Abstract
The nuclear functions of NF-κB p50/RelA heterodimers are regulated in part by posttranslational modifications of its RelA subunit, including phosphorylation and acetylation. Acetylation at lysines 218, 221, and 310 differentially regulates RelA's DNA binding activity, assembly with IκBα, and transcriptional activity. However, it remains unclear whether the acetylation is regulated or simply due to stimulus-coupled nuclear translocation of NF-κB. Using anti-acetylated lysine 310 RelA antibodies, we detected p300-mediated acetylation of RelA in vitro and in vivo after stimulation of cells with tumor necrosis factor alpha (TNF-α). Coexpression of catalytically inactive mutants of the catalytic subunit of protein kinase A/mitogen- and stress-activated kinase 1 or IKK1/IKK2, which phosphorylate RelA on serine 276 or serine 536, respectively, sharply inhibited RelA acetylation on lysine 310. Furthermore, phosphorylation of RelA on serine 276 or serine 536 increased assembly of phospho-RelA with p300, which enhanced acetylation on lysine 310. Reconstitution of RelA-deficient murine embryonic fibroblasts with RelA S276A or RelA S536A decreased TNF-α-induced acetylation of lysine 310 and expression of the endogenous NF-κB-responsive E-selectin gene. These findings indicate that the acetylation of RelA at lysine 310 is importantly regulated by prior phosphorylation of serines 276 and 536. Such phosphorylated and acetylated forms of RelA display enhanced transcriptional activity.
The NF-κB/Rel family of transcription factors plays a key role in regulating inflammatory and immune responses and other programs of cell growth and survival. The five known mammalian Rel genes encode seven Rel-related proteins: RelA/p65; p105 and its processing product, p50; p100 and its processing product, p52; c-Rel; and RelB. Each contains an N-terminal Rel homology domain (∼300 amino acids) that mediates DNA binding, dimerization, and interaction with the IκB family of NF-κB/Rel inhibitors. RelA, c-RelA, and RelB contain C-terminal transactivation domains, but p50 and p52 do not. Each NF-κB/Rel protein forms different homo- or heterodimers with other members of the family, which may contribute to the activation of specific target genes (1, 5).
The prototypical NF-κB complex is a p50/RelA heterodimer. NF-κB is largely sequestered in the cytoplasm through its association with an IκB inhibitor. Nuclear NF-κB expression is induced by various stimuli, including proinflammatory cytokines, growth factors, DNA-damaging agents, and viral proteins (13). The activation of NF-κB can be divided into two phases. The first phase involves cytoplasmic events culminating in the activation of the IκB kinases (IKK1 and IKK2). These kinases promote N-terminal phosphorylation of serines 32 and 36 in IκBα, leading to its polyubiquitylation and proteasome-mediated degradation. The liberated NF-κB complex rapidly translocates to the nucleus, ending the first phase (13). The second phase occurs primarily in the nucleus and involves posttranslational modification of the NF-κB transcription factor complex or relevant histones surrounding NF-κB target genes (5). These modifications determine both the strength and duration of the NF-κB-mediated transcriptional response (5).
One of the nuclear events is the reversible acetylation of RelA (4). Endogenous RelA is acetylated in a stimulus-coupled manner after activation of cells with tumor necrosis factor alpha (TNF-α), phorbol myristate acetate, or other stimuli at multiple sites, including lysines 122, 123, 218, 221, and 310 (4, 17). The acetyltransferases p300 and CBP appear to play a major role in the in vivo acetylation of RelA (6, 17). Site-specific acetylation of RelA regulates discrete biological actions of the NF-κB complex (5, 6). For example, acetylation of lysine 221 by p300/CBP increases the DNA binding affinity of RelA for the κB enhancer and, together with acetylation of lysine 218, impairs assembly of RelA with newly synthesized IκBα, which shuttles in and out of the nucleus. Acetylation of lysine 310 does not modulate DNA binding or IκBα assembly but markedly enhances the transcriptional activity of NF-κB. Deacetylation of lysine 310 by histone deacetylase 3 (HDAC3) or SIRT1 inhibits the transcriptional activity of RelA and augments cellular apoptosis in response to TNF-α (6, 32). While it is clear that signal-coupled acetylation of RelA participates in the nuclear regulation of NF-κB action (4, 17), many unanswered questions remain. Chief among these is how the acetylation of RelA is regulated.
RelA is also subject to phosphorylation by kinases that modify different sites (5). For example, serine 276 in the Rel homology domain of RelA is phosphorylated by the catalytic subunit of protein kinase A (PKAc) and mitogen- and stress-activated kinase 1 (MSK-1), which are activated by lipopolysaccharide and TNF-α, respectively (29, 34, 35). Phosphorylation of RelA at serine 276 enhances the recruitment of coactivator p300/CBP, leading to increased transcriptional activation involving acetylation of histones surrounding NF-κB-responsive genes (29, 34, 35). Serine 536 in the RelA transactivation domain is phosphorylated by IKKs (23, 26, 31) or by ribosomal subunit kinase 1 (3). This modification also enhances the transcriptional activity of NF-κB (26, 30), although the underlying mechanism remains unclear.
Phosphorylation regulates the acetylation of histones and some nonhistone proteins. For example, phosphorylation of serine 10 in the tail of histone H3 increases acetylation at lysine 14 (7, 20, 21). Phosphorylation at serine 15 of p53 stimulates subsequent acetylation, likely reflecting the ability of phospho-serine 15 to promote increased binding of CBP/p300 to p53 (19, 25). Prior studies showing that phosphorylation of RelA at serine 276 enhances the binding of p300/CBP certainly could provide a mechanism for a contingent linkage between these two posttranslational modifications (35). In this study, we have investigated whether a similar link between phosphorylation and acetylation exists for RelA and how these modifications regulate the transcription of NF-κB target genes.
MATERIALS AND METHODS
Cell lines, recombinant proteins, and plasmids.
Cell lines stably expressing wild-type, S276A, and S536A RelA have been described previously (23). A cell line stably expressing K310R was generated as described previously (23). RelA-deficient mouse embryonic fibroblasts (MEFs) were kindly provided by A. Beg (Columbia University), and MSK-1/MSK-2-deficient MEFs were provided by J. S. Arthur (University of Dundee, United Kingdom). Recombinant RelA proteins corresponding to wild-type, K310R, KR (K218/221/310R), S276A, and S536A RelA were prepared from baculovirus-infected Sf9 insect cells (Pharmingen) or from plasmid-transduced Escherichia coli as described previously (29). RelA S276A, S276E, S536A, and S536E containing a T7 epitope tag were generated by site-directed mutagenesis (Stratagene); all mutations were confirmed by DNA sequencing. Gal4-p300, hemagglutinin (HA)-p300, and MSK-1(D565A) expression vectors were kindly provided by Y. Shi (Harvard University), Q.-H. Zhang (Vollum Institute), and G. Haegeman (Ghent University), respectively.
Generation of anti-acetylated lysine 310 RelA antibodies.
Rabbits were immunized with a peptide sequence containing acetylated lysine 310 and 13 flanking RelA amino acid residues [RTYETF(Ac)KSIMKKSP] after conjugation to a keyhole limpet hemocyanin carrier protein. Sera were screened for reactivity and selectivity by enzyme-linked immunosorbent assay and immunoblotting. To deplete this serum of antibodies reacting with unmodified RelA, positive sera were collected, pooled, and negatively selected by affinity chromatography with a column containing a nonacetylated version of the lysine 310 peptide.
Preparation of mammalian p300 complexes.
293T cells at 80 to 90% confluence in 100-mm dishes were transfected with 15 μg of HA-p300 plasmid DNA by the calcium phosphate method. After 36 h, the cells were lysed in 2 ml of lysis buffer (50 mM HEPES, pH 7.4, 250 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride) containing a protease inhibitor cocktail (Roche). Supernatants were immunoprecipitated as previously described (4), with 100 μl of a 50% slurry of anti-HA agarose (Covance). After immunoprecipitation, agarose beads were washed twice with ice-cold lysis buffer and resuspended in 200 μl of HAT assay buffer (250 mM Tris base, pH 8.0, 50% glycerol, 0.5 mM EDTA, and 5 mM dithiothreitol). Resuspended beads were aliquoted and stored at −80°C.
In vitro acetylation assays.
Recombinant RelA protein (1 μg) was incubated with 10 μl of agarose beads from the p300 immunoprecipitates, 2 μl of [14C]acetyl coenzyme A (50 nCi/μl) (Amersham), and 4 μl of 5× HAT assay buffer at 30°C for 1 h.
Whole-cell extracts.
After stimulation with TNF-α, 293T cells or MEFs cultured in 100-mm dishes were harvested, centrifuged, and incubated in 100 μl of lysis buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 25% glycerol, 0.01% NP-40, 1 mM EDTA, 2.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) for 30 min on ice. The sample was vigorously vortexed for 30 s, frozen in liquid nitrogen, and thawed on ice. After vortexing and centrifugation at 4°C, the supernatant was isolated and used as a whole-cell extract.
Chromatin immunoprecipitation (ChIP) assays.
HeLa cells (7 × 106 cells) in 100-mm dishes were fixed in 1% formaldehyde for 10 min, washed twice with cold phosphate-buffered saline, and resuspended in a solution containing 1% sodium dodecyl sulfate (SDS), 50 mM Tris, and 10 mM EDTA. Cells were sonicated under conditions optimized to generate DNA fragments with an average size of 1 kilobase. Samples were precleared with protein A agarose beads (Roche) and incubated overnight with anti-RelA (catalog number sc-109; Santa Cruz Biotechnology), anti-acetylated lysine 310 RelA, anti-phosphorylated serine 276, or no antibody. Protein A agarose beads were added. After 6 h, the beads were washed exhaustively, and proteins were eluted in elution buffer (25 mM Tris-Cl, pH 7.5, 10 mM EDTA, 0.5% SDS) for 15 min at 60°C. Eluates were de-cross-linked by incubation at 65°C for 12 h and digested for 1 h with pronase (Sigma). DNA was isolated by QIAGEN QuickSpin and analyzed by PCR. Two primers were used for PCR amplification of interleukin-8 (IL-8) promoter DNA: IL-8-5 (5′-GGGCCATCAGTTGCAAATC-3′) and IL-8-3 (5′-TTCCTTCCGGTGGTTTCTTC-3′). To detect control β-actin DNA sequences, two primers specific for a 239-bp region in the β-actin gene were used: β-actin-5 (5′-GTCGACAACGGCTCCGGC-3′) and β-actin-3 (5′-GGTGTGGTGCCAGATTTTCT-3′). DNA was amplified for 40 cycles with Taq polymerase (QIAGEN), and the products were analyzed on 2.5% agarose gels. Images were acquired with an EagleEye digital camera.
Quantitative real-time PCR.
Samples from the ChIP assays were analyzed by real-time quantitative PCR with QIAGEN Cyber-green PCR kits. The PCR primers were the same as those in the ChIP assays. Expression of E-selectin was detected with QIAGEN QuantiTect probe reverse transcription-PCR (RT-PCR) kits. TaqMan probes for E-selectin were obtained from QIAGEN, and the PCR was performed according to the manufacturer's protocol with an ABI 7700 sequence detection system (Applied Biosystems).
Immunoprecipitation and immunoblotting.
Transfected 293T cells cultured in 6-well plates were lysed in 300 μl of lysis buffer (50 mM HEPES, pH 7.4, 250 mM NaCl, 1% NP-40, 1 mM EDTA) for 10 min at 4°C. Cell lysates from two wells were immunoprecipitated for 2 h at 4°C with 20 μl of a slurry containing 50% anti-T7-conjugated agarose beads (Novagen). After two washes in lysis buffer, the immunoprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (12%), transferred onto nitrocellulose membranes, and immunoblotted with various antibodies. Immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham).
RESULTS
RelA is acetylated at lysine 310 by p300 in vitro.
Our previous studies indicated that p300/CBP played a key role in the acetylation of RelA in vivo and further indicated that a p300 HAT-deficient mutant dominantly interfered with the acetylation of RelA (6). To determine if p300 can acetylate RelA in vitro, HA-p300 was immunoprecipitated from transfected 293T cells and used with [14C]acetyl coenzyme A in an in vitro acetylation assay employing RelA or glutathione S-transferase (GST)-p53 proteins as substrates. RelA and GST-p53 were acetylated at similar levels by p300 in vitro (Fig. 1A).
FIG. 1.
RelA is acetylated on lysine 310 by p300 in vitro. (A) p300 immunoprecipitated from 293T cells acetylates RelA in vitro. 293T cells were cotransfected with HA-p300 expression vector DNA. p300 was immunoprecipitated from the cells and incubated with 1 μg of recombinant RelA (lane 2) or GST-p53 (lane 3) in an in vitro acetylation assay. Autoradiograms showing the acetylation of RelA and p53 and the autoacetylation of p300 are shown in the top panel. Levels of p300, RelA, and GST-p53 detected by Coomassie blue staining are shown in the bottom panel. (B) Anti-acetylated lysine 310 RelA antibodies specifically recognize acetylated lysine 310. Recombinant proteins (1 μg) of wild-type RelA, K310R, or RelA-KR were incubated with immunoprecipitated HA-p300 in an in vitro acetylation assay. Reaction products were separated by SDS-polyacrylamide gel electrophoresis, and acetylation (Ac) was detected by immunoblotting with anti-acetylated lysine 310 antibodies (top). Levels of RelA, RelA-K310, and RelA-KR are shown in the bottom panels. (C). p300 acetylates RelA at lysine 310 in vivo. 293T cells were cotransfected with expression vector DNA encoding wild-type RelA, K310R, or RelA-KR (0.5 μg) and p300 (2 μg). Acetylation was detected by immunoblotting of anti-T7 immunoprecipitates with anti-acetylated lysine 310 antibodies (top). Levels of T7-RelA and the various RelA mutants are shown in the bottom panel. IB, immunoblotting; IP, immunoprecipitation.
We next used a rabbit polyclonal antibody that selectively reacts with acetylated lysine 310 to determine if p300 immunoprecipitates acetylate RelA in vitro. Immunoblotting revealed reactivity of this antibody with wild-type RelA in the presence of p300 (Fig. 1B, lane 2). In contrast, no acetylation signal was observed when lysine 310 was substituted with arginine either singly (K310R) or in combination with lysines 218 and 221 (RelA-KR) (Fig. 1B, lanes 3 and 4).
Next, we tested the reactivity of the antibody in 293T cells cotransfected with expression vectors encoding wild-type RelA, RelA-K310R, or RelA-KR mutants and p300. Wild-type RelA was acetylated on lysine 310 in the presence of p300; however, the K310R and RelA-KR mutants displayed markedly reduced reactivity with this antibody (Fig. 1C). The faint residual signals obtained with these mutants likely reflect very weak binding of this antibody to RelA or to RelA that is acetylated at sites other than lysine 310. These results confirm that p300 acetylates RelA on lysine 310 both in vitro and in vivo and that the anti-acetylated lysine 310 antibodies selectively react with this acetylated form of RelA.
Stimulus-coupled acetylation of lysine 310 in endogenous RelA.
We next investigated whether this antibody could be used to evaluate changes in the acetylation of endogenous RelA after cellular stimulation. When 293T cells were activated with TNF-α for 0 to ∼60 min, acetylated RelA was first detected at 10 min and at increasing levels at 30 to ∼60 min (Fig. 2A, top). IκBα was degraded in 10 min, and its resynthesis was detectable within 20 min (Fig. 2A, bottom). These findings indicate that the anti-acetylated lysine 310 antibody can be employed to monitor changes in the acetylation status of the endogenous RelA protein and that this posttranslational modification occurs in an inducible manner.
FIG. 2.
Stimulus-coupled acetylation of endogenous RelA on lysine 310 in vivo. (A) TNF-α induces lysine 310 acetylation of RelA in vivo. 293T cells were stimulated with TNF-α (20 ng/ml) for the indicated periods of time. Whole-cell extracts were prepared, and the acetylation (Ac) of endogenous RelA was detected by immunoblotting (IB) with anti-acetylated lysine-310 antibodies (top). Levels of RelA and IκBα are shown in the middle and bottom panels, respectively. (B) TNF-α induces the binding of lysine 310-acetylated RelA, and phospho-276 RelA binds to the κB enhancer of the IL-8 gene. HeLa cells were treated with TNF-α for the indicated periods. ChIP assays were performed using anti-RelA (α-RelA), anti-acetylated lysine 310 (α-Ac-310), or anti-phosphorylated serine 276 antibodies or in the absence of antibodies (No Ab) and probed for the IL-8 promoter sequences spanning the κB binding sites (left panels) or for nonspecific control β-actin DNA (right panels). (C) Quantification of ChIP binding using RT-PCR and cyber green. Samples presented as described above (B) were analyzed as described in Materials and Methods. Data represent the average of two independent experiments. (D) TNF-α-induced expression of E-selectin, an endogenous NF-κB target gene, is impaired in RelA-deficient MEFs reconstituted with the RelA-K310R mutant. RelA-deficient MEFs reconstituted with either wild-type (WT) RelA or RelA-K310R were stimulated with TNF-α for 0 or 6 h. Total RNA from cells was isolated, and expression levels of E-selectin mRNA were measured by quantitative real-time PCR. Results represent the averages of two independent experiments.
We next employed ChIP assays to assess whether RelA acetylated on lysine 310 effectively binds to DNA. HeLa cells were stimulated with TNF-α for 0, 30, or 60 min. In the absence of stimulation, no binding of acetylated RelA or RelA to the IL-8 promoter in coimmunoprecipitated DNA was detected in PCR (Fig. 2B, lane 1). After stimulation with TNF-α for 30 or 60 min, IL-8 κB enhancer DNA but not β-actin DNA was amplified in immunoprecipitates obtained with either anti-acetylated lysine 310 RelA or RelA antibodies (Fig. 2B), demonstrating the specificity of the DNA immunoprecipitation. Of note, anti-phosphorylated serine 276 antibodies also immunoprecipitated κB enhancer DNA from the IL-8 promoter (Fig. 2B). Quantitation of these ChIP assays by real-time PCR showed more promoter-bound acetylated RelA at 60 min than at 30 min after TNF-α stimulation (Fig. 2C). These findings indicate that lysine 310 of endogenous RelA protein undergoes inducible acetylation after TNF-α stimulation and that this acetylated form of RelA effectively binds to the native κB enhancer of a recognized NF-κB-responsive gene in living cells.
Acetylation of lysine 310 is required for the full transcriptional activity of RelA (6). We next evaluated whether acetylated lysine 310 RelA was required for full TNF-α activation of E-selectin, an endogenous NF-κB target gene. TNF-α increased E-selectin mRNA levels more than ninefold in RelA-deficient MEFs reconstituted with wild-type RelA but only threefold in RelA-K310R cells (Fig. 2D). These findings further support the notion that acetylation of lysine 310 is required for the full transcriptional activity of NF-κB.
Phosphorylation of serine 276 promotes the acetylation of lysine 310.
Phosphorylation of serine 276 is mediated by either PKAc or MSK-1 (29, 35). To investigate the potential role of this modification in RelA acetylation, we first cotransfected 293T cells with RelA and enzymatically inactive forms of PKAc (K73M) and MSK-1 (D565A). Both of these enzymatically inactive kinases impaired p300-mediated acetylation of RelA at lysine 310 (Fig. 3A). Next, substitution mutants of RelA where serine 276 was replaced with alanine or glutamic acid were tested for effects on p300-mediated acetylation. Replacement of serine 276 with either alanine or glutamic acid markedly reduced p300-mediated acetylation of RelA, although a residual acetylation signal was detected with both mutants (Fig. 3B). These findings argue that phosphorylation of serine 276 may markedly facilitate RelA acetylation on lysine 310, although the failure of S276E to mimic phosphorylation raises the issue that substitution at this site may alter overall conformation of the protein.
FIG. 3.
Phosphorylation of serine 276 is critical for the acetylation of RelA. (A) Enzymatically inactive forms of PKAc and MSK-1 inhibit acetylation (Ac) of RelA at lysine 310. 293T cells were cotransfected with the expression plasmid DNA encoding T7-RelA, p300, and graded doses of PKAc(K47M) or MSK-1(D656A), as indicated. Acetylation of RelA was detected by immunoblotting (IB) of the anti-T7 (α-T7) immunoprecipitates (IP) with anti-acetylated lysine 310 antibodies (top). Levels of T7-RelA present in each of the lysates are shown in the bottom panel. (B) Alanine or glutamic acid substitutions at serine 276 block the acetylation of RelA at lysine 310. 293T cells were transfected with wild-type (WT) RelA, S276A, or S276E expression vectors (0.5 μg) together with p300 (2 μg). Levels of acetylation of each protein were detected by immuno-blotting of anti-T7 immunoprecipitates with anti-acetylated lysine 310 antibodies. Levels of RelA, S276A, and S276E are shown in the bottom panel. (C) Phosphorylation of RelA at serine 276 by PKAc enhances the acetylation of RelA in vitro. Recombinant proteins (1 μg) of wild-type or S276A RelA were phosphorylated with recombinant PKAc (0.5 μl) (Promega) in an in vitro kinase assay. Reactions were then incubated with p300 immunoprecipitates in an in vitro acetylation assay. Levels of acetylation of RelA were detected by immunoblotting with anti-acetylated lysine 310 antibodies. Levels of RelA in each reaction are shown in the bottom panel. (D) Failure of TNF-α to induce acetylation of RelA on lysine 310 in MSK-1/MSK-2-deficient MEFs. MEFs from MSK-1/MSK-2-deficient mice were stimulated with TNF-α for the indicated time periods. Acetylation and phosphorylation levels of RelA were analyzed by immunoblotting with anti-acetylated lysine 310 antibodies (top) or with anti-phosphorylated serine 276 antibodies (α-P-276) (middle). The levels of RelA present in each of the cell extracts are also shown in the bottom panel.
To further test the effect of phosphorylation of serine 276 on RelA acetylation, we used phosphorylated RelA as substrate in the in vitro acetylation assay. Prior phosphorylation of RelA on serine 276 by PKAc clearly enhanced the subsequent acetylation of lysine 310 (Fig. 3C, lane 3). Consistent with the results presented in Fig. 3B, the RelA S276A mutant was also acetylated, albeit to a lesser extent than wild-type RelA, but prior phosphorylation did not increase lysine 310 acetylation.
Since MSK-1 participates in the phosphorylation of RelA at serine 276 after TNF-α stimulation (29), we investigated TNF-α-induced acetylation of endogenous RelA on lysine 310 in MSK-1-deficient MEFs. MSK-1/MSK-2-deficient MEFs were employed to avoid a possible redundant function of these two kinases (29). TNF-α induced acetylation of lysine 310 and phosphorylation of serine 276 in wild-type MEFs (Fig. 3D, lanes 1 to 3). In contrast, little or no TNF-α-inducible acetylation of lysine 310 or phosphorylation of serine 276 was detected in MSK-1/MSK-2-deficent MEFs (Fig. 3D, lanes 4 to 6).
Phosphorylation of serine 536 also facilitates the acetylation of RelA on lysine 310.
Since stimulus-induced phosphorylation of serine 536 by IKK1 or IKK2 has been linked to enhanced transcriptional activity (24, 26, 31), we assessed its effect on RelA acetylation on lysine 310. In 293T cells coexpressing p300 and catalytically inactive forms of IKK1 or IKK2, acetylation of RelA on lysine 310 was reduced (Fig. 4A). These results suggest a potential role for the IKKs in the regulation of RelA acetylation.
FIG. 4.
Phosphorylation of RelA at serine 536 facilitates the acetylation of RelA on lysine 310. (A) Kinase-deficient mutants of IKK1 and IKK2 inhibit the acetylation (Ac) of RelA at lysine 310. 293T cells were cotransfected with the indicated expression plasmid DNA encoding T7-RelA (0.5 μg), p300 (2 μg), IKK1(K47M) (0.5 μg), and IKK2(K47A) (0.5 μg). Acetylation of RelA was detected by immunoblotting (IB) of anti-T7 (α-T7) immunoprecipitates (IP) with anti-acetylated lysine 310 (α-Ac-K310) antibodies (top). Levels of T7-RelA present in each of the lysates are shown in the bottom panel. (B) Alanine or glutamic acid substitutions at serine 536 impair or enhance, respectively, the acetylation of RelA at lysine 310. 293T cells were transfected with wild-type (WT) RelA, S536A, or S536E expression vectors (0.5 μg) together with p300 (2 μg). Levels of acetylation on each protein were detected by immunoblotting of anti-T7 immunoprecipitates with anti-acetylated lysine 310 antibodies. Levels of RelA, S536A, and S536E are shown in the bottom panel. (C) Phosphorylation of RelA at serine 536 by IKK2 enhances the acetylation of RelA in vitro. Recombinant RelA (1 μg) was phosphorylated with recombinant IKK2 (0.5 μl) (P-RelA) in an in vitro kinase assay. Reactions were then incubated with p300 immunoprecipitates in an in vitro acetylation assay as described in the legend to Fig. 1A. Levels of acetylation of RelA were detected by immunoblotting with anti-acetylated lysine 310 antibodies. Levels of RelA in each reaction are shown in the bottom panel. (D) RelA-deficient MEFs were transfected with E-selectin-luciferase reporter plasmid DNA (0.5 μg) and the various S276A, S276E, S536A, and S536E mutants of RelA (0.1 μg). After 36 h, cells were treated with or without TNF-α (20 ng/ml) for 6 h. Luciferase activity was measured as described previously (4). Expression levels of RelA and various RelA mutants in each transfection are shown in the bottom panel. Results represent the averages of three independent experiments ± standard deviations.
Next, we tested p300-mediated acetylation of lysine 310 with different serine 536 mutants. Serine 536 was substituted with either alanine (S536A) or glutamic acid (S536E). p300-mediated acetylation of RelA on lysine 310 was impaired with the S536A mutant but enhanced with the phosphomimetic S536E mutant (Fig. 4B, lanes 3 and 4). Acetylation of lysine 310 does not obligately require phosphorylation of serine 536, since the S536A mutant is still acetylated, albeit at lower levels than wild-type RelA.
Since IKKs appear to play an important role in phosphorylation of serine 536 (24, 26, 31), we next examined the effect of IKK2-mediated phosphorylation of serine 536 on RelA acetylation in vitro. RelA was first phosphorylated by IKK2 in vitro, and the phosphorylated RelA was then used as a substrate in the in vitro acetylation assay. While RelA was clearly acetylated on lysine 310 by p300, the level of RelA acetylation was enhanced by prior IKK2-mediated phosphorylation of RelA on serine 536 by IKK2 (Fig. 4C).
When the transcriptional activity of the various serine mutants of RelA was tested in 293T cells in the presence of a cotransfected κB-luciferase reporter plasmid, the transcriptional activities of S276A, S276E, and S536A were sharply impaired, while the activity of S536E was greater than that of wild-type RelA (Fig. 4D). Interestingly, although additional stimulation with TNF-α produced minimal effects on the transcriptional activities of S276A, S276E, and S536A, it consistently enhanced the activity of S536E (Fig. 4D). The transcriptional potential of each RelA mutant in the presence or absence of TNF-α correlated closely with the level of acetylation achieved with each mutant (Fig. 3B and 4B). These results further underscore the role of RelA phosphorylation on serine 536 in the ensuing acetylation on lysine 310 as key events needed for full transcriptional activation of RelA.
Phosphorylation of serines 276 and 536 enhances the association of RelA with p300.
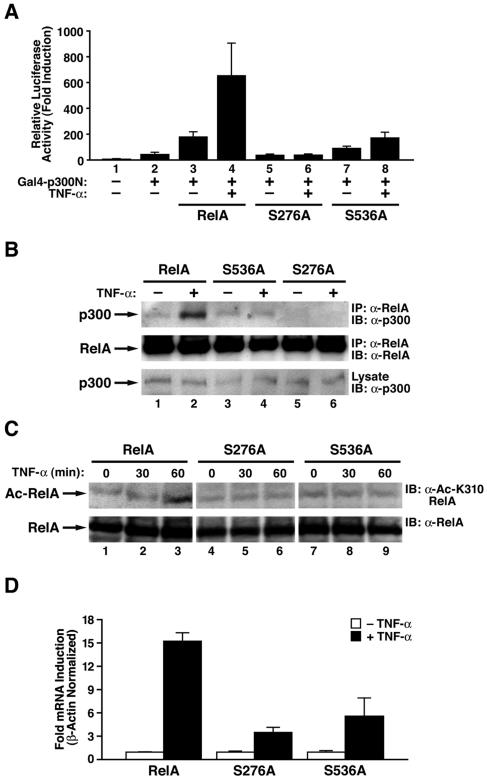
Next, we investigated whether the serine phosphorylation-induced increase in RelA acetylation reflected increased assembly of RelA and p300/CBP. First, we examined the binding of p300 to RelA and the S276A and S536A mutants of RelA in a mammalian one-hybrid system. For these assays, the N-terminal region of p300 fused to a GAL4 DNA binding domain was coexpressed with wild-type, S276A, or S536A RelA in RelA-deficient cells. Binding of RelA and p300 triggers transcriptional activation of a cotransfected GAL4-responsive reporter (Fig. 5A). TNF-α stimulation increased the transcriptional activation of wild-type RelA but had little or no effect on the activation of S276A and S536A mutants (Fig. 5A). When similarly tested, the S276E mutant induced essentially no response, whereas the S536E mutant induced a much greater response than wild-type RelA (data not shown). These findings further support the conclusion that phosphorylation of serines 276 and 536 promotes enhanced interaction of p300 with RelA.
FIG. 5.
Phosphorylation of RelA at serines 276 and 536 enhances the association of RelA with p300. (A) Alanine substitution of serines 276 and 536 impairs RelA interaction with p300 in a mammalian one-hybrid assay. RelA-deficient MEFs were transfected with expression plasmids (1 ng) encoding RelA, S276A, or S536A and GAL4-p300N (50 ng) together with a GAL4 enhancer-luciferase reporter (0.1 μg) (pFR-luc). After 24 h, the cells were stimulated with TNF-α for 5 h. Luciferase activity was measured as previously described (4). Results represent the averages of three independent experiments ± standard deviations. (B) TNF-α stimulates the interaction of wild-type RelA, but not the S276A and S536A mutants, with p300. MEFs stably expressing the wild type or the S276A or S536A mutant were stimulated with TNF-α for 60 min. Whole-cell lysates were then immunoprecipitated (IP) with anti-RelA (α-RelA) antibodies (catalog number F-6; Santa Cruz Biotechnology) followed by immunoblotting (IB) with anti-p300 (α-p300) (catalog number SC-584; Santa Cruz Biotechnology). Levels of p300 in the lysates and immunoprecipitated RelA are shown in the bottom two panels. (C) TNF-α induced acetylation of lysine 310 in reconstituted cells with wild-type RelA. MEFs reconstituted with wild-type, S276A, and S536A RelA were stimulated with TNF-α (20 ng/ml) for the indicated periods of time, whole-cell extracts were prepared, and the acetylation of RelA (Ac-RelA) was detected by immunoblotting with anti-acetylated lysine 310 (α-Ac-K310) antibodies (top). Levels of RelA are shown in the bottom panel. (D) TNF-α-induced activation of the endogenous E-selectin gene is impaired in RelA-deficient MEFs reconstituted with the RelA S276A or S536A mutant. RelA-deficient MEFs reconstituted with either wild-type RelA, RelA S276A, or RelA S536A were stimulated with TNF-α for 0 or 6 h. Total RNA from cells was isolated, and expression levels of E-selectin mRNA were measured by quantitative real-time PCR. Results represent the averages of two independent experiments.
The role of phosphorylation of serines 276 and 536 in the recruitment of p300 was also evaluated in coimmunoprecipitation experiments with RelA-deficient cells stably reconstituted with wild-type, S276A, or S536A RelA. After treatment with TNF-α, RelA interacted with p300 in cells expressing wild-type RelA, but no enhanced interaction was observed in cells expressing S536A or S276A mutants of RelA (Fig. 5B). Next, we investigated the effects of TNF-α on acetylation of lysine 310 in these reconstituted cells. Acetylation of lysine 310 was readily detectable in cells expressing wild-type RelA but not in cells expressing the RelA mutants (Fig. 5C). Taken together, these various results support a model in which phosphorylation of serine 276 and/or serine 536 enhances the effective assembly of RelA with the p300 coactivator. This phosphorylation-dependent enhancement of RelA-p300 binding could certainly underlie the contingent nature of RelA phosphorylation and acetylation at lysine 310.
Finally, we evaluated TNF-α-induced expression of the endogenous E-selectin gene by RT-PCR in RelA-deficient MEFs reconstituted with wild-type or mutant RelA. Cells reconstituted with the S276A and S536A mutants consistently displayed blunted E-selectin responses after TNF-α stimulation compared with cells reconstituted with wild-type RelA (Fig. 5D). These findings indicate that TNF-α-induced phosphorylation of RelA at serine 276 as well as serine 536 contributes to the acetylation of RelA on lysine 310, leading in turn to a heightened transcriptional response by an endogenous NF-κB-inducible cellular gene.
DISCUSSION
In this study, we used anti-acetylated lysine 310 antibodies to demonstrate that RelA is acetylated on lysine 310 both in vitro by p300 and in vivo in response to TNF-α stimulation. We further find that phosphorylation of RelA at either serine 276 or serine 536 facilitates the acetylation of lysine 310, likely by enhancing the binding of RelA to p300. Finally, the importance of these sequential posttranslational modifications of RelA for full transcriptional activation of an endogenous NF-κB target gene is shown.
Although p300 acetylates RelA in vivo (4, 6), recombinant p300 fails to acetylate RelA in vitro despite employing conditions where the acetylation of p53 and histones by recombinant p300 is readily detected (22, 27, 35). This result raises the possibility that a p300 cofactor may be required for RelA acetylation. Consistent with this notion, RelA is readily acetylated in vitro when p300 immunoprecipitates from 293T cells are used as the acetyltransferase (Fig. 1A). It has been reported that p300 requires human immunodeficiency virus Tat protein for the in vitro acetylation of another subunit of NF-κB, p50 (11). Although certainly not a viral factor, p300 appears to require an additional cellular factor for the effective in vitro acetylation of RelA. The nature of this putative cofactor is currently under study. p300/CBP is not the only HAT that mediates acetylation of RelA. PCAF displays such activity, and other HATs have been implicated as coactivators of NF-κB (11). It is possible that different HATs selectively target different lysine residues and that other NF-κB coactivators with HAT activity, such as SRC-1 and SRC-3, play a role, independently or in conjunction with p300/CBP, in the acetylation of different sites in RelA.
Our studies of RelA acetylation have been facilitated by the use of specific anti-acetylated lysine 310 antibodies. Using this antibody, we now demonstrate that lysine 310 in the endogenous RelA protein is acetylated in vivo in a stimulus-coupled manner. Although the exact kinetics and stoichiometry of this acetylation reaction are not precisely known, the acetylation of lysine 310 is temporarily delayed relative to the phosphorylation of RelA. Specifically, acetylation of lysine 310 occurs 10 min after TNF-α stimulation in 293T cells (Fig. 2A), while phosphorylation occurs as early as 5 min after stimulation (23, 31; data not shown). In addition, RelA is phosphorylated in both the cytoplasm and the nucleus (23, 31, 34), whereas acetylation appears restricted to the nucleus, consistent with the nuclear localization of p300/CBP (14). In vitro assays of RelA acetylation further support a contingent relationship between acetylation and phosphorylation. Although recombinant RelA can be forcibly acetylated by p300 in vitro in the absence of phosphorylation (Fig. 1A), prior phosphorylation of RelA markedly enhances its acetylation at lysine 310 in vitro. TNF-α-induced acetylation of RelA is also severely compromised in RelA-deficient cells reconstituted with RelA S276A or RelA S536A (Fig. 5C). Of note, TNF-α-induced phosphorylation of RelA at serines 276 and 536 remains unchanged in RelA-deficient cells reconstituted with RelA-K310R compared with cells reconstituted with wild-type RelA (see Fig. S1 in the supplemental material). Together, these various lines of evidence strongly support a model where the acetylation of RelA at lysine 310 is enhanced by prior phosphorylation of RelA at serine 276 or serine 536. It should be noted that RelA is also acetylated at other sites (5). Unfortunately, site-specific anti-acetylated lysine antibodies are not yet available for their study. It will be interesting to determine whether these other acetylation sites similarly undergo signal-dependent acetylation that is regulated by prior phosphorylation.
In ChIP assays, we demonstrated that RelA acetylated on lysine 310 is effectively recruited to the endogenous IL-8 promoter in vivo in a stimulus-coupled manner (Fig. 2B). Of note, acetylation of lysine 221 improves DNA binding of RelA, while acetylation of lysine 310 does not. However, due to the lack of the appropriate site-specific antibody, we were unable to evaluate the status of lysine 221 acetylation in these ChIP assays. Nevertheless, these findings further strengthen the notion that acetylation of lysine 310 in RelA plays important roles in modeling the overall transcriptional response elicited by NF-κB. However, how acetylation of lysine 310 regulates the overall transcriptional response of NF-κB is not yet clear. It is possible that acetylation of lysine 310 might recruit an unidentified factor which is required for the full transactivation of NF-κB.
In ChIP assays, we also detected the recruitment of phospho-276 RelA to the κB enhancer of the IL-8 promoter after TNF-α stimulation (Fig. 2B). We suspect that some of the bound RelA proteins contain both phosphorylated and acetylated forms of RelA, although sequential ChIP assays will be required to confirm this supposition. We also suspect that phospho-536 RelA is effectively recruited to endogenous κB enhancers after TNF-α stimulation, but this has been technically challenging to demonstrate because, in our hands, the currently available anti-phospho-536 RelA antibody generates a very high nonspecific background signal in the ChIP assays.
The observed link between RelA phosphorylation and acetylation is not unique. Specifically, the phosphorylation of both histones and p53 enhances their acetylation. This link also appears due to improved phosphorylation-dependent recruitment of coactivators such as p300 (7-9, 18, 21, 35) as is the case for phospho-RelA (Fig. 5B and C). Zhong et al. have similarly described that phosphorylation of RelA at serine 276 enhances its assembly with p300 (35). However, our study now extends this initial finding showing that one important consequence of this improved interaction is the acetylation of a key lysine in RelA that potentiates its overall transcriptional activity.
Phosphorylation of serine 276 also enhances the recruitment of coactivator p300/CBP to and displaces transcriptionally repressive p50-HDAC complexes from the promoter region of several NF-κB target genes, thereby facilitating the acetylation of histones surrounding the gene (29, 33, 35). As such, the final overall transcriptional output mediated by phosphorylation of serine 276 likely reflects the combined effects produced by modifying RelA, including its increased acetylation and changes in the chromatin structure surrounding the promoter region of NF-κB target genes. Recent studies also identified phosphorylation of serine 276 as essential for protecting the cells from TNF-α-induced cell death (23). In this regard, deacetylation of lysine 310 by SIRT1 or HDAC3 inhibits the transactivation potential of RelA and similarly augments cellular apoptosis in response to TNF-α (6, 32). Thus, many of the biological events ascribed to phosphorylation of serine 276 may in fact be linked to acetylation of lysine 310.
Of note, while the introduction of a phosphomimetic S536E mutation in RelA led to enhanced acetylation of lysine 310, the corresponding S276E mutation did not (Fig. 3B). This finding suggests that mutation of serine 276 may somehow change the overall conformation of the RelA protein “defeating” the potential enhancing effects of the phosphomimetic substitution. Nevertheless, it is clear that phosphorylation of serine 276 enhances the acetylation of lysine 310 (Fig. 3C) and that the level of acetylation achieved by these mutants closely correlates with their transcriptional activity (Fig. 3B and 4D).
Consistent with the notion that IKK appears to be the major kinase mediating phosphorylation of serine 536 in response to different stimuli (24, 26, 31), we find that expression of a kinase-deficient form of both IKK1 and IKK2 inhibits the acetylation of RelA at lysine 310 (Fig. 4A). Furthermore, phosphorylation of RelA at serine 536 by IKK2 enhances the acetylation of lysine 310 (Fig. 4C). These results highlight the importance of phosphorylation of serine 536 in the regulation of RelA acetylation. Phosphorylation of serine 536 can be mediated by many kinases in response to many stimuli. For example, IKK1, but not IKK2, phosphorylates serine 536 in response to ligation of the lymphotoxin β receptor (16) or after stimulation with the human T-cell leukemia virus type 1 (HTLV-1) Tax oncoprotein (24). IKK2 plays an essential role in the phosphorylation of RelA on serine 536 induced by lipopolysaccharide or TNF-α (31). Serine 536 is also phosphorylated by ribosomal subunit kinase 1 after p53 activation and plays a key role in the NF-κB response elicited by tumor suppressor. We suspect that these various kinases in fact play an important role in regulating the overall acetylation status of lysine 310.
While phosphorylation of serine 536 is clearly linked to the transcriptional activation of NF-κB, the exact mechanism underlying this response was not clear. We now demonstrate that phosphorylation of serine 536 stimulates the acetylation of lysine 310 again by enhancing the recruitment of p300 to RelA. Both the N-terminal and C-terminal regions of RelA have been shown to interact with p300 (12, 27). Phosphorylation at serine 536 may change the conformation of the C-terminal region of RelA promoting its interaction with p300.
In addition to serines 276 and 536, RelA is also phosphorylated at serine 311 by protein kinase C ζ (10) and at serine 529 by casein kinase II (30). Whether phosphorylation of RelA at these serines similarly regulates the acetylation of RelA remains unknown. Phosphorylation of serine 311 has been shown to lead to the more effective recruitment of p300/CBP to RelA and to the promoter region of NF-κB target genes, leading to the hyperacetylation of histones and the activation of transcription (10). It seems likely that phosphorylation of this serine may also influence the acetylation of RelA. Consistent with this hypothesis, we have observed diminished levels of RelA acetylation when serine 311 is substituted with alanine (data not shown).
In summary, our findings indicate that both phosphorylation and acetylation of RelA are required for NF-κB to achieve its full transcriptional activity and that acetylation is largely contingent on prior phosphorylation (Fig. 6). Various posttranslational modifications of the histone tails, including phosphorylation and acetylation, contribute to the construction of a specialized “histone code” within localized regions of the chromatin that appears to regulate the association or disassociation of different cofactors. These modifications, alone or in combination, create specific marks defining the actual or potential transcriptional state of the target genes (2, 15, 28). The different posttranslational modifications of the RelA subunit of NF-κB at specific sites may also create specific marks for the recruitment or divestment of select factors and thus correspond to a “transcription factor code” that dictates specific biological responses of NF-κB in response to different stimuli.
FIG. 6.
Schematic model for the role of phosphorylation of serines 276 and 536 in the regulation of RelA acetylation. PKAc- or MSK-1-mediated phosphorylation of serine 276 or IKK-mediated phosphorylation of serine 536 (A) leads to more effective recruitment of p300 (B), which in turn mediates the acetylation (Ac) of lysine 310 (C). Phosphorylated and acetylated forms of RelA then exhibit potential transcription of NF-κB-dependent genes (D).
Supplementary Material
Acknowledgments
We thank A. Beg, Y. Shi, G. Haegeman, J. S. Arthur, and Q.-H. Zhang for the gift of reagents and Z.-Q. Zhang (Massachusetts General Hospital) for advice on the purification of recombinant RelA. We also thank J. Carroll and C. Goodfellow for assistance in the preparation of figures and S. Ordway and G. Howard for editorial assistance.
L.-F.C. is the recipient of an Arthritis Foundation Investigator Award. This work was supported in part by a National Institutes of Health training grant (AI07305) to L.-F.C., by a National Institutes of Health grant (RO1 CA89001-02) to W.C.G., and by funds from the J. David Gladstone Institutes and Pfizer, Inc., and benefited from core facilities provided through the UCSF-GIVI Center for AIDS Research (National Institutes of Health grant P30 MH59037).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baldwin, A. S., Jr. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 3.Bohuslav, J., L. F. Chen, H. Kwon, Y. Mu, and W. C. Greene. 2004. p53 Induces NF-κB activation by an IκB kinase-independent mechanism involving RSK1 phosphorylation of p65. J. Biol. Chem. 279:26115-26125. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L. F., W. Fischle, E. Verdin, and W. C. Greene. 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293:1653-1657. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L. F., and W. C. Greene. 2004. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 5:392-401. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. F., Y. Mu, and W. C. Greene. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21:6539-6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 8.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 9.Dumaz, N., and D. W. Meek. 1999. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18:7002-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duran, A., M. T. Diaz-Meco, and J. Moscat. 2003. Essential role of RelA Ser311 phosphorylation by ζPKC in NF-κB transcriptional activation. EMBO J. 22:3910-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furia, B., L. Deng, K. Wu, S. Baylor, K. Kehn, H. Li, R. Donnelly, T. Coleman, and F. Kashanchi. 2002. Enhancement of NF-κB acetylation by coactivator p300 and HIV-1 Tat proteins. J. Biol. Chem. 277:4973-4980. [DOI] [PubMed] [Google Scholar]
- 12.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 15.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, X., N. Takahashi, N. Matsui, T. Tetsuka, and T. Okamoto. 2003. The NF-κB activation in lymphotoxin beta receptor signaling depends on the phosphorylation of p65 at serine-536. J. Biol. Chem. 278:919-926. [DOI] [PubMed] [Google Scholar]
- 17.Kiernan, R., V. Bres, R. W. Ng, M. P. Coudart, S. El Messaoudi, C. Sardet, D. Y. Jin, S. Emiliani, and M. Benkirane. 2003. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758-2766. [DOI] [PubMed] [Google Scholar]
- 18.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 19.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 21.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 22.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki, T., S. Sakon, T. Sasazuki, H. Sakurai, T. Doi, H. Yagita, K. Okumura, and H. Nakano. 2003. Phosphorylation of serine-276 is essential for p65 NF-κB subunit-dependent cellular responses. Biochem. Biophys. Res. Commun. 300:807-812. [DOI] [PubMed] [Google Scholar]
- 24.O'Mahony, A. M., M. Montano, K. Van Beneden, L. F. Chen, and W. C. Greene. 2004. Human T-cell lymphotropic virus type 1 Tax induction of biologically active NF-κB requires IκB kinase-1-mediated phosphorylation of RelA/p65. J. Biol. Chem. 279:18137-18145. [DOI] [PubMed] [Google Scholar]
- 25.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakurai, H., H. Chiba, H. Miyoshi, T. Sugita, and W. Toriumi. 1999. IκB kinases phosphorylate NF-κB p65 subunit on serine-536 in the transactivation domain. J. Biol. Chem. 274:30353-30356. [DOI] [PubMed] [Google Scholar]
- 27.Sheppard, K. A., D. W. Rose, Z. K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M. G. Rosenfeld, C. K. Glass, and T. Collins. 1999. Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol. 19:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 29.Vermeulen, L., G. De Wilde, P. Van Damme, W. Vanden Berghe, and G. Haegeman. 2003. Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, D., S. D. Westerheide, J. L. Hanson, and A. S. Baldwin, Jr. 2000. Tumor necrosis factor alpha-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 275:32592-32597. [DOI] [PubMed] [Google Scholar]
- 31.Yang, F., E. Tang, K. Guan, and C. Y. Wang. 2003. IKKβ plays an essential role in the phosphorylation of RelA/p65 on serine-536 induced by lipopolysaccharide. J. Immunol. 170:5630-5635. [DOI] [PubMed] [Google Scholar]
- 32.Yeung, F., J. E. Hoberg, C. S. Ramsey, M. D. Keller, D. R. Jones, R. A. Frye, and M. W. Mayo. 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, H., M. J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9:625-636. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, H., H. SuYang, H. Erdjument-Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89:413-424. [DOI] [PubMed] [Google Scholar]
- 35.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.