTLs on Chr 2, 3, 5, 8, and 19 contribute to the high expression of both Ucp1 and Pparα in backcross mice, depending on whether the mice are fed an LF or an HF diet. Furthermore, an HF diet causes a twofold increase in Pparα mRNA levels in RP tissue, and finally we show that treatment of A/J and B6 mice with a PPARα agonist can enhance Ucp1 mRNA expression. This combination of in vivo and in vitro molecular and genetic data provides compelling evidence in support of the idea that PPARα is an important regulator of Ucp1 expression. On the other hand, how do we now reconcile this litany of data supporting the hypothesis that PPARα is a QTL for Ucp1 induction with the finding that there was no reduction in Ucp1 expression in the Pparα KO mice and no evidence for compensation by the Pparγ or -δ gene in the mutant mice (data not shown)? It has also previously been shown that Pparα KO mice are resistant to cold and have normal levels of Ucp1 expression in interscapular BAT (24). We think that our study indicates that for complex quantitative genetic traits involving epistatic interactions and transgressive expression, the simple gene KO experiment may not be capable of evaluating the potential role of the gene in the phenotype. It is necessary to evaluate the effects of the differences in genetic background and, perhaps more importantly, to understand the adaptation adipose tissue may undergo when an important transcription factor like PPARα has been absent throughout the development of the mouse.
The rationale for investigating mechanisms for induction of brown adipocytes in white fat depots of adult animals is that this could be a thermogenic mechanism for the regulation of body weight. No such mechanism has been identified either in humans or in experimental animal models that has been designed for this purpose. We have speculated that the metabolic purpose for induction of brown adipocytes in traditional white fat depots of “adult animals” is to maintain a normal body composition. Accordingly, if the expression of brown adipocytes in white fat is related to the maintenance of energy balance, then an environment that perturbs the energy balance should affect the regulation of brown adipocyte induction. To test this idea, we conducted a QTL analysis of mice fed an HF diet from weaning until they were sacrificed at the end of the cold induction period. The data indicate that the effects of an HF diet on parameters of brown fat induction are profound. Except for Ucp1 expression in fat depots of A/J and (A/J × B6)F1 mice, which does not change significantly, the HF diet enhanced expression of Ucp1, Pgc-1α, Dio2, and Pparα in all other genotypes. The effects were particularly strong for Dio2 in backcross progeny, where the mean expression in the presence of an HF diet was sixfold higher than it was in backcross mice fed an LF diet. However, this higher level of Dio2 in the backcross progeny or in A/J mice did not result in higher levels of Ucp1 expression for the backcross population as a whole, even though a high correlation was maintained between Ucp1 and Dio2 (Fig. 3 and Table 2 in reference 13). We interpret this to indicate that the level of T3 that can be generated by cold induction and an LF diet is sufficient to support the observed levels of Ucp1 expression. Whether higher levels of T3 can enhance other aspects of brown adipogenesis, such as mitochondrial biogenesis or Ucp1 expression under different environmental conditions, remains to be determined. In general, similar patterns of expression following an HF diet are also observed for PPARα. Since the expression of Ucp1 and Dio2 in adipose tissue is specific for brown adipocytes (6, 23, 32), the differential effects of an HF diet on Ucp1 and Dio2 suggest that the regulation of these genes in brown adipocytes is clearly different and may indicate that Dio2 has a role in brown adipocyte energy balance that is distinct from Ucp1. Thus, the induction of Dio2 in brown adipocytes of white fat by adrenergic stimulation and an HF diet, as we demonstrate here for the first time, may act synergistically to maintain the energy balance with thermogenic mechanisms independent of UCP1.
A major dilemma in our analysis of Ucp1 induction has been our inability to determine the identity and function of the QTLs that have been mapped with genetic approaches. Congenic lines for regions of Chr 2, 3, 8, and 19 and a consomic line for Chr 15 have failed to show significant effects on the induction of Ucp1 for A/J-derived QTLs on a B6 background (data not shown). We interpret the lack of phenotypic effects as due to an inability to establish essential selective allelic interactions with QTLs on other Chr. In addition, stocks with targeted and spontaneous mutations of genes with putative roles in Ucp1 expression have failed to provide evidence for function. In short, with the exception of PPARα, any attempt to measure the degree of Ucp1 induction by allelic changes in a single QTL or candidate gene has failed. The regulatory features of this complex multifactorial trait are based upon changes in gene expression from the analysis of the parental A/J and B6 strains, backcross progeny, and A × B and B × A recombinant inbred lines. The predominant and reoccurring theme in Ucp1 genetics is that its induction depends upon specific interactions among chromosomal regions that generate transgressive phenotypes. This has been clearly shown in both recombinant inbred strains and backcross progeny (28). It is also apparent from the genetic analysis performed with mice fed an HF diet that the genes involved in the process are highly influenced by diet, as well as the environmental temperature. Experiments described in this paper in which the levels of expression of signaling molecules and transcription factors in A/J and B6 mice were determined suggest a mechanism of transcriptional synergy that is consistent with the importance of epistatic interactions and transgressive phenotypes elucidated by genetic experiments. The experiments on signaling and transcription molecules at the protein level point to key roles for PKA, p38 MAP kinase, and CREB. Exactly where or how the allelic variation between A/J and B6 mice affects PKA signaling remains unknown. Evidence from other laboratories indicates that the absence of the retinoblastoma protein in Rb−/− mouse embryo fibroblasts, which can be induced to differentiate into brown adipocytes, enhances FoxC2 expression to modulate the composition of PKA regulatory isoforms, thereby increasing the sensitivity of cAMP signaling. However, the absence of changes in either FoxC2 expression or the isoform composition of PKA regulatory isoforms in our studies does not support this as a rate-limiting mechanism for induction of Ucp1 expression in RP fat of mice. Finally, except for Pparα and Ucp1, the chromosomal locations of the molecules present in Fig. 7 do not map to the locations of the QTLs shown in Fig. 4 and 5.
In summary, genetic variation in the expression of the Ucp1 gene in brown adipocytes during their induction by adrenergic stimulation in the RP tissue of mice is controlled by a minimum of nine QTLs on eight Chr. Strong evidence has been presented that Pparα, located on Chr 15, is one of these QTLs. The identities and functions of the other QTLs remain to be determined. The considerable knowledge of signaling and transcription molecules involved in adrenergic regulation of Ucp1 expression has enabled us to evaluate, at the mRNA and protein levels, how the allelic variation affects the expression of the components of the regulatory network. The earliest differences occur in the phosphorylation of CREB and p38 MAP kinase and then downstream on the levels of mRNAs for PPARγ, PPARα, PGC-1α, and type 2 deiodinase. However, compared to the 80-fold difference in Ucp1 expression at the mRNA and protein levels in the A/J and B6 parental strains and even larger differences among recombinant inbred strains of mice (21, 28), the two- to fourfold differences in the expression of the regulatory components are very modest. We propose that the small differences in the levels of transcriptional components of the Ucp1 enhanceosome interact synergistically to achieve large differences in Ucp1 expression.
Acknowledgments
We thank Christie Bearden for managing the mouse colony and Michael Morris for genotyping.
This research was supported by grant R01 DK58152 from the NIH.
REFERENCES
- 1.Aoyama, T., J. M. Peters, N. Iritani, T. Nakajima, K. Furihata, T. Hashimoto, and F. J. Gonzalez. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARα). J. Biol. Chem. 273:5678-5684. [DOI] [PubMed] [Google Scholar]
- 2.Ashwell, M., D. Stirling, S. Freeman, and B. R. Holloway. 1987. Immunological, histological and biochemical assessment of brown adipose tissue activity in neonatal, control and beta-stimulant-treated adult dogs. Int. J. Obes. 11:357-365. [PubMed] [Google Scholar]
- 3.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. [DOI] [PubMed] [Google Scholar]
- 4.Barbera, M. J., A. Schluter, N. Pedraza, R. Iglesias, F. Villarroya, and M. Giralt. 2001. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 276:1486-1493. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar, J. S., A. Chatterjee, L. W. Castellani, D. A. Ross, J. Ohmen, J. Cavalcoli, C. Wu, K. M. Dains, J. Catanese, M. Chu, S. S. Sheth, K. Charugundla, P. Demant, D. B. West, P. de Jong, and A. J. Lusis. 2002. Positional cloning of the combined hyperlipidemia gene Hyplip1. Nat. Genet. 30:110-116.11753387 [Google Scholar]
- 6.Bouillaud, F., D. Ricquier, G. Mory, and J. Thibault. 1984. Increased level of mRNA for the uncoupling protein in brown adipose tissue of rats during thermogenesis induced by cold exposure or norepinephrine infusion. J. Biol. Chem. 259:11583-11586. [PubMed] [Google Scholar]
- 7.Cao, W., K. W. Daniel, J. Robidoux, P. Puigserver, A. V. Medvedev, X. Bai, L. M. Floering, B. M. Spiegelman, and S. Collins. 2004. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 24:3057-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, W., A. V. Medvedev, K. W. Daniel, and S. Collins. 2001. Beta-adrenergic activation of p38 MAP kinase in adipocytes. cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 276:27077-27082. [DOI] [PubMed] [Google Scholar]
- 9.Cassard-Doulcier, A. M., M. Larose, J. C. Matamala, O. Champigny, F. Bouillaud, and D. Ricquier. 1994. In vitro interactions between nuclear proteins and uncoupling protein gene promoter reveal several putative transactivating factors including Ets1, retinoid X receptor, thyroid hormone receptor, and a CACCC box-binding protein. J. Biol. Chem. 269:24335-24342. [PubMed] [Google Scholar]
- 10.Cederberg, A., L. M. Gronning, B. Ahren, P. Tasken, P. Carlsson, and S. Enerback. 2001. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 106:563-573. [DOI] [PubMed] [Google Scholar]
- 11.Champigny, O., and D. Ricquier. 1996. Evidence from in vitro differentiating cells that adrenoceptor agonists can increase uncoupling protein mRNA level in adipocytes of adult humans: an RT-PCR study. J. Lipid Res. 37:1907-1914. [PubMed] [Google Scholar]
- 12.Collins, S., K. W. Daniel, A. E. Petro, and R. S. Surwit. 1997. Strain-specific response to beta 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology 138:405-413. [DOI] [PubMed] [Google Scholar]
- 13.Coulter, A. A., C. M. Bearden, X. Liu, R. A. Koza, and L. P. Kozak. 2003. Dietary fat interacts with QTLs controlling induction of Pgc-1α and Ucp1 during conversion of white to brown fat. Physiol. Genomics 14:139-147. [DOI] [PubMed] [Google Scholar]
- 14.Cousin, B., S. Cinti, M. Morroni, S. Raimbault, D. Ricquier, L. Pénicaud, and L. Casteilla. 1992. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J. Cell Sci. 103:931-942. [DOI] [PubMed] [Google Scholar]
- 15.Cummings, D. E., E. P. Brandon, J. V. Planas, K. Motamed, R. L. Idzerda, and G. S. McKnight. 1996. Genetically lean mice result from targeted disruption of the RIIβ subunit of protein kinase A. Nature 382:622-626. [DOI] [PubMed] [Google Scholar]
- 16.Digby, J. E., C. T. Montague, C. P. Sewter, L. Sanders, W. O. Wilkison, S. O'Rahilly, and J. B. Prins. 1998. Thiazolidinedione exposure increases the expression of uncoupling protein 1 in cultured human preadipocytes. Diabetes 47:138-141. [DOI] [PubMed] [Google Scholar]
- 17.Enerback, S., A. Jacobsson, E. M. Simpson, C. Guerra, H. Yamashita, M. E. Harper, and L. P. Kozak. 1997. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387:90-94. [DOI] [PubMed] [Google Scholar]
- 18.Esterbauer, H., H. Oberkofler, Y. M. Liu, D. Breban, E. Hell, F. Krempler, and W. Patsch. 1998. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J. Lipid Res. 39:834-844. [PubMed] [Google Scholar]
- 18a.Fekete, C., G. Legradi, E. Mihaly, Q. H. Huang, J. B. Tatro, W. M. Rand, C. H. Emerson, and R. M. Lechan. 2000. α-Melanocyte-stimulating hormone is contained in nerve terminals innervating thyrotropin-releasing hormone-synthesizing neurons in the hypothalamic paraventricular nucleus and prevents fasting-induced suppression of prothyrotropin-releasing hormone gene expression. J. Neurosci. 20:1550-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghorbani, M., and J. Himms-Hagen. 1997. Appearance of brown adipocytes in white adipose tissue during CL 316,243-induced reversal of obesity and diabetes in Zucker fa/fa rats. Int. J. Obes. 21:465-475. [DOI] [PubMed] [Google Scholar]
- 20.Graves, R. A., P. Tontonoz, and B. M. Spiegelman. 1992. Analysis of a tissue-specific enhancer: ARF6 regulates adipogenic gene expression. Mol. Cell. Biol. 12:1202-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerra, C., R. A. Koza, H. Yamashita, K. Walsh, and L. P. Kozak. 1998. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Investig. 102:412-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, J. B., C. Jorgensen, R. K. Petersen, P. Hallenborg, R. De Matteis, H. A. Boye, N. Petrovic, S. Enerback, J. Nedergaard, S. Cinti, H. te Riele, and K. Kristiansen. 2004. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl. Acad. Sci. USA 101:4112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsson, A., U. Stadler, M. A. Glotzer, and L. P. Kozak. 1985. Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. J. Biol. Chem. 260:16250-16254. [PubMed] [Google Scholar]
- 24.Kersten, S., J. Seydoux, J. M. Peters, F. J. Gonzalez, B. Desvergne, and W. Wahli. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Investig. 103:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1:119-129. [DOI] [PubMed] [Google Scholar]
- 26.Kopecky, J., G. Clarke, S. Enerback, B. Spiegelman, and L. P. Kozak. 1995. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Investig. 96:2914-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopecky, J., Z. Hodny, M. Rossmeisl, I. Syrovy, and L. P. Kozak. 1996. Reduction of dietary obesity in aP2-Ucp transgenic mice: physiology and adipose tissue distribution. Am. J. Physiol. 270:E768-E775. [DOI] [PubMed] [Google Scholar]
- 28.Koza, R. A., S. M. Hohmann, C. Guerra, M. Rossmeisl, and L. P. Kozak. 2000. Synergistic gene interactions control the induction of the mitochondrial uncoupling protein (Ucp1) gene in white fat tissue. J. Biol. Chem. 275:34486-34492. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, U. C., J. Kopecky, J. Teisinger, S. Enerback, B. Boyer, and L. P. Kozak. 1994. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol. 14:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Laemmli, U. K. 1970. Cleavage of structural proteins during the formation of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lean, M. E., W. P. James, G. Jennings, and P. Trayhurn. 1986. Brown adipose tissue uncoupling protein content in human infants, children and adults. Clin. Sci. 71:291-297. [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 32.Leonard, J. L., S. A. Mellen, and P. R. Larsen. 1983. Thyroxine 5′-deiodinase activity in brown adipose tissue. Endocrinology 112:1153-1155. [DOI] [PubMed] [Google Scholar]
- 33.Linhart, H. G., K. Ishimura-Oka, F. DeMayo, T. Kibe, D. Repka, B. Poindexter, R. J. Bick, and G. J. Darlington. 2001. C/EBPα is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 98:12532-12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loncar, D. 1991. Convertible adipose tissue in mice. Cell Tissue Res. 266:149-161. [DOI] [PubMed] [Google Scholar]
- 35.Loncar, D., L. Bedrica, J. Mayer, B. Cannon, J. Nedergaard, B. A. Afzelius, and A. Svajger. 1986. The effect of intermittent cold treatment on the adipose tissue of the cat. Apparent transformation from white to brown adipose tissue. J. Ultrastruct. Mol. Struct. Res. 97:119-129. [DOI] [PubMed] [Google Scholar]
- 36.MacDougald, O. A., and M. D. Lane. 1995. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64:345-373. [DOI] [PubMed] [Google Scholar]
- 37.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 38.Rim, J. S., B. Xue, B. Gawronska-Kozak, and L. P. Kozak. 2004. Sequestration of thermogenic transcription factors in the cytoplasm during development of brown adipose tissue. J. Biol. Chem. 279:25916-25926. [DOI] [PubMed] [Google Scholar]
- 39.Rosen, E. D., P. Sarraf, A. E. Troy, G. Bradwin, K. Moore, D. S. Milstone, B. M. Spiegelman, and R. M. Mortensen. 1999. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4:611-617. [DOI] [PubMed] [Google Scholar]
- 40.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 41.Schneider, M. J., S. N. Fiering, S. E. Pallud, A. F. Parlow, D. L. St Germain, and V. A. Galton. 2001. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol. Endocrinol. 15:2137-2148. [DOI] [PubMed] [Google Scholar]
- 42.Sears, I. B., M. A. MacGinnitie, L. G. Kovacs, and R. A. Graves. 1996. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 16:3410-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva, J. E. 2001. The multiple contributions of thyroid hormone to heat production. J. Clin. Investig. 108:35-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soloveva, V., R. A. Graves, M. M. Rasenick, B. M. Spiegelman, and S. R. Ross. 1997. Transgenic mice overexpressing the β1-adrenergic receptor in adipose tissue are resistant to obesity. Mol. Endocrinol. 11:27-38. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, T., N. Yoshida, T. Kishimoto, and S. Akira. 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and C/EPBδ gene. EMBO J. 16:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiraby, C., G. Tavernier, C. Lefort, D. Larrouy, F. Bouillaud, D. Ricquier, and D. Langin. 2003. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 278:33370-33376. [DOI] [PubMed] [Google Scholar]
- 47.Tontonoz, P., E. Hu, and, S. B. M. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 48.Tsukiyama-Kohara, K., F. Poulin, M. Kohara, C. T. DeMaria, A. Cheng, Z. Wu, A. C. Gingras, A. Katsume, M. Elchebly, B. M. Spiegelman, M. E. Harper, M. L. Tremblay, and N. Sonenberg. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7:1128-1132. [DOI] [PubMed] [Google Scholar]
- 49.Yubero, P., C. Manchado, A. M. Cassard-Doulcier, T. Mampel, O. Vinas, R. Iglesias, M. Giralt, and F. Villarroya. 1994. CCAAT/enhancer binding proteins alpha and beta are transcriptional activators of the brown fat uncoupling protein gene promoter. Biochem. Biophys. Res. Commun. 198:653-659. [DOI] [PubMed] [Google Scholar]