Abstract
A key step in homologous recombination is the loading of Rad51 onto single-stranded DNA to form a nucleoprotein filament that promotes homologous DNA pairing and strand exchange. Mediator proteins, such as Rad52 and Rad55-Rad57, are thought to aid filament assembly by overcoming an inhibitory effect of the single-stranded-DNA-binding protein replication protein A. Here we show that mediator proteins are also required to enable fission yeast Rad51 (called Rhp51) to function in the presence of the F-box DNA helicase Fbh1. In particular, we show that the critical function of Rad22 (an orthologue of Rad52) in promoting Rhp51-dependent recombination and DNA repair can be mostly circumvented by deleting fbh1. Similarly, the reduced growth/viability and DNA damage sensitivity of an fbh1− mutant are variously suppressed by deletion of any one of the mediators Rad22, Rhp55, and Swi5. From these data we propose that Rhp51 action is controlled through an interplay between Fbh1 and the mediator proteins. Colocalization of Fbh1 with Rhp51 damage-induced foci suggests that this interplay occurs at the sites of nucleoprotein filament assembly. Furthermore, analysis of different fbh1 mutant alleles suggests that both the F-box and helicase activities of Fbh1 contribute to controlling Rhp51.
Homologous recombination is a fundamental process of DNA metabolism. It promotes the repair of DNA damage, including double-strand breaks, lesion-containing single-stranded gaps, and broken replication forks, and is critical for the correct segregation of homologous chromosomes during meiosis I. A central stage of recombination is the loading of Rad51 onto single-stranded DNA (ssDNA) to form a nucleoprotein filament that promotes homologous DNA pairing and strand exchange (36). The ssDNA-binding protein replication protein A (RPA) both aids and inhibits these processes. For example, it can aid nucleofilament assembly by removing secondary structures in the DNA. However, its abundance and efficiency of binding means that it also out-competes Rad51 for binding to the ssDNA (11, 36). Nucleofilament assembly therefore depends on mediator proteins that enable Rad51 to bind to ssDNA that is coated with RPA. In yeast these mediators include Rad52 and heterodimeric Rad55-Rad57 (11, 36). Rad52 is an ssDNA-binding protein, which interacts both with Rad51 and RPA, and is essential for Rad51-mediated recombination (23, 33, 34, 38). It is thought to bind to RPA-coated ssDNA and act as a nucleation site for the loading of Rad51 (33). Rad55-Rad57 also interacts with Rad51 and may promote the stability of the growing nucleofilament (10, 35).
Despite its beneficial functions, inappropriate recombination can result in genome instability. The evolution of an efficient “recombinosome” has therefore had to go hand in hand with enzymes that temper its action. In humans the tumor suppressor protein BRCA2 may help to control when and where Rad51 loads onto DNA (44). In yeast inappropriate loading of Rad51 can be reversed by the Srs2 DNA helicase, which dissociates Rad51 nucleofilaments (17, 43). Members of the widely conserved RecQ DNA helicase family, which includes Sgs1 and Rqh1 in budding and fission yeasts, respectively, and BLM and WRN in humans, can direct the way in which recombination junctions are resolved (7, 12, 13, 45). Defects in BLM and WRN result in the cancer-prone diseases Blooms syndrome and Werners syndrome, respectively, illustrating the importance of this activity.
Our studies of homologous recombination and its control have made use of the fission yeast Schizosaccharomyces pombe as a model system. In S. pombe there are two Rad52-like proteins, Rad22 and Rti1 (also known as Rad22B) (37, 41, 42). These were thought to be functionally redundant (38). However, we showed recently that Rad22 alone is essential for Rhp51-dependent recombination (Rhp51 is the homologue of Rad51 in S. pombe) (8). Rad22 is therefore just as important for recombination in S. pombe as Rad52 is in Saccharomyces cerevisiae. Previous studies had failed to detect the importance of Rad22 because of suppressor mutations in the rad22 mutant strains that were used. Intriguingly, suppressors of rad52 have not been identified in S. cerevisiae (38). Here we reveal the identity of the suppressor as the F-box DNA helicase Fbh1 and show that it plays an important role in controlling the action of Rhp51. Fbh1 is conserved in humans but is absent in S. cerevisiae and several other model eukaryotic systems (15). Its presence in S. pombe therefore provides a unique opportunity for its study in a genetically tractable system.
MATERIALS AND METHODS
Media and genetic methods.
Media and general genetic methods for S. pombe have been described previously (21). The complete medium was yeast extract with supplements (YES). The minimal medium was Edinburgh minimal medium or yeast nitrogen base supplemented with appropriate amino acids. The medium used to select Ade+ recombinants in plating assays was YES lacking adenine and supplemented with 200 mg/liter guanine to prevent uptake of residual adenine. Where appropriate, thiamine was added to media at a final concentration of 4 μM. Fbh1 was cloned by complementing the methyl methanesulfonate (MMS) and camptothecin (CPT) sensitivities of an fbh1-1 strain (MCW1585) with a pUR19-based S. pombe genomic library (4).
Strains and plasmids.
The S. pombe strains used in this study are listed in Table 1. The fbh1::kanMx6 mutant contains an insertion of a kanR marker between the two SalI sites in fbh1, whereas the fbh1Δ::kanMx6 mutant contains a replacement of the entire fbh1+ open reading frame by a kanR marker and was constructed by PCR-based gene targeting (3). Construction of the ECFP-rhp51+-kanR strain will be described elsewhere. The fbh1+ open reading frame was cloned into pREP41-EYFP to give plasmid pMW651, expressing N-terminally tagged EYFP-fbh1+ from the thiamine-repressible nmt1 promoter. Plasmid pMW637 is a pREP41 derivative that expresses wild-type fbh1+ from an nmt1 promoter. The F-box (Fbh1L14A/P15A) mutant and helicase-defective (Fbh1D485N) versions of this plasmid were derived from pMW637 by using a QuikChange site-directed mutagenesis kit (Stratagene), and the changes were confirmed by nucleotide sequencing. In order to construct the fbh1L14A/P15A and fbh1D485N mutant strains, the fbh1L14A/P15A and fbh1D485N mutant genes were subcloned into plasmid pFA6a-KanMx6 (3) upstream of the kanR marker. The region of genomic DNA (∼600 bp) that lies immediately downstream of fbh1 was then cloned into these plasmids downstream of the kanR marker to make plasmids pMW695 (containing fbh1L14A/P15A) and pMW696 (containing fbh1D485N). NdeI-plus-BamHI digestion of these plasmids results in a linear fragment, which consists of mutant fbh1 and downstream genomic DNA flanking a kanR marker. This was used to transform strain MCW1221. Geneticin-resistant transformants were screened for successful replacement of fbh1 by the fbh1 mutant-kanR cassette by PCR. The L14A/P15A and D485N mutations in these strains were confirmed by nucleotide sequencing.
TABLE 1.
S. pombe strains
| Strain | Genotype | Source or reference |
|---|---|---|
| MCW1221 | h+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1489 | h+fbh1::kanRura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1490 | h+fbh1Δ::kanRura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1585 | h−smt-0 fbh1-1 ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1285 | h+rad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | To be described elsewhere |
| MCW1553 | h+fbh1Δ::kanRrad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1222 | h+fbh1-1 rad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This studya |
| MCW1686 | h+rad22Δ::ura4+rti1Δ::LEU2 ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1512 | h+fbh1Δ::kanRrti1Δ::LEU2 ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1677 | h+fbh1Δ::kanRrad22Δ::ura4+rti1Δ::LEU2 ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1088 | h+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1587 | h+fbh1Δ::kanRrhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1588 | h+rad22Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1589 | h+fbh1Δ::kanRrhp51Δ::arg3+rad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW429 | h+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | Lab strain |
| MCW1494 | h+rad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | Lab strain |
| MCW1495 | h+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | Lab strain |
| MCW1496 | h+rad22Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | Lab strain |
| MCW1504 | h+fbh1Δ::kanRura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | This study |
| MCW1506 | h+fbh1Δ::kanRrad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | This study |
| MCW1508 | h+fbh1Δ::kanRrhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | This study |
| MCW1510 | h+fbh1Δ::kanRrad22Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 int::pUC8/his3+/ade6-L469 | This study |
| YA177 | h−smt-0 swi5Δ::his3+ura4-D18 leu1-32 his3-D1 arg3-D4 | 2 |
| MCW1590 | h+fbh1Δ::kanRswi5Δ::his3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1231 | h+rhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1568 | h+fbh1Δ::kanRrhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1679 | h+rad22Δ::ura4+rhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1681 | h+fbh1Δ::kanRrad22Δ::ura4+rhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1643 | h+rad22Δ::ura4+swi5Δ::his3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1645 | h+fbh1Δ::kanRrad22Δ::ura4+swi5Δ::his3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1017 | h+srs2Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1570 | h+fbh1Δ::kanRsrs2Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1571 | h+srs2Δ::ura4+rhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1568 | h+fbh1Δ::kanRsrs2Δ::ura4+rhp55Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1804 | h+rqh1Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1240 | h+rqh1Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1591 | h+fbh1Δ::kanRrqh1Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1099 | h+srs2Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1803 | h+fbh1Δ::kanRsrs2Δ::ura4+rhp51Δ::arg3+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1548 | h+ura4-D18 leu1-32 his3-D1 arg3-D4 carrying pMW651 | This study |
| MCW1487 | h+ECFP-rhp51+-KanRura4-D18 leu1-32 his3-D1 arg3-D4 | To be described elsewhere |
| MCW1556 | h+ECFP-rhp51+-KanRura4-D18 leu1-32 his3-D1 arg3-D4 carrying pMW651 | This study |
| MCW1718 | h+fbh1D485N-kanRura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1767 | h+fbh1D485N-kanRrad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1768 | h+fbh1L14A/P15A-kanRura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
| MCW1769 | h+fbh1L14A/P15A-kanRrad22Δ::ura4+ura4-D18 leu1-32 his3-D1 arg3-D4 | This study |
The parent of this strain is RGL6 (41), which acquired the fbh1-1 allele at some point before we received it.
Microscopy.
Early-log-phase cells growing in liquid media or arrested cells grown for 4 h in liquid media containing 10 μM CPT were used for microscopic examination. Cells were harvested, washed in water, and used live or after fixation in 70% ethanol. Cells were stained with the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) where appropriate. Cells were visualized with a 100× objective on a Olympus BX50 microscope equipped with a cooled charge-coupled-device camera (Princeton). Enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein (ECFP) foci were visualized using 31044v2 and 41028 filters (Chroma). Images were taken and analyzed using MetaMorph software.
Plasmid loss assay.
Wild-type and fbh1Δ cells harboring vector pREP41 (containing the LEU2 marker, which complements the leu1-32 auxotrophic marker in the parental strains) were grown for 24 h in nonselective YES liquid medium. Cells were harvested, washed in water, appropriately diluted, and plated on YES medium. Colonies were formed after 4 days at 30°C, and retention or loss of the pREP41 plasmid was determined by replica plating onto Edinburgh minimal medium lacking leucine. Three independent wild-type (pREP41) and fbh1Δ (pREP41) transformants were assayed for plasmid loss. The percent plasmid loss per generation (ρ) was determined by the formula ρ = {1−e[1/n × ln(Rn/R0]} × 100, where R0 and Rn are the percentages of cells that retain the plasmid at generations 0 and n, respectively.
Spot assays.
Exponentially growing cells from liquid cultures were harvested, washed, and resuspended in water at a density of 1 × 107 to 1 × 103 cells/ml. Aliquots (10 μl) of the cell suspensions were spotted onto solid media containing various concentrations of chemical genotoxins (MMS, hydroxyurea [HU], or CPT) or irradiated with various doses of UV light using a Stratalinker (Stratagene). In each spot assay, the undiluted spot (designated 1 in each of the figures) represents 1 × 105 cells, whereas the 0.1, 0.01, and 0.001 spots represent 1 × 104, 1 × 103, and 1 × 102 cells, respectively. For each spot assay four different doses of each genotoxin was used. However, for most figures only a single dose is displayed for simplicity. Plates were incubated at 30°C for up to 6 days and were normally photographed on each day, starting from day 2, in order to monitor differences in growth rates between the strains. The lengths of incubation for the spot assays shown in the figures are indicated in the relevant figure legends. All spot assays were repeated at least once to ensure reproducibility. The spot assays reveal relative differences in survival and growth following a single (acute) exposure to UV or prolonged (chronic) exposure to MMS, HU, and CPT.
MMS, CPT, and UV each generate DNA lesions that result in replication fork blockage or breakage. Specifically, UV and MMS generate base damage that can block replication fork progression, whereas CPT inhibits the religation step during the topoisomerase I reaction cycle, which results in increased numbers of single-strand breaks in the DNA at which replication forks collapse (26). As mentioned in the introduction, homologous recombination is used to tolerate and/or repair blocked and broken replication forks (8). HU inhibits ribonucleotide reductase, which results in a decrease in deoxynucleoside triphosphate pools and subsequent replication fork stalling. Chronic exposure to HU results in DNA breakage, which is repaired by homologous recombination (29).
Doubling time and plating efficiency.
Doubling times were determined by measuring the optical density at 600 nm during exponential growth in liquid YES at 30°C. Mean values are from at least four independent cultures. Plating efficiency was determined by plating a known number of cells from cultures in liquid YES during exponential growth at 30°C onto YES agar. Cells were counted using a hemocytometer. Mean values are from at least four independent cultures.
Quantitative UV survival assays.
Dilutions of cells growing exponentially were plated in triplicate onto YES plates and UV irradiated using a Stratalinker (Stratagene). Plates were incubated at 30°C for 6 days before colonies were counted. All data points represent the mean value from three independent cultures.
Recombination assays.
Mitotic recombination was assayed by the recovery of Ade+ recombinants from strains containing the intrachromosomal recombination substrate shown in Fig. 4D, as described previously (7). Recombinant frequencies represent the mean value from at least 15 colonies for each strain.
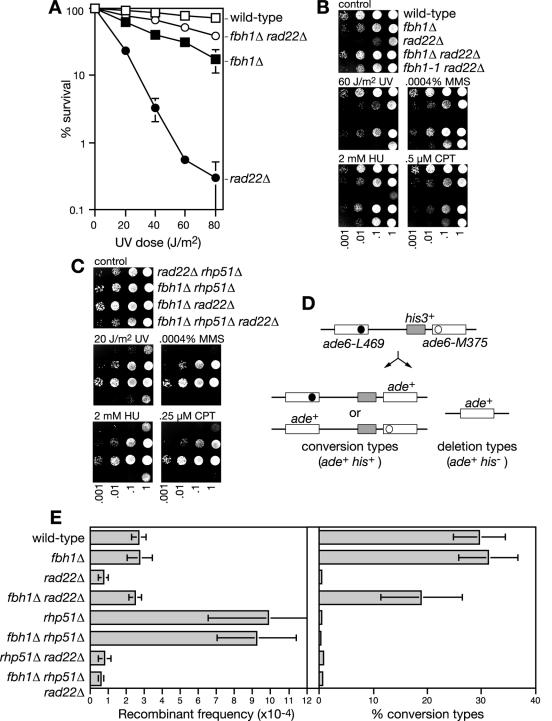
FIG. 4.
Suppression of rad22Δ mutant phenotypes by fbh1Δ. (A) UV survival curves of wild-type (MCW1221), fbh1Δ (MCW1490), rad22Δ (MCW1285), and fbh1Δ rad22Δ (MCW1553) strains. The error bars represent the standard deviations about the mean values. (B) Spot assay comparing the sensitivities of the indicated strains to different genotoxins. The strains are the same as in panel A, with the addition of the fbh1-1 rad22Δ strain (MCW1222). The plates were photographed after 3 days of incubation. (C) Spot assay showing that rad22Δ suppression of genotoxin sensitivity by fbh1Δ is dependent on Rhp51. The strains are rad22Δ rhp51Δ (MCW1588), fbh1Δ rhp51Δ (MCW1587), fbh1Δ rad22Δ (MCW1553), and fbh1Δ rhp51Δ rad22Δ (MCW1589). The plates were photographed after 5 days of incubation. (D) Schematic of the recombination substrate and potential recombinant products. The solid and open circles indicate the position of the point mutations in ade6-L469 and ade6-M375, respectively. (E) Bar charts showing the recombinant frequencies and percentages of recombinants that are conversion types for the indicated strains. The strains are wild type (MCW429), fbh1Δ (MCW1504), rad22Δ (MCW1494), fbh1Δ rad22Δ (MCW1506), rhp51Δ (MCW1495), fbh1Δ rhp51Δ (MCW1508), rhp51Δ rad22Δ (MCW1496), and fbh1Δ rhp51Δ rad22Δ (MCW1510). Error bars are the standard deviations about the mean of each value.
RESULTS
Identification of a rad22 suppressor.
We reported recently that three different rad22 mutant strains, which we had obtained from other laboratories, grew much better and were more resistant to genotoxins than a de novo rad22 deletion strain made in our laboratory (8). One possible explanation for this discrepancy was that the published rad22 mutant strains each carried a suppressor mutation. To test this, we backcrossed the putative suppressed rad22 mutant strains to a wild-type strain and obtained rad22− segregants that were as sensitive to genotoxins as our de novo rad22Δ strain (not shown). We also found that our de novo rad22Δ strain readily acquires faster-growing colonies, which are more resistant to genotoxins (not shown). These data indicate that rad22 mutant strains can acquire suppressor mutations.
To identify the suppressor in one of the rad22 mutant strains, we backcrossed it to a wild-type strain and obtained non-rad22 mutant segregants that grew more slowly than the wild type and exhibited hypersensitivity to the genotoxins MMS and CPT (for an explanation of the different genotoxins used in this study, see “Spot assays” in Materials and Methods). In preliminary experiments (data not shown), we demonstrated 1:1 segregation of these phenotypes through further crosses, which showed that they were likely to be due to a single-gene mutation. Moreover, when the non-rad22 mutant segregants were crossed with a nonsuppressed rad22Δ strain, half of the rad22Δ progeny were suppressed, consistent with these segregants carrying the suppressor mutation.
The suppressor was cloned by complementation of its mutant phenotypes by using a genomic DNA library. Two complementing clones were isolated from approximately 20,000 transformants. Each contained a different but overlapping piece of genomic DNA with a single common gene encoding the homologue of the human F-box DNA helicase 1 (Fbh1) (not shown). Nucleotide sequencing of fbh1 in the suppressed rad22Δ strain, and in the suppressor mutant strain derived from it, revealed a frameshift mutation at codon 568 (an A insertion), which would result in a truncated protein lacking key helicase motifs (Fig. 1A), and a T-to-G transversion resulting in a F735C change. We have named this fbh1 allele fbh1-1.
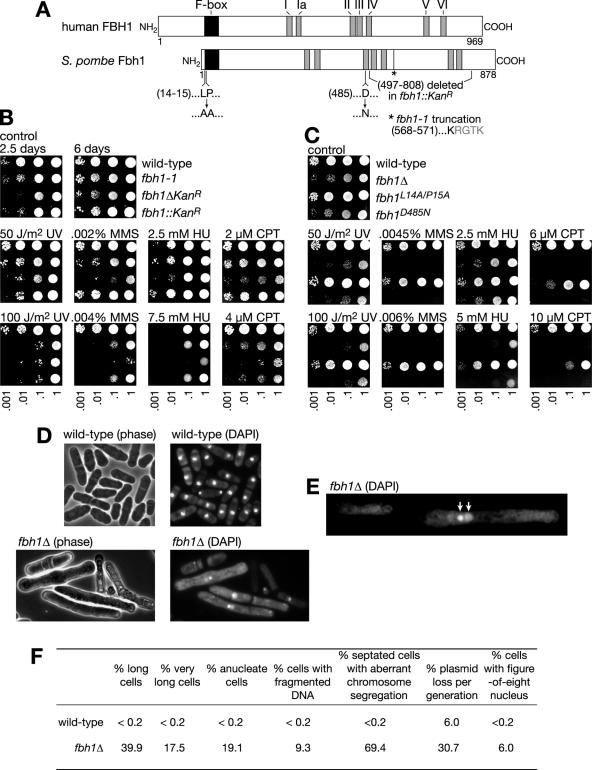
FIG. 1.
Mutant phenotypes of fbh1. (A) Schematic of human FBH1 and S. pombe Fbh1, showing their respective lengths (in amino acids) and positions of F-box and helicase motifs. Also shown are the terminal amino acids in the truncated Fbh1 expressed by the fbh1-1 mutant (the point of truncation is indicated by the asterisk), the region deleted in the fbh1::kanR mutant, and the mutations introduced to investigate the importance of F-box and helicase activities. (B) Spot assay comparing the growth and sensitivity of wild-type (MCW1221), fbh1-1 (MCW1585), fbh1Δ (MCW1490), and fbh1::kanR (MCW1489) strains to different genotoxins. The plates were photographed after 6 days of incubation. The control plate is also shown after 2.5 days incubation. (C) Spot assay comparing the sensitivity of wild-type (MCW1221), fbh1Δ (MCW1490), fbh1L14A/P15A (MCW1768), and fbh1D485N (MCW1718) strains to different genotoxins. The plates were photographed after 5 days of incubation. (D) Phase-contrast and fluorescence microscopic images of wild-type and fbh1Δ cells stained with DAPI. (E) Fluorescence microscopic image of a DAPI-stained fbh1Δ cell with a figure-eight nucleus. The arrows indicate the two circular nuclear domains. (F) Aberrant cell morphology and DNA segregation defects of an fbh1Δ mutant growing at 30°C in minimal medium. Long cells are 14 to 28 μm in length, and very long cells are >28 μm. Septated cells were scored as having aberrant chromosome segregation if they displayed an unequal distribution of DNA between the two daughter cells or if the DNA was fragmented or bisected by the septum (“cut” phenotype). For each strain ∼500 cells were assessed.
Fbh1 promotes DNA repair.
Fbh1 was first identified from a biochemical screen for DNA helicases, and it is conserved in S. pombe and mammals but absent in budding yeast, fruit fly, frog, fish, and plants (15, 24). In S. pombe Fbh1 was originally named Fdh1 (24). However, this is also the name of a formate dehydrogenase gene from the methylotrophic yeast Candida boidinii (30). Hence, we have decided to call S. pombe Fdh1 Fbh1 after its human homologue. Fbh1 contains the seven conserved motifs of a superfamily 1 DNA helicase (Fig. 1A) and unwinds DNA with 3′-to-5′ directionality, and its close relatives include UvrD, Rep, and Srs2. Interestingly it is the only known DNA helicase that contains an F-box motif. F-box proteins are substrate recognition components of SCF (Skp, Cullin, F-box) ubiquitin-ligase complexes, which catalyze the polyubiquitination of proteins to target them for degradation (6). In the case of human Fbh1, it is known to form a bone fide SCF complex, although its target(s) remains unknown (16).
To characterize Fbh1, we made two strains, one containing a replacement of the entire fbh1 gene with a geneticin resistance marker (fbh1ΔkanR) and a second one containing a replacement of amino acids 497 to 808 with a geneticin resistance marker (fbh1::kanR) (Fig. 1A). A comparison of these strains with the fbh1-1 strain revealed that the fbh1ΔkanR strain grows slower (Fig. 1B, compare growth after 2.5 and 6 days) and is more sensitive to chronic exposure to HU, MMS, and CPT than either the fbh1-1 or fbh1::kanR strain. All three fbh1 mutant strains display similar levels of survival following acute exposure to UV, and the growth and genotoxin hypersensitivities of the fbh1-1 and fbh1::kanR strains are equivalent (Fig. 1B). These data show that Fbh1 promotes the tolerance/repair of DNA damage and is required for normal growth. Furthermore, as both fbh1-1 and fbh1::kanR mutants would express truncated forms of Fbh1 missing key helicase motifs (see Fig. 1A), we suspect that Fbh1 with an intact F-box, but little or no helicase activity, retains some ability to promote growth and DNA repair.
To investigate the relative importance of Fbh1's DNA helicase and putative ubiquitin ligase activities in promoting DNA repair, we constructed strains containing mutations in the F-box (L14A/P15A) and helicase (D485N in motif II) domains of Fbh1 (Fig. 1A) and compared them to the fbh1Δ strain for growth and genotoxin resistance (Fig. 1C). Equivalent mutations in other proteins have disabled F-box and helicase functions, respectively (25, 28). We also confirmed by Western blot analysis that Fbh1L14A/P15A and Fbh1D485N were expressed at levels comparable to those of the wild-type protein (not shown). The fbh1D485N strain exhibits growth and sensitivity to genotoxins similar to those of the fbh1Δ strain, whereas fbh1L14A/P15A displays only modest hypersensitivity to CPT (Fig. 1C). These data indicate that Fbh1's DNA helicase activity is critical for promoting DNA repair. However, this contradicts data obtained with the fbh1-1 and fbh1::kanR strains, which suggest that Fbh1 can promote some DNA repair without its DNA helicase activity (see above). One possible explanation for this discrepancy is that fbh1D485N has a dominant negative effect, which masks any residual activity it might have in promoting DNA repair. In accord with this, overexpression of Fbh1D485N, from the nmt1 promoter in the plasmid pREP41, does have a dominant negative effect on the growth and genotoxin resistance of a wild-type strain (not shown). With respect to the importance of the F-box in Fbh1, our data with fbh1L14A/P15A suggest that it plays only a minor role in promoting DNA repair. However, we have no direct evidence that the F-box activity in Fbh1 is destroyed by the L14A/P15A mutation.
Fbh1 promotes proper chromosome segregation.
The doubling time of the fbh1Δ mutant strain growing in liquid culture is approximately twice that of the wild type (Table 2). At least part of this slow growth can be explained by a reduction in cell viability as judged by the plating efficiency of the fbh1Δ strain (Table 2). Cytological examination revealed that more than half of fbh1Δ cells growing in culture are elongated, with ∼17% greater than twice the normal haploid cell division length (∼14 μm) (Fig. 1D and F). Moreover, staining with the DNA-specific dye DAPI showed that ∼30% of fbh1Δ cells are anucleate or contain fragmented DNA and that ∼70% of septated cells contain DNA segregating unevenly between the two daughters (Fig. 1D and F). These phenotypes are indicative of a defect in chromosome segregation. Consistent with this, fbh1Δ cells exhibit a ∼5-fold-higher rate of plasmid loss than the wild type (Fig. 1F). DAPI staining also revealed a significant number of cells with an unusual “figure-eight” nuclear region, where the majority of DAPI-stained material is in one circular domain, which is closely associated with, or connected to, a second, larger circular domain that is less stained (Fig. 1E and F). The significance of this phenotype is currently uncertain.
TABLE 2.
Doubling times and plating efficiencies of wild-type and mutant strains
| Relevant genotype | Strain | Doubling time (min)a | Relative plating efficiencyab |
|---|---|---|---|
| Wild type | MCW1221 | 172 (6) | 1.0 |
| fbh1Δ | MCW1490 | 332 (52) | 0.47 (0.19) |
| rhp51Δ | MCW1088 | 287 (24) | 0.67 (0.20) |
| fbh1Δ rhp51Δ | MCW1587 | 323 (28) | 0.80 (0.18) |
| rad22Δ | MCW1285 | 373 (50) | 0.39 (0.10) |
| fbh1Δ rad22Δ | MCW1553 | 216 (5) | 1.0 (0.06) |
| rad22Δ rhp51Δ | MCW1588 | 341 (49) | 0.46 (0.16) |
| fbh1Δ rad22Δ rhp51Δ | MCW1589 | 328 (29) | 0.42 (0.12) |
| fbh1D485Nrad22Δ | MCW1767 | 228 (8) | Not determined |
Mean values with standard deviations in parentheses.
values are relative to that for the wild type.
Fbh1 functions in an Rhp51-dependent pathway of DNA repair.
The ability of fbh1-1 to suppress rad22Δ mutant phenotypes suggested that Fbh1 might function in an Rhp51-dependent pathway for DNA repair. To see if this was true, fbh1Δ and rhp51Δ single and double mutant strains were first compared for growth and cell viability (Table 2). The doubling time of an fbh1Δ rhp51Δ double mutant is similar to that of an fbh1Δ single mutant, which in turn is longer than that of an rhp51Δ single mutant. However, the plating efficiency of an fbh1Δ single mutant is slightly less than those of both the rhp51Δ and fbh1Δ rhp51Δ mutants (Table 2). These data indicate an epistatic relationship between fbh1 and rhp51 for promoting normal growth and cell viability.
We next compared the fbh1Δ, rhp51Δ, and fbh1Δ rhp51Δ strains for genotoxin sensitivity (Fig. 2A). The fbh1Δ rhp51Δ double mutant exhibits no increase in sensitivity to acute UV exposure compared to an rhp51Δ single mutant (Fig. 2A). In contrast, the double mutant appears to be slightly more sensitive to chronic MMS, HU, and CPT exposure than either of the single mutants (Fig. 2A). However, this is due to the double mutant growing slower than the rhp51Δ single mutant in the presence of genotoxins, rather than to an actual reduction in viability (not shown). Together these data indicate that Fbh1 functions in an Rhp51-dependent pathway for DNA repair.
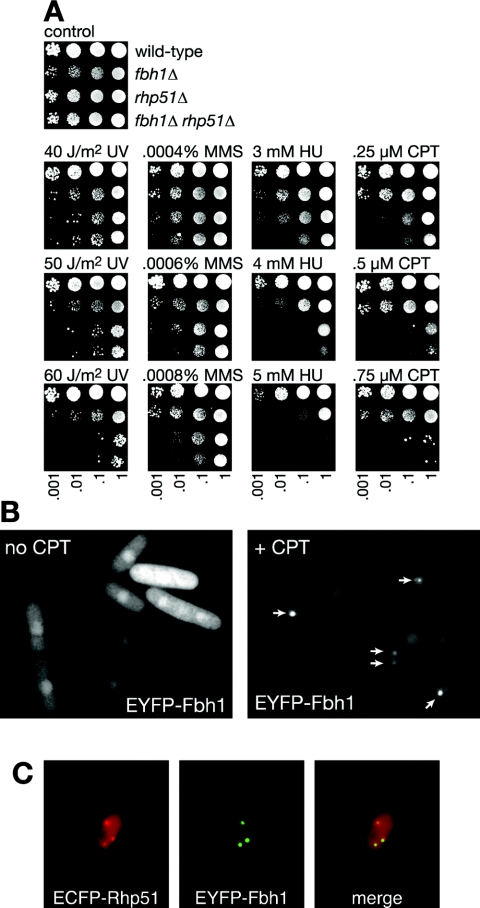
FIG. 2.
Fbh1 functions in an Rhp51-dependent DNA repair pathway. (A) Spot assay comparing the genotoxin sensitivities of wild-type (MCW1221), fbh1Δ (MCW1490), rhp51Δ (MCW1088), and fbh1Δ rhp51Δ (MCW1587) strains. The plates were photographed after 5 days of incubation. (B) Detection of EYFP-Fbh1 foci by fluorescence microscopy. Strain MCW1548, which expresses EYFP-Fbh1 from plasmid pMW651, was grown at 30°C with and without 10 μM CPT for 4 h as indicated. The arrows in the right panel indicate the position of foci. (C) Colocalization of Fbh1 and Rhp51 at damage-induced foci. Foci were induced by CPT as for panel B. The strain is MCW1556, which expresses ECFP-Rhp51 from the endogenous rhp51 promoter and EYFP-Fbh1 from pMW651.
Fbh1 forms DNA damage-induced nuclear foci that colocalize with Rhp51.
Both Rhp51 and Rad22 are known to localize at discrete foci within the nucleus following DNA damage (18, 20). These foci are believed to be centers of DNA repair. To see if Fbh1 also localizes to these sites, we fused EYFP to its N terminus and expressed it from the nmt1 promoter in the plasmid pREP41. This tagged construct was able to partially complement the hypersensitivity to genotoxins of an fbh1Δ strain, indicating that it retained at least some functionality (not shown). When expressed in a wild-type strain in the absence of an exogenous genotoxin, the majority of cells exhibit EYFP-Fbh1 as faint fluorescence throughout the nucleus (Fig. 2B). However, a few cells (<1%) contain a single EYFP-Fbh1 focus within the nucleus (not shown), and following exposure to 10 μM CPT for 4 h, >30% of cells exhibit one or more foci (similar results were obtained with HU, MMS, and mitomycin C) (Fig. 2B, see Fig. 7, and data not shown). To see if these foci colocalize with Rhp51 foci, EYFP-Fbh1 was expressed in a strain in which Rhp51 is fused to ECFP. Following DNA damage, >90% of EYFP-Fbh1 and ECFP-Rhp51 foci colocalize (Fig. 2C and data not shown). These data are consistent with Fbh1 functioning together with Rhp51 at sites of DNA damage.
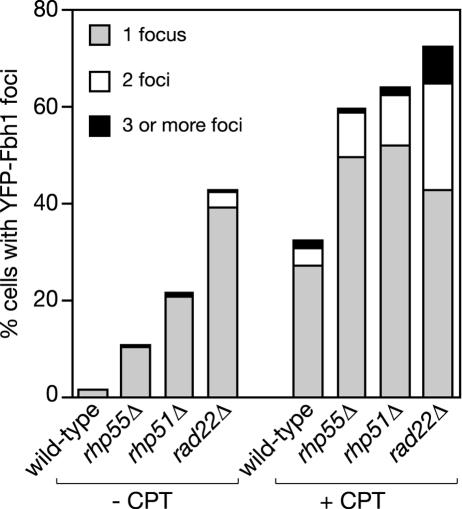
FIG. 7.
Spontaneous and CPT-induced EYFP-Fbh1 focus formation in different recombination mutants. Strains were grown at 30°C with and without 10 μM CPT for 4 h as indicated. The strains are wild type (MCW1221), rhp55Δ (MCW1231), rhp51Δ (MCW1088), and rad22Δ (MCW1285), each carrying pMW651. Values for the percentage of cells containing foci are based on the assessment of 500 cells in each case.
Genetic interactions with Rqh1 and Srs2.
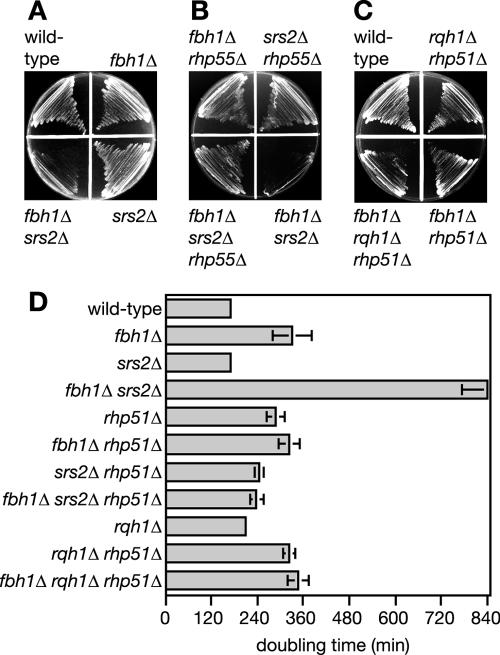
The DNA helicases Rqh1 and Srs2 function in separate but overlapping pathways for processing Rhp51-dependent recombination intermediates (9, 18, 19, 22). To see if Fbh1 functions in either of these pathways, an fbh1Δ mutant was crossed with both rqh1Δ and srs2Δ mutant strains. Each gene deletion was marked with a different selectable marker so that the genotypes of the viable progeny could readily be distinguished. For the fbh1Δ × rqh1Δ cross, >100,000 viable progeny from several independent crosses were analyzed for the presence of a rqh1Δ fbh1Δ double mutant (not shown). However, only wild-type, rqh1Δ, and fbh1Δ progeny were recovered, indicating that the double mutant is unviable. In contrast, fbh1Δ srs2Δ double mutants were readily recovered from the fbh1Δ × srs2Δ cross. However, a comparison of the fbh1Δ srs2Δ double mutant with its single mutants reveals a more-than-additive reduction in growth (Fig. 3A and D). Both the nonviability of an fbh1Δ rqh1Δ double mutant and the poor growth of an fbh1Δ srs2Δ double mutant are rescued by deleting either rhp51 or its mediator rhp55 (Fig. 3B, C, and D). This indicates either that Fbh1 shares an overlapping role with both Srs2 and Rqh1 for processing Rhp51-dependent recombination intermediates or that it prevents Rhp51 action that would necessitate subsequent processing by Srs2 and Rqh1.
FIG. 3.
Suppressing the poor viability of the fbh1Δ srs2Δ mutant and nonviability of the fbh1Δ rqh1Δ mutant by rhp55Δ and rhp51Δ mutations. (A to C) A comparison of the growth on agar plates of wild-type (MCW1221), fbh1Δ (MCW1490), srs2Δ (MCW1017), fbh1Δ srs2Δ (MCW1570), fbh1Δ rhp55Δ (MCW1568), srs2Δ rhp55Δ (MCW1571), fbh1Δ srs2Δ rhp55Δ (MCW1568), rqh1Δ rhp51Δ (MCW1240), fbh1Δ rhp51Δ (MCW1587), and fbh1Δ rqh1Δ rhp51Δ (MCW1591) strains. Strains were streaked onto complete medium plates and grown for 3 days at 30°C before being photographed. (D) Doubling times of wild-type (MCW1221), fbh1Δ (MCW1490), srs2Δ (MCW1017), fbh1Δ srs2Δ (MCW1570), rhp51Δ (MCW1088), fbh1Δ rhp51Δ (MCW1587), srs2Δ rhp51Δ (MCW1099), fbh1Δ srs2Δ rhp51Δ (MCW1803), rqh1Δ (MCW1804), rqh1Δ rhp51Δ (MCW1240), fbh1Δ rhp51Δ (MCW1587), and fbh1Δ rqh1Δ rhp51Δ (MCW1591) strains. The error bars represent the standard deviations about the mean values.
Suppression of rad22 mutant phenotypes by fbh1Δ.
Having characterized some of the basic phenotypes of an fbh1Δ mutant, we next looked at its genetic interaction with rad22. While we already knew that fbh1-1 partially suppressed the genotoxin hypersensitivity of a rad22Δ mutant, it was important to establish that the same was true for the fbh1Δ mutant. To this end, we constructed an fbh1Δ rad22Δ double mutant and compared it to its respective single mutants and to an fbh1-1 rad22Δ double mutant. Both the poor growth and genotoxin sensitivity of a rad22Δ mutant are suppressed by fbh1Δ (Fig. 4A and B and Table 2). Greater suppression is seen with fbh1Δ than with the fbh1-1 allele (Fig. 4B). This suggests that truncated Fbh1, which would probably be devoid of helicase activity, retains some ability to block repair in the absence of Rad22. Another important observation is that the fbh1Δ rad22Δ double mutant grows better and is more viable than an fbh1Δ single mutant and is also more resistant to UV light (Table 2 and Fig. 4A). Cosuppression in the double mutant indicates that both Rad22 and Fbh1 have a negative effect on growth and DNA repair in the absence of the other.
To see if rad22 suppression by fbh1Δ depends on the other Rad52-like protein in S. pombe, Rti1, the genotoxin sensitivities of rad22Δ rti1Δ, fbh1Δ rad22Δ, and fbh1Δ rti1Δ double mutants were compared to those of an fbh1Δ rad22Δ rti1Δ triple mutant (not shown). An rti1Δ single mutant exhibits no detectable hypersensitivity to UV, HU, MMS, or CPT, and this mutation has no effect on the hypersensitivity of a rad22Δ mutant (not shown). Likewise, it has no effect on the suppression of rad22Δ by fbh1Δ as judged by the similar sensitivities of fbh1Δ rad22Δ and fbh1Δ rad22Δ rti1Δ mutant strains.
An analysis similar to that described above was performed to see if rad22Δ suppression depends on Rhp51. The rad22Δ rhp51Δ double mutant exhibits the same hypersensitivity to genotoxins as a rad22Δ single mutant (8), but, unlike for the rad22Δ strain, these phenotypes are not suppressed by the deletion of fbh1 (Fig. 4C). Likewise, the poor growth and viability of the rad22Δ rhp51Δ double mutant are not suppressed by deleting fbh1 (Table 2). This absolute dependence on rhp51+ for the suppression of rad22Δ mutant phenotypes suggests that Fbh1 inhibits the action of Rhp51 in the absence of Rad22.
Most homologous recombination depends on Rad22 in S. pombe (8). To see if this role is also suppressed by deleting fbh1, we constructed relevant mutant strains containing a direct repeat of ade6− mutant alleles integrated at the ade6 locus on chromosome III (Fig. 4D). Inter- or intrachromatid recombination between the ade6− mutant alleles results in ade+ recombinants that retain (conversion types) or lose (deletion types) the intervening his3+ gene. In the wild-type strain the frequency of spontaneous ade+ recombinants is ∼3 × 10−4 (Fig. 4E). Approximately 30% of these recombinants are conversion types. Deletion of fbh1 has no effect on spontaneous direct repeat recombination as measured by this assay (Fig. 4E). It also does not affect the frequency of UV-induced recombinants or the hyper-deletion-type recombination in an rhp51Δ mutant (Fig. 4E and data not shown). In the rad22Δ strain the recombinant frequency drops to <1 × 10−4, and all of these recombinants are deletion types (Fig. 4E). Recombination is restored to wild-type levels by deleting fbh1, although the percentage of recombinants that are conversion types is less than in the wild type (Fig. 4E). As with the suppression of genotoxin hypersensitivity, the improvement in recombinant formation in a rad22Δ mutant, by deleting fbh1, is dependent on Rhp51. This is evident from the fact that an fbh1Δ rhp51Δ rad22Δ triple mutant exhibits the same low frequency of recombination as both a rad22Δ single mutant and rhp51Δ rad22Δ double mutant (Fig. 4E). With the data above, these results show that much of Rad22's role in promoting Rhp51-dependent recombination and DNA repair can be circumvented by the removal of Fbh1.
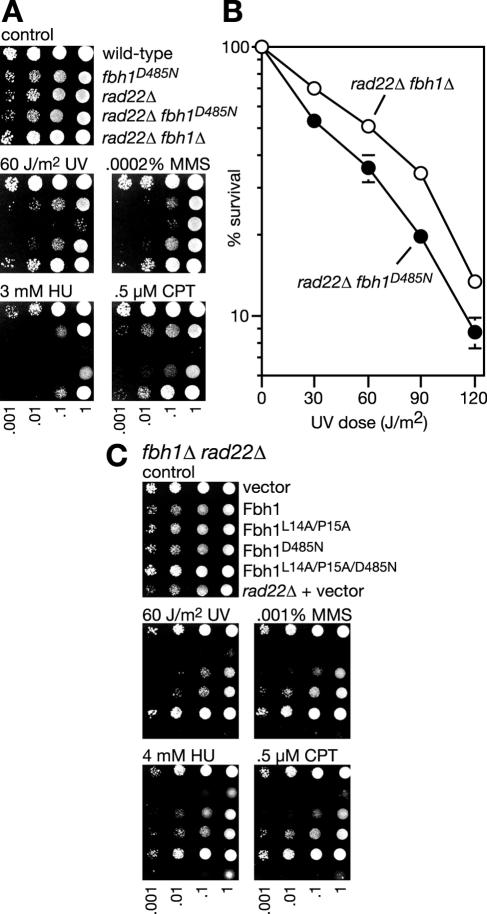
Both the F-box and helicase domains of Fbh1 contribute to the inhibition of Rhp51-dependent DNA repair in the absence of Rad22.
As mentioned above, the failure of fbh1-1 to suppress rad22Δ genotoxin hypersensitivity as much as fbh1Δ suggests that Fbh1 inhibits repair by means other than just its DNA helicase activity. To test this further, we compared the abilities of fbh1Δ and fbh1D485N to suppress the poor growth and genotoxin sensitivity of a rad22Δ mutant. Both mutant alleles reduce the doubling time of a rad22Δ mutant, growing in liquid culture, to similar levels (Table 2). However, an fbh1D485N rad22Δ double mutant does not grow as well as an fbh1Δ rad22Δ double mutant on solid media in the presence of genotoxins, and this effect is due to a combination of slower growth and reduced survival (Fig. 5A and B). This provides further evidence that Fbh1 does not depend solely on its DNA helicase activity to inhibit Rhp51-dependent repair in the absence of Rad22. To see whether Fbh1's F-box is important here, we tested whether fbh1L14A/P15A could suppress rad22Δ. However, neither the poor growth nor genotoxin hypersensitivity of a rad22Δ mutant was suppressed by fbh1L14A/P15A (not shown). We also tested whether overexpression of fbh1L14A/P15A and fbh1D485N, from the nmt1 promoter in pREP41, could complement the improved genotoxin resistance of an fbh1Δ rad22Δ double mutant (i.e., make an fbh1Δ rad22Δ double mutant as sensitive to genotoxins as a rad22Δ single mutant) (Fig. 5C). Wild-type Fbh1 fully complements this mutant to the sensitivity of a rad22Δ single mutant, whereas both Fbh1L14A/P15A and Fbh1D485N only partially complement. To confirm that both protein domains are independently contributing to Fbh1 action, an fbh1L14A/P15A/D485N mutant was constructed and shown not to complement the fbh1Δ rad22Δ double mutant at all (Fig. 5C). Overall, these data indicate that Fbh1's DNA helicase activity is most critical for controlling Rhp51 in the absence of Rad22. However, the F-box also appears to have a role here, which suggests that Fbh1-directed ubiquitination may play some part in the control of Rhp51.
FIG. 5.
The importance of F-box and helicase activities to Fbh1's ability to inhibit DNA repair in the absence of Rad22. (A) Spot assay comparing the abilities of fbh1D485N and fbh1Δ to suppress rad22Δ genotoxin hypersensitivity. The strains are wild type (MCW1221), fbh1D485N (MCW1718), rad22Δ (MCW1285), fbh1D485N rad22Δ (MCW1767), and fbh1Δ rad22Δ (MCW1553). The plates were photographed after 5 days of incubation. (B) UV survival curves of fbh1D485N rad22Δ (MCW1767) and fbh1Δ rad22Δ (MCW1553) strains. The error bars represent the standard deviations about the mean values. (C) Abilities of wild-type and mutant Fbh1 proteins to complement the genotoxin resistance of an fbh1Δ rad22Δ strain (MCW1553). Vector refers to empty pREP41. The bottom row of each panel shows results for the rad22Δ strain (MCW1285) carrying pREP41. The medium was yeast nitrogen base, and plates were photographed after 4 days of incubation.
Genetic interactions between fbh1 and other mediator proteins.
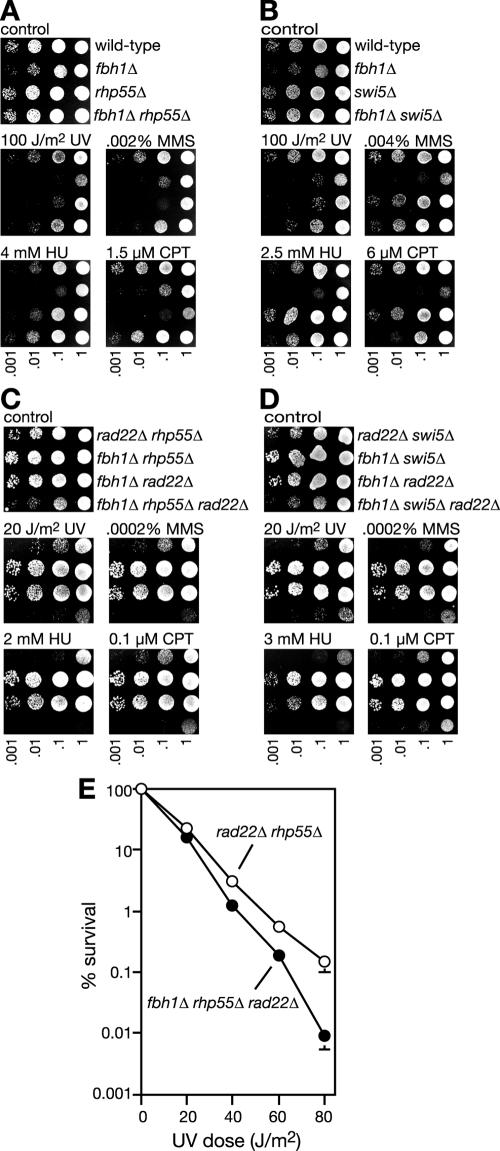
Like Rad22 the Rhp55-Rhp57 heterodimer (Rad55-Rad57 in budding yeast) is believed to act as a mediator aiding the assembly of Rhp51 onto DNA in the presence of RPA (14, 39, 40). In S. pombe a second protein complex, consisting of Swi5 and Srf1, appears to function in a separate pathway from Rhp55-Rhp57 to promote Rhp51 action (2). To see if the deletion of fbh1 would suppress the need for either Rhp55-Rhp57 or Swi5-Srf1, fbh1Δ rhp55Δ and fbh1Δ swi5Δ double mutant strains were constructed and compared to their respective single mutants for genotoxin sensitivity. In the case of the fbh1Δ rhp55Δ mutant, reciprocal suppression of genotoxin sensitivity was observed such that, depending on the genotoxin used, the double mutant is as resistant or almost as resistant as the wild-type strain (Fig. 6A). A similar result was seen for the fbh1Δ swi5Δ mutant with UV (Fig. 6B). Furthermore, swi5Δ, which exhibits relatively little hypersensitivity to HU, MMS, or CPT, suppresses fbh1Δ sensitivity to these agents. These data show that the need for Rhp55/Swi5 and Fbh1 for DNA repair is largely negated by their pairwise removal. Importantly, the poor growth of an fbh1Δ strain is also suppressed by deleting Rhp55 or Swi5 (not shown). Taken together, these data indicate that the mediator proteins are important for promoting Rhp51 action in the presence of Fbh1. Furthermore, where Rhp51 action is attenuated by the removal of a mediator protein, the requirement for Fbh1 to promote growth and DNA repair is reduced.
FIG. 6.
Genetic interactions between fbh1 and rhp55 or swi5. (A and B) Spot assays comparing the genotoxin sensitivities of wild-type (MCW1221), fbh1Δ (MCW1490), rhp55Δ (MCW1231), fbh1Δ rhp55Δ (MCW1568), swi5Δ (YA177), and fbh1Δ swi5Δ (MCW1590) strains. The plates were photographed after 3 days of incubation. (C and D) Spot assays assessing the dependence on rhp55 and swi5 for the suppression of rad22Δ genotoxin sensitivity by fbh1Δ. The strains are rad22Δ rhp55Δ (MCW1679), fbh1Δ rhp55Δ (MCW1568), fbh1Δ rad22Δ (MCW1553), fbh1Δ rhp55Δ rad22Δ (MCW1681), rad22Δ swi5Δ (MCW1643), fbh1Δ swi5Δ (MCW1590), and fbh1Δ swi5Δ rad22Δ (MCW1645). The plates were photographed after 5 days of incubation.(E) UV survival curves of rad22Δ rhp55Δ (MCW1679) and fbh1Δ rhp55Δ rad22Δ (MCW1681) strains. The error bars represent the standard deviations about the mean values.
Both Rhp55 and Swi5 are required for the suppression of rad22Δ by fbh1Δ.
To see if deleting fbh1 would suppress the need for mediator proteins altogether, fbh1Δ rhp55Δ rad22Δ and fbh1Δ swi5Δ rad22Δ triple mutants were constructed and compared to their respective double mutants for genotoxin sensitivity (Fig. 6C and D). In both cases the triple mutant was at least as sensitive as the most sensitive double mutant (i.e., rad22Δ rhp55Δ and rad22Δ swi5Δ, respectively), which in turn was as sensitive as a rad22Δ single mutant. These data show that the deletion of fbh1 cannot suppress the need for more than one mediator protein at a time. In fact, an fbh1Δ rhp55Δ rad22Δ triple mutant is slightly more sensitive to genotoxins than a rad22Δ rhp55Δ double mutant (Fig. 6C and E). This suggests that Fbh1 may be required to process any residual loading of Rhp51 onto DNA that occurs in the absence of both Rad22 and Rhp55.
Fbh1 subnuclear localization in the absence of recombination proteins.
To see if the formation of Fbh1 nuclear foci depends on recombination proteins, we expressed EYFP-Fbh1 in rhp55Δ, rhp51Δ, and rad22Δ mutants, in the presence and absence of CPT, and determined the percentage of cells with foci. Rather than being required for focus formation, each of the recombination mutants exhibited a greater percentage of cells with spontaneous foci and CPT-induced foci, including cells with multiple foci (Fig. 7). This suggests that Fbh1 may target sites of DNA damage directly rather than the recombination proteins that load at these sites. In the absence of recombination proteins, sites of spontaneous or induced damage would persist, resulting in the increased numbers of Fbh1 foci. Furthermore, the removal of one recombination protein may result in the inefficient or aberrant loading of the other recombination proteins at sites of DNA damage, and this may also result in increased Fbh1 focus formation.
DISCUSSION
We have shown that Fbh1 is a new member of a group of proteins that control recombinases. A central finding here is that Fbh1 prevents Rhp51-dependent recombination in the absence of Rad22. This shows that Rhp51 can form functional nucleofilaments in vivo in the presence of RPA without what was thought to be the essential mediator function of Rad22. It also provides a significant clue as to what Fbh1's precise role in vivo might be (see below).
Genetic analyses show that any double combination of fbh1, srs2, and rqh1 mutations results in a dramatic reduction or total loss of viability, which can be remedied by removing Rhp51 (9, 19). This suggests that these DNA helicases share an overlapping function for processing Rhp51-dependent recombination intermediates. In accord with their function in controlling recombination, both srs2 and rqh1 mutants exhibit elevated levels of direct repeat recombination (7, 9). In contrast, direct repeat recombination at ade6 is normal in an fbh1 mutant. This may mean that Fbh1 controls recombination only at specific loci or that the recombination intermediates that are not processed by Fbh1 result in cell death.
Based on our analysis of DAPI-stained cells, an fbh1Δ mutant exhibits a defect in chromosome segregation, which presumably underlies its reduced growth and viability. Rqh1 mutants also have impaired chromosome segregation, especially following exposure to agents that cause replication fork arrest (7, 32). Here chromosome segregation is improved by the expression of the bacterial Holliday junction resolvase RusA (7). This suggests that an accumulation of unprocessed Holliday junctions between sister chromatids perturbs chromosome segregation. Unlike the case for rqh1 mutants, neither chromosome segregation nor viability of an fbh1Δ mutant is improved by RusA (not shown). This suggests that Fbh1 is not required for processing Holliday junctions. Instead, we suspect that Fbh1 controls recombination by dissociating recombinases from DNA while translocating along it. This would be analogous to the action of its close relative Srs2 (17, 43). Evidence that active translocation would be required comes from our analysis of the fbh1D485N mutant, which contains a mutation of a conserved residue within helicase motif II that is known to be required for ATP hydrolysis in other DNA helicases. This mutant exhibits essentially a null phenotype with respect to genotoxin sensitivity, and the mutation suppresses a rad22Δ mutant almost as well as a complete deletion of fbh1. Exactly which recombinases might be dissociated is not known, and Fbh1's attenuation of Rhp51 activity could be by direct removal of the Rhp51 nucleofilament and/or the removal of the mediator proteins.
If Fbh1 does indeed function in a manner analogous to that of Srs2, why should the cell require two similar DNA helicases? As mentioned above, genetic analyses indicate that there is an overlap between the Fbh1 and Srs2 pathways, but it also shows that they have distinct functions. In S. cerevisiae Srs2 is important for channeling DNA repair away from recombination to the postreplication repair pathway (1, 5, 27, 31), and it may fulfill a similar function in S. pombe (9). Fbh1 may also be required for channeling lesions away from recombination, but whether this is to direct them into a particular repair pathway is not clear at present.
A distinguishing feature of Fbh1 is its F-box; however, our analyses of the fbh1L14A/P15A mutant suggest that this may play a relatively minor role in Fbh1 function. Alternatively, the L14A/P15A mutation may not fully inactivate the F-box, in which case it remains a possibility that this domain is critical for Fbh1's function. It is therefore tempting to speculate that Fbh1 may promote ubiquitin-mediated degradation to curb recombination. We are currently investigating this possibility.
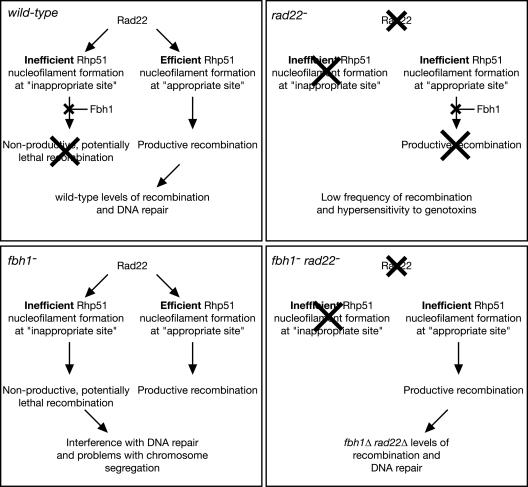
Another distinction between Fbh1 and either Srs2 or Rqh1 is that deletion of Fbh1 reduces significantly the need for certain mediator proteins. In fact, double mutant strains combining fbh1Δ with rad22Δ, rhp55Δ, or swi5Δ each show at least some improvement in growth/viability and/or genotoxin resistance compared to their respective single mutants. This reciprocal suppression, which is not restricted to one particular mediator protein, leads us to suggest the following model (summarized in Fig. 8). Combinations of Rad22, Rhp55-Rhp57, and Swi5-Sfr1 might make Rhp51 nucleofilament assembly fast and efficient but may also increase the chances of “inappropriate” filament assembly. Exactly what might constitute an “inappropriate” filament is unknown; however, possibilities include filament assembly at inappropriate sites on the DNA and/or at inappropriate times during the cell cycle. In our model we make the assumption that inappropriate filament assembly is generally less efficient than the appropriate assemblies, possibly due to a paucity of recombination proteins, accessibility of the DNA substrate, or competition with other proteins. We suspect that Rhp51 nucleofilaments that form inefficiently are targeted by Fbh1 for removal. These filaments may be more susceptible to dissociation by Fbh1 because they are less extensive/robust and/or engage in strand invasion with slower kinetics. In either a rad22 or rhp55 mutant filament, assembly is made less efficient, thereby making appropriate Rhp51 assemblages susceptible to Fbh1 action. At the same time, the inappropriate events would also be made less efficient such that they may not occur at all, removing the need for Fbh1. Thus, in either a rad22 or rhp55 mutant the removal of Fbh1 would redress the balance, enabling the appropriate events to occur without the danger of the inappropriate ones. However, if both Rad22 and Rhp55 are missing, then nucleofilament assembly might be so disabled that Fbh1 is needed to remove any Rhp51 that manages to load onto DNA. Residual amounts of loading would be insufficient for recombination and, by interfering with other processes, could be toxic. This could explain our observation that an fbh1Δ rad22Δ rhp55Δ triple mutant is slower growing and more sensitive to genotoxins than a rad22Δ rhp55Δ double mutant.
FIG. 8.
Hypothetical model to explain the genetic interactions between fbh1 and rad22. Crosses indicate pathways that are blocked. See the text for further details.
In conclusion, we suggest that in fission yeast Rhp51 is controlled by a balance between the action of its mediator proteins and that of Fbh1. Fbh1 appears to curb inappropriate recombinase action that might otherwise block alternative DNA repair enzymes, cause cell cycle arrest, and interfere with chromosome segregation. We suspect that FBH1 in humans plays a similar controlling role.
Acknowledgments
We thank Jong Sook Ahn for help with the growth experiments, Hiroshi Iwasaki for the swi5Δ mutant, and Hideo Shinagawa for communicating results prior to publication.
This work was supported by a project grant (065278/Z/01/Z) and Senior Fellowship in Basic Biomedical Research from The Wellcome Trust awarded to M.C.W.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa, and H. Iwasaki. 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100:15770-15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 4.Barbet, N., W. J. Muriel, and A. M. Carr. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59-66. [DOI] [PubMed] [Google Scholar]
- 5.Broomfield, S., and W. Xiao. 2002. Suppression of genetic defects within the RAD6 pathway by srs2 is specific for error-free post-replication repair but not for damage-induced mutagenesis. Nucleic Acids Res. 30:732-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshaies, R. J. 1999. SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 7.Doe, C. L., J. Dixon, F. Osman, and M. C. Whitby. 2000. Partial suppression of the fission yeast rqh1(−) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19:2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doe, C. L., F. Osman, J. Dixon, and M. C. Whitby. 2004. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 32:5570-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doe, C. L., and M. C. Whitby. 2004. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 32:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortin, G. S., and L. S. Symington. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 21:3160-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasior, S. L., H. Olivares, U. Ear, D. M. Hari, R. Weichselbaum, and D. K. Bishop. 2001. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98:8411-8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 13.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khasanov, F. K., G. V. Savchenko, E. V. Bashkirova, V. G. Korolev, W. D. Heyer, and V. I. Bashkirov. 1999. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics 152:1557-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J., J. H. Kim, S. H. Lee, D. H. Kim, H. Y. Kang, S. H. Bae, Z. Q. Pan, and Y. S. Seo. 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277:24530-24537. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. H., J. Kim, D. H. Kim, G. H. Ryu, S. H. Bae, and Y. S. Seo. 2004. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 32:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 18.Laursen, L. V., E. Ampatzidou, A. H. Andersen, and J. M. Murray. 2003. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 23:3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maftahi, M., J. C. Hope, L. Delgado-Cruzata, C. S. Han, and G. A. Freyer. 2002. The severe slow growth of Deltasrs2 Deltarqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 30:4781-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meister, P., M. Poidevin, S. Francesconi, I. Tratner, P. Zarzov, and G. Baldacci. 2003. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 31:5064-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 22.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.New, J. H., T. Sugiyama, E. Zaitseva, and S. C. Kowalczykowski. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407-410. [DOI] [PubMed] [Google Scholar]
- 24.Park, J. S., E. Choi, S. H. Lee, C. Lee, and Y. S. Seo. 1997. A DNA helicase from Schizosaccharomyces pombe stimulated by single-stranded DNA-binding protein at low ATP concentration. J. Biol. Chem. 272:18910-18919. [DOI] [PubMed] [Google Scholar]
- 25.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pommier, Y., C. Redon, V. A. Rao, J. A. Seiler, O. Sordet, H. Takemura, S. Antony, L. Meng, Z. Liao, G. Kohlhagen, H. Zhang, and K. W. Kohn. 2003. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532:173-203. [DOI] [PubMed] [Google Scholar]
- 27.Rong, L., F. Palladino, A. Aguilera, and H. L. Klein. 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, I. D., A. S. Grancell, and P. K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saintigny, Y., F. Delacote, G. Vares, F. Petitot, S. Lambert, D. Averbeck, and B. S. Lopez. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai, Y., A. P. Murdanoto, T. Konishi, A. Iwamatsu, and N. Kato. 1997. Regulation of the formate dehydrogenase gene, FDH1, in the methylotrophic yeast Candida boidinii and growth characteristics of an FDH1-disrupted strain on methanol, methylamine, and choline. J. Bacteriol. 179:4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama, T., and S. C. Kowalczykowski. 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277:31663-31672. [DOI] [PubMed] [Google Scholar]
- 34.Sung, P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194-28197. [DOI] [PubMed] [Google Scholar]
- 35.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 36.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 37.Suto, K., A. Nagata, H. Murakami, and H. Okayama. 1999. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol. Biol. Cell 10:3331-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsutsui, Y., F. K. Khasanov, H. Shinagawa, H. Iwasaki, and V. I. Bashkirov. 2001. Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics 159:91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Bosch, M., K. Vreeken, J. B. Zonneveld, J. A. Brandsma, M. Lombaerts, J. M. Murray, P. H. Lohman, and A. Pastink. 2001. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461:311-323. [DOI] [PubMed] [Google Scholar]
- 42.van den Bosch, M., J. B. Zonneveld, K. Vreeken, F. A. de Vries, P. H. Lohman, and A. Pastink. 2002. Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res. 30:1316-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 44.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]