Abstract
Although it has been well established that histone acetyltransferases (HATs) are involved in the modulation of chromatin structure and gene transcription, there is only little information on their developmental role in higher organisms. Gcn5 was the first transcription factor with HAT activity identified in eukaryotes. Here we report the isolation and characterization of Drosophila melanogaster dGcn5 mutants. Null dGcn5 alleles block the onset of both oogenesis and metamorphosis, while hypomorphic dGcn5 alleles impair the formation of adult appendages and cuticle. Strikingly, the dramatic loss of acetylation of the K9 and K14 lysine residues of histone H3 in dGcn5 mutants has no noticeable effect on larval tissues. In contrast, strong cell proliferation defects in imaginal tissues are observed. In vivo complementation experiments revealed that dGcn5 integrates specific functions in addition to chromosome binding and acetylation. Surprisingly, a dGcn5 variant protein with a deletion of the bromodomain, which has been shown to recognize acetylated histones, appears to be fully functional. Our results establish dGcn5 as a major histone H3 acetylase in Drosophila which plays a key role in the control of specific morphogenetic cascades during developmental transitions.
Gene expression in eukaryotes has to accommodate the presence of nucleosomes and the packaging of DNA into higher-order chromatin structures. Nucleosomes are composed of octamers of histone proteins H2a, H2b, H3, and H4, whose N-terminal tails project outward from the nucleosomal core and are subjected to covalent modifications such as acetylation, methylation, phosphorylation, and ubiquitination. The variety of these modifications and their association with distinct states of gene transcription suggested that they may act as a combinatorial code to specify downstream events such as the recruitment of transcription factors or modifications of the chromatin structure (23, 58).
The acetylation of lysine residues is one of the most studied histone modifications and has long been linked to gene activation. For instance, a twofold up-regulation of transcription from the male X chromosome in Drosophila melanogaster is correlated with histone hyperacetylation (60), while gene silencing in heterochromatin or X chromosome inactivation in mammals are correlated with histone hypoacetylation (27). Numerous sequence-specific activators, such as the nuclear receptors MyoD and CREB, have been shown to recruit coactivator complexes with histone acetyltransferase (HAT) activity, while transcriptional repressors have been found associated with corepressor complexes with histone deacetylase activity (13). HAT activity is also associated with more general transcription factors, such as TATA-binding protein-associated factor 1 (TAF1) and yeast elongation factor 3 (44, 63). Collectively, these data point to a causal role of histone acetylation in transcriptional activation. In support of this hypothesis, the acetylation of lysine 8 in histone H4 (H4-AcK8) and lysine 14 in histone H3 (H3-AcK14) has been implicated in the sequential recruitment of transcription factors leading to the activation of the human beta interferon gene in vitro (1), and distinct patterns of histone acetylation have been associated with groups of coexpressed genes in genome-wide studies (36, 52).
The yeast adaptor Gcn5 was the first transcription factor identified as a bona fide HAT (14, 35). It defines a family of evolutionarily conserved Gcn5-related N-acetyltransferases (GNATs) whose members were purified as essential subunits of ADA and SAGA complexes in yeast and of PCAF, STAGA, and TFTC complexes in mammals. The GNAT complexes contain Ada transcriptional adapters, Spt proteins, and a set of TATA-binding protein-associated factors, and they are structurally related, suggesting that they perform similar functions in transcription (13, 65). In vitro, Gcn5 acetylates lysine 14 of free, but not nucleosomal, histone H3. In contrast, Gcn5 acetylates an expanded set of lysines of nucleosomal histone H3 when copurified with native ADA or SAGA complexes (26), suggesting that one function of these complexes in vivo is to modulate the activity and specificity of Gcn5.
Two distinct genes, hGCN5 and hPCAF, encode Gcn5 homologues in humans. Both hGCN5 isoforms, GCN5S and GCN5L, and hPCAF (P300/CBP-associated factor) share a close similarity with the complete yeast Gcn5 sequence, including more matches within the HAT catalytic domain as well as the bromodomain, which has been shown to bind acetylated histone lysines (21, 30-32, 46, 47). PCAF and GCN5L share an additional N-terminal domain (Pcaf homology domain) of about 350 amino acids involved in binding to CBP/p300 and to several nuclear receptors. While the GCN5 gene knockout results in early embryonic lethality, the PCAF gene knockout has no detectable consequences on mouse development (66). However, GCN5-PCAF double mutants die earlier than single GCN5 mutants, indicating that PCAF and GCN5 functions are not completely redundant.
Although the Gcn5 HAT has been clearly involved in the control of Arabidopsis growth, development, and homeostasis (8, 61), its contribution to the control of specific morphogenetic events during animal development remains poorly understood. There is only one Gcn5 homologue in Drosophila (56), which thus provides a simplified model for the study of the function of a GNAT in the context of a whole organism. Drosophila Gcn5 (dGcn5) has been isolated in at least two GNAT complexes that contain distinct Ada2 variants (37, 45). A 1.8-MDa SAGA-like complex includes the Ada2b variant, the Ada3 and Spt3 homologues, and several TAFs. An Ada2b loss-of-function mutation is lethal and suppresses the histone H3 acetylation of polytene chromosomes (49), indicating that the SAGA-like complex plays an essential role in gene expression in Drosophila. In addition, dGcn5 associates with the Ada2a variant and with Ada3 in a 440-kDa non-SAGA-like complex. Interestingly, Ada2a and Ada2b mainly localize to different bands on polytene chromosomes, suggesting that GNAT complexes may play distinct functions depending on their composition.
In order to characterize the function of dGcn5 and GNAT complexes during Drosophila development, we have undertaken two complementary approaches. We isolated dGcn5 loss-of-function mutants from a genetic screen, and we performed in vivo targeting of dGcn5-specific RNA interference using inverted repeat transgenes. Here we show that dGcn5 is the major HAT for the lysine residues K9 and K14 of histone H3, while the acetylation of histone H4 involves other HATs. Our data indicate that dGcn5 is required for larva-to-adult metamorphosis and suggest an essential function of dGcn5 in the control of the cell cycle in imaginal tissues. An analysis of dGcn5 variant proteins revealed that the Pcaf homology domain, the domain of interaction with the Ada proteins, and the catalytic domain are all required for the function of the dGcn5 protein. In contrast, the bromodomain appears to be dispensable.
MATERIALS AND METHODS
DNA constructs.
An EcoRV-KpnI fragment encompassing the dGcn5 and CG14121 genes was subcloned from the bacterial artificial chromosome BACR48G06 into the pBluescript vector. Clones containing this fragment were selected by colony hybridization with dGcn5 cDNA excised from the LD17356 expressed-sequence-tag clone (BDGP). The PL genomic rescue construct was then generated by subcloning the CG14121-dGcn5 region as a 4.7-kb KpnI-NotI fragment into the pCaSper-4 transformation vector.
pUAS-Gcn5 and its derivatives were all made by subcloning the dGcn5 cDNA from the LD17356 expressed-sequence-tag clone into the pUAST vector. Deletions were generated by the excision of dGcn5 regions and cloning of the appropriate PCR-amplified fragments. Gcn5 variant constructs contained the following deletions: pUAS-Gcn5ΔPcaf, deletion of the first 361 amino acids of the dGcn5 peptide; pUAS-Gcn5ΔHAT, deletion of amino acids M554 to K595; pUAS-Gcn5ΔAda, deletion of amino acids F635 to P699; and pUAS-Gcn5ΔBromo, deletion of amino acids S703 to T788. The structure of all Gcn5 variant constructs was confirmed by DNA sequencing. The cloning procedures and PCR primers used are available upon request.
Generation of dGcn5 mutants.
Sixteen Df(3L)iro-2 noncomplementing P elements on chromosome III (a generous gift from Peter Maroy) were mapped by inverse PCR. A P-element insertion, 1479/10, that was localized in the CG14121 gene, 521 bp upstream of the dGCN5 transcription start site, is homozygous lethal at puparium formation. This P element was mobilized by crossing with a strain expressing transposase. Derived lines with both a lost white marker gene and early embryonic lethality associated with chromosome III were analyzed using combinations of PCR primers. The flanking regions of the sex204 deficiency were PCR amplified and sequenced.
Males isogenic for an FRT79D ebony marked chromosome were treated with 35 mM ethyl methanesulfonate (EMS) and mated en masse to TM3SerGFP/TM6b virgin females at 25°C. F1 males (FRT79D ebony/TM6b or FRT79D ebony/TM3SerGFP) were then pair mated with females with the deficiency sex204, and crosses were scored for the absence of the F2 progeny class that was heterozygous for the mutagenized chromosome and the deficiency. If this class was absent, stocks were established from FRT79D ebony/TM3SerGFP or TM6b flies. We recovered 11 lethal mutations that failed to complement the sex204 deficiency from 6,000 fertile pair matings scored. Each of these mutations was placed in complementation groups by complementation tests with all other mutations and with the P-element insertions 1479/10 (a CG14121 allele) and KG01697 (a Citron allele from BDGP). Four dGcn5 alleles were rescued in crosses with a PL/PL; sex204/TM3TbSb line homozygous for the PL genomic transgene (Fig. 1) and in crosses with a UAS-Gcn5/UAS-Gcn5; sex204/da-GAL4/TM3TbSb line which constitutively expresses the dGcn5 cDNA. Six mutations were genetically mapped in Citron. The remaining allele was mapped in CG14121 by noncomplementation with the P-element 1479/10 and was rescued with a short genomic transgene encompassing only the CG14121 locus. We did not isolate a mutation in CG10686 in our genetic screen because the loss of function of this gene is likely not lethal. Six pairs of primers were used to PCR amplify the dGcn5 coding region from flies heterozygous for dGcn5 mutations and a balancer chromosome. We identified the dGcn5 alleles by comparison of the sequence from mutant heterozygotes to that obtained from the homozygous parental strain upon which the mutations were induced. The primer sequences used for this study are available upon request.
FIG. 1.
dGcn5 is required for Drosophila metamorphosis. (A) Genetic map of the dGcn5 region. The Drosophila dGcn5 gene with its two introns (white boxes) is depicted in black. The deletion sex204 was generated by imprecise excision of the 1479/10 P element (black triangle) inserted in the CG14121 gene. dGcn5 alleles are indicated, as well as the position of the genomic fragment included in the PL transgenic construct. (B) Impaired metamorphosis in Gcn5E333st mutants. (Left side) Homozygous Gcn5E333st animals failed to formal normal puparium compared to Gcn5E333st/TM3 control animals. Salivary glands from control or Gcn5E333st late-third-instar larvae (top) were immunostained with a dGcn5 antibody. (Right side) Squashed salivary glands from wild-type (control) and homozygous Gcn5E333st (E333st) late-third-instar larvae. Brackets indicate chromosomal regions corresponding to 2B (top) and 74EF-75B (bottom) early puffs, respectively. (C) Gcn5E333st/Gcn5C137T animals mostly died as pharate adults with abnormally elongated metathoracic legs (black arrowhead), strong defects in abdominal cuticle deposition (open arrowhead), and rough eyes. Note that the eye pigmentation was stronger in Gcn5E333st/Gcn5C137T animals than in Gcn5C137T/TM3 control animals because of the presence of a FRT79D white+ marker on the mutagenized chromosomes. A metathoracic twisted and crooked leg (black arrow) from a Gcn5E333st/Gcn5C137T adult escaper is shown to the right. The same defects and lethality were observed for the Gcn5E333st/Gcn5ΔT280-F285 heteroallelic combination (not shown). (D) Structures of the wild-type and variant dGcn5 proteins expressed from pUAST-derived transgenic constructs. The N-terminal domain conserved in vertebrate Pcaf, the catalytic HAT domain, the Ada domain, and the bromodomain are indicated as shaded boxes.
Germ line transformations and fly strains.
Transgenic lines were established as described previously (11). The GAL4-UAS system (10) was used to express dGcn5, dGcn5 variants, and the green fluorescent protein (GFP) reporter. The da-GAL4 (GAL4daG32) driver line has been described previously (64). The lio-GAL4P15 line (55) was provided by Jean-Maurice Dura, IGH, Montpellier, France. The engrailed-GAL4 (en-GAL4) and UAS-GFP lines were provided by Jean-Paul Vincent, MRC, London, United Kingdom. The vestigial-GAL4 (vg-GAL4) driver line was provided by Fréderic Bernard, IJM, Paris, France. The distal-less-GAL4 (dll-Gal4) and patch-GAL4 (ptc-GAL4) driver lines were obtained from the Bloomington Stock Center. The escargot-GAL4 (esg-GAL4, or NP5153) line was obtained from Drosophila Genetic Resources, Japan (http://www.shigen.nig.ac.jp/fly/nigfly/).
Immunohistochemistry and Western blotting.
Immunostaining of imaginal disks dissected from mid-third-instar larvae was performed as described previously (20). Rabbit anti-dGcn5 (50) and anti-CBP (a gift from A. Mazo, Kimmel Cancer Center, Philadelphia, Pa.) were used at a 1:1,000 dilution. Antibodies against the histone tail modifications were obtained from Upstate and were used at the follow dilutions: anti-Ac-H3K9, 1:1,000; anti-Ac-H3K14, 1:200; anti-Ph-H3S10, 1:1,000; anti-Ac-H4K8, 1:1,000; and anti-Ac-H3K9/K14, 1:1,000. Secondary antibodies were used at a 1/200 dilution (Jackson Laboratories). Preparations were mounted in Citifluor AF1, and imaging was carried out using a Leica TCS-SP confocal microscope.
Western blots were performed as described previously (11), using primary antibodies at dilutions of 1:5,000 (anti-dGcn5), 1:1,000 (anti-Ac-H3K9), 1:1,000 (anti-Ac-H3K14), 1:1,000 (anti-Ac-H4K8), and 1:50,000 (anti-MBF-1; a gift of Marek Jindra, Institute of Entomology, CAS, Ceske Budejovice, Czech Republic). A horseradish peroxidase-conjugated anti-rabbit secondary antibody (Vector) was used at a 1:5,000 dilution and detected using the SuperSignal West Pico chemiluminescent substrate (Pierce).
Polytene chromosome staining.
Glands from third-instar larvae raised at 18°C were dissected in phosphate-buffered saline (PBS), fixed for 10 min in 50% acetic acid-3.7% formaldehyde, and squashed on glass slides. Immunostaining of polytene chromosomes was performed essentially as described previously (67). The primary antibodies used were mouse anti-GFP (1:200) from Roche to reveal the H2b-YFP marker protein (4) and anti-Ac-H3K9 (1:100), anti-Ac-H3K14 (1:100), anti-Ac-H3K9/K14 (1:100), anti-Ph-H3S10 (1:100), and anti-Ac-H4K8 (1:100) from Upstate. The anti-HP1 antibody (a gift from S. Elgin, Washington University, St. Louis, Mo.) was used at a 1:500 dilution. The secondary antibodies were Cy3-conjugated anti-rabbit (1:200) and fluorescein isothiocyanate-conjugated anti-mouse (1:200) from Jackson Laboratories. Slides were mounted in Vectashield (Vector Laboratories) with 1.5 μg/ml DAPI (4,6-diamidino-2-phenylindole), and imaging was carried out using a Zeiss Axiovert 200 M microscope with a Roper Scientific Coolsnap HQ camera.
BrdU incorporation and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) experiments.
For the BrdU (5-bromo-2′-bromodeoxyuridine) incorporation procedure, imaginal disks from third-instar larvae were dissected in Schneider medium and incubated for 15 min in 75 μg/ml BrdU at room temperature. Disks were washed twice with PBS and fixed for 30 min at 4°C in PBS with 4% formaldehyde. Disks were then washed three times for 10 min each time with 1× PBS-0.3% Triton X-100 (PBT). DNAs were denatured in 3 N HCl for 30 min and washed three times for 10 min each time with PBT. The disks were blocked (three times for 10 min each time) in 1× PBS-0.3% Triton X-100-1% bovine serum albumin (PBTA), incubated overnight at 4°C with an anti-BrdU monoclonal antibody (Upstate) according to the manufacturer's instructions, and washed with PBTA. The disks were then incubated for 1 h at room temperature with a Cy3-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch) in PBTA, washed with PBT (three times for 10 min each time), and mounted in Vectashield (Vector Laboratories).
TUNEL assays were performed using a TUNEL apoptosis detection kit (Upstate). Wing disks from wandering third-instar larvae were dissected and fixed in PBS with 4% formaldehyde for 20 min at 4°C. The disks were washed with PBS for 10 min, blocked in 1× PBS-0.05% Tween 20-0.2% bovine serum albumin for 10 min, and treated for the TUNEL reaction according to the manufacturer's instructions. The disks were washed three times with PBS and mounted in Vectashield (Vector Laboratories).
RESULTS
Isolation of dGcn5 mutant alleles.
We determined the insertion sequences of several P elements which cytologically mapped close to the 69C4 position of dGcn5 on polytene chromosomes and were lethal over the Df(3L)iro-2 deficiency (P. Maroy, personal communication). The 1479/10 P element was found inserted 521 bp upstream of the dGcn5 transcription start site, in the coding sequence of the CG14121 gene (Fig. 1A). We used this insertion to generate deletions via transposase-mediated P-element excision and recovered the lethal deficiency sex204 that removes a part of the CG14121 coding sequence, the dGcn5 and CG10686 genes, and the transcription start site of the citron gene (Fig. 1A). To obtain dGcn5-specific alleles, we screened for EMS-induced dGcn5 mutants by noncomplementation of the sex204 deficiency. We recovered 11 mutants that failed to complement the sex204 deficiency for viability. Five mutants were rescued by a PL transgenic construct encompassing the dGcn5 and CG14121 genes (Fig. 1A). Four of these alleles were fully rescued by a UAS-Gcn5 cDNA transgene (Fig. 1D) expressed under the control of the ubiquitous da-GAL4 driver, indicating that these alleles specifically impair dGcn5 function. The remaining allele is a mutation of the CG14121 gene (see Materials and Methods). We sequenced the dGcn5 mutations and found that they all lie within the first dGcn5 exon. Two of them are localized in the Pcaf homology domain and result in a cysteine-to-tyrosine substitution (C137T) or a small deletion from tyrosine 280 to phenylalanine 285 (ΔT280-F285). The two other mutations generate stop codons (Q186st and E333st). Gcn5E333st mutants were outcrossed and then rescued as homozygous by the PL transgene, indicating that they do not bear other irrelevant EMS-induced lethal mutations.
dGcn5 is required for metamorphosis and oogenesis.
To determine the lethal phase associated with the dGcn5 loss of function, we maintained the Gcn5E333st allele over a GFP-expressing balancer chromosome. About 26% of eggs laid by females from this stock did not reach the first larval instar, a number attributable to embryonic lethality from the homozygous balancer chromosome. Nonfluorescent homozygous Gcn5E333st larvae developed similarly to their GFP-expressing fluorescent heterozygous siblings until the end of the third larval instar, despite the absence of detectable dGcn5 protein in mutant tissues at this stage (Fig. 1B and data not shown). However, Gcn5E333st mutant larvae did not form their puparium at the normal time and continued to wander for 4 to 5 additional days. They eventually stopped moving but failed to evert spiracles, formed abnormally long prepupae, and died with partial separation of the posterior part of the prepupa from the pupal case (Fig. 1B). Similar developmental arrest and defects were observed with the heteroallelic null combination Gcn5Q186st/Gcn5E333st (not shown).
Puparium formation is triggered by a pulse of 20-hydroxyecdysone at the end of the third larval instar. This involves the coordinated induction of a small number of early puff genes, whose products in turn regulate the expression of a larger set of late puff genes (3). Homozygous Gcn5E333st mutants were taken at various times during their extended wandering stage, and polytene chromosomes from their salivary glands were squashed (Fig. 1B, right panels). We observed a strong reduction of the size of the early puffs 2B, 74EF, and 75B in these animals, indicating a failure of the ecdysone-controlled genetic program in dGcn5 mutants.
Heteroallelic Gcn5C137T/Gcn5E333st and Gcn5E333st/Gcn5ΔT280-F285 mutant larvae formed their puparium normally, indicating that the Gcn5C137T and Gcn5ΔT280-F285 variant proteins retain partial function. However, these mutants failed to elongate their metathoracic legs correctly, had rough eyes, and displayed a partial to complete absence of abdominal cuticle deposition (Fig. 1C). Both heteroallelic combinations gave rise to rare adult escapers with misshapen wings, rough eyes, and crooked and twisted metathoracic legs (Fig. 1C and data not shown). These escapers died a few hours after eclosion. Altogether, these data point to an essential function of dGcn5 at the onset of and during metamorphosis.
Since an important stock of dGcn5 protein is detected in oocytes and presyncytial embryos (data not shown), this maternal contribution of dGcn5 may be sufficient to allow embryonic and larval development. To generate embryos lacking a maternal contribution, we took advantage of the absence of expression of the pUAST-derived construct UAS-Gcn5 in the female germ line (10). We generated UAS-Gcn5/+; Gcn5E333st/sex204 da-GAL4 rescued adults and found oogenesis in females to be arrested at stages 5 and 6 (data not shown). This effect was due to the lack of UAS-Gcn5 expression in the germ line, since control females rescued by the dGcn5 genomic construct (PL/+; Gcn5E333st/sex204) were fertile. With dGcn5 being required for oogenesis, we were not able to analyze its contribution to embryonic development.
dGcn5 is required for cell proliferation.
Imaginal disks from homozygous Gcn5E333st mutant third-instar larvae are misshapen and severely reduced in size (Fig. 2A), suggesting that a dGcn5 loss of function impairs the proliferation of imaginal cells during larval instars. In a previous report, we made use of the inverted-repeat transgenic construct UAS-IR[Gcn5] to target RNA interference (RNAi) against dGcn5 (50). To further analyze the role of dGcn5 in the cell proliferation of imaginal tissues, a UAS-IR[gcn5] transgenic line was crossed with en-GAL4 UAS-GFP individuals expressing the GAL4 activator. The en-GAL4 driver induced both GFP expression and specific dGcn5 depletion in the posterior (P) compartments of imaginal disks of third-larval-instar progeny (compare Fig. 3A and A′). In a control experiment, silencing of the unrelated CBP protein was not observed (Fig. 3B). The dGcn5-depleted P compartments often had a reduced size and appeared flatter than anterior compartments, suggesting that cell proliferation is slowed down in these compartments (most visible in Fig. 3B). Although dGcn5 RNAi in the P compartments of imaginal disks induced strong lethality during late pupal development, few animals survived until the adult stage. The posterior part of their wings was reduced in size and displayed abnormal veins and cross veins. In the most dramatic cases, a bubble indicative of abnormal adhesion between the dorsal and ventral wing epithelial sheets was observed (Fig. 2B). The cell density was not significantly changed in the silenced compartment (data not shown), indicating that the size reduction is due to a reduction in the cell number. A vg-GAL4 driver triggered dGcn5 RNAi in the wing margin and induced a strong reduction of this structure in silenced adults (Fig. 2C), while the induction of dGcn5 RNAi using a dll-GAL4 driver led to a reduction in the size of the distal part of the tarsus and wings (not shown). Finally, the induction of dGcn5 RNAi by an esg-GAL4 driver, which is strongly expressed in imaginal histoblasts, resulted in lethality of pharate adults, with a partial to complete absence of the abdominal cuticle (Fig. 2D). Collectively, these data strongly suggested that dGcn5 RNAi limits cell proliferation.
FIG. 2.
dGcn5 is required for cell proliferation in imaginal tissues. (A) Reduced and misshapen imaginal wing disks from homozygous Gcn5E333st mutants compared to a wing disk from a Gcn5E333st/TM3 control third-instar larva. (B) Wings from control animals (+/+) and en-GAL4/UAS-IR[Gcn5] adult escapers with vein and cross-vein defects (black arrows) in the smaller posterior compartment. (C) Wings from vg-GAL4/UAS-IR[Gcn5] adult escapers. (D) Complete absence of abdominal adult cuticle in esg-GAL4/UAS-IR[Gcn5] pharate adults compared to a control animal (+/+).
FIG. 3.
dGcn5 loss of function induces cell cycle defects and apoptosis. The results of a confocal analysis of GFP (green) and dGcn5 or CBP (red) expression in wing disks from en-GAL4 UAS-GFP/UAS-IR[Gcn5] (A and B) or en-GAL4/UAS-GFP (A′) late-third-instar larvae are shown. BrdU incorporation experiments (C) and anti-phospho(S10)-histone H3 immunostaining (D) revealed a greater proportion of cells in S phase and at mitosis, respectively, in the dGcn5 silenced compartment from en-GAL4 UAS-IR[Gcn5] wing disks. The results of a TUNEL analysis of Gcn5E333st/Gcn5E333st (E), Gcn5E333st/TM6 Tb (F), en-GAL4/UAS-IR[Gcn5] (G), and ptc-GAL4/UAS-IR[Gcn5] (H) wing disks are also shown. UAS-IR[GFP] transgenes did not induce apoptosis in control experiments (not shown).
Surprisingly, however, a greater proportion of cells in S phase (Fig. 3C) as well as a significantly larger number of cells undergoing mitosis (Fig. 3D) was observed in the P compartments of wing disks silenced by UAS-IR[Gcn5] upon activation by en-GAL4. Notably, we also detected a high level of apoptosis in imaginal disks from third-instar dGcn5 mutant larvae (Fig. 3E) as well as in response to dGcn5 RNAi triggered by either en-GAL4 (Fig. 3G) or ptc-GAL4 (Fig. 3H). Together, these data suggest that the net effect of the dGcn5 loss of function on the compartment size results from a deregulation of the cell cycle coupled to cell death.
Histone H3 acetylation of polytene chromosomes depends on dGcn5.
To investigate the contribution of dGcn5 to histone acetylation, we immunostained polytene chromosomes from larval salivary glands using antibodies specific for the acetylation of various lysine residues of histones H3 and H4. In order to compare their acetylation levels, polytene chromosomes from wild-type and mutant larvae were squashed and stained together on the same slide. The expression of a histone 2B yellow fluorescent fusion protein (H2b-YFP) in wild-type larvae allowed us to distinguish their chromosomes from dGcn5 mutant chromosomes (Fig. 4B and data not shown). We detected the acetylation of the H3 K14 and K9 residues in numerous loci of wild-type polytene chromosomes (Fig. 4A and C). In striking contrast, the acetylation of H3 K14 was undetectable and the acetylation of H3 K9 was barely detectable in dGcn5 mutant chromosomes. Antibodies against the acetylated H4 K8 (Fig. 4D) and H4 K16 residues (data not shown) revealed unchanged acetylation of these residues in dGcn5 mutants compared to that in wild-type animals. We then examined whether the loss of H3 K9 and K14 acetylation in dGcn5 mutants affects other modifications of histone H3 residues. Like H3 K9 and H3 K14 acetylation, the phosphorylation of H3 S10 and di- and trimethylation of H3 K4 have been associated with the transcriptional activation of target loci. We found that global levels of these modifications are not affected in dGcn5 mutant polytene chromosomes (Fig. 4E and F). On the other hand, methylation of the H3 K9 residue acts as a marker for the recruitment of the heterochromatin protein HP1 and transcriptional silencing. The loss of H3 K9 acetylation in dGcn5 mutants could have favored ectopic H3 K9 methylation and the subsequent delocalization of HP1. However, we found that both H3 K9 methylation levels and HP1 localization in the pericentromeric regions of polytene chromosomes were unchanged in dGcn5 mutants (Fig. 4G and H).
FIG. 4.
dGcn5 is required for acetylation of histone H3 K9 and H3 K14 residues in polytene chromosomes. Polytene chromosomes from a Gcn5E333st/sex204 mutant and a lio-GAL4/UAS-H2b-YFP wild-type animal were squashed together and costained with DAPI (blue), anti-GFP (green), and an antibody (red) against acetylated H3 K14 (A), acetylated H3 K9 (C), acetylated H4 K8 (D), phosphorylated H3 S10 (E), dimethylated H3 K4 (F), dimethylated H3 K9 (G), or HP1 (H). Panels A and B show the same field through the red channel (acetylated H3 K14) and the green channel (H2b-YFP), respectively. The H2b-YFP signal in the green channel is not shown for immunostaining of the other histone modifications.
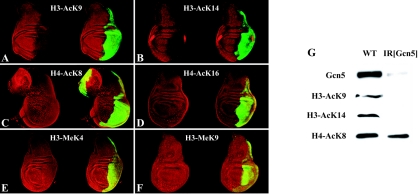
We also analyzed the contribution of dGcn5 to histone acetylation in imaginal tissues by immunostaining of imaginal disks in which dGcn5 was silenced in the P compartment by UAS-IR[gcn5] under the control of en-GAL4. The acetylation of H3 K9 and H3 K14 residues was strongly reduced in the nuclei of the dGcn5-depleted imaginal cells (Fig. 5A and B), while the acetylation of H4 K8 and H4 K16 residues was not affected (Fig. 5C and D). As seen with polytene chromosomes of dGcn5 mutants, dGcn5 depletion did not affect the level of H3 K4 or H3 K9 methylation in the nuclei of imaginal disks (Fig. 5E and F). When expression of the UAS-IR[Gcn5] transgene was driven by the ubiquitous da-GAL4 driver, animals arrested their development at the onset of metamorphosis. Western blot analysis of such late-third-instar larvae showed a strong depletion of histone H3 acetylated at the K9 and K14 residues, while the level of histone H4-AcK8 remained unchanged (Fig. 5G). Hence, the effect of the dGcn5 loss of function on polytene chromosomes, together with the results of our dGcn5 RNAi studies with imaginal disks and larval tissues, points to dGcn5 as the major histone H3 K9 and H3 K14 acetyltransferase in Drosophila.
FIG. 5.
dGcn5 RNA interference depletes imaginal disks of acetylated H3 K9 and H3 K14 residues. The results of a confocal analysis of GFP (green) and acetylated H3 K9 (A), acetylated H3 K14 (B), acetylated H4 K8 (C), acetylated H4 K16 (D) dimethylated H3 K4 (E), or dimethylated H3 K9 (F) in wing disks from en-GAL4 UAS-GFP/UAS-IR[Gcn5] late-third-instar larvae are shown. (G) Western blot of da-GAL4/UAS-IR[Gcn5] (IR[Gcn5]) and +/UAS-IR[Gcn5] (WT) late-third-instar larvae probed with antibodies against dGcn5, H3-AcK9, H3-AcK14, and H4-AcK8, as indicated.
Functional analysis of dGcn5 domains.
To analyze the functional requirement of the evolutionarily conserved domains of the dGcn5 protein, we established lines transgenic for dGcn5 variant genes under the control of the UAS promoter (Fig. 1D). The variant genes were designed in order to express dGcn5 with a deletion of the Pcaf homology domain (ΔPcaf), the catalytic domain for acetylation (ΔHAT), the conserved domain shown in yeast to be involved in interaction with the Ada2 protein (ΔAda), or the bromodomain (ΔBromo). We verified by Western blot analysis that the variant transgenes are expressed in the presence of da-GAL4 at levels comparable to that of the UAS-Gcn5 wild-type transgene (Fig. 6K) and then tested their ability to rescue the dGcn5 function. In contrast to the UAS-Gcn5 wild-type construct, the UAS-Gcn5ΔHAT construct did not rescue the lethal phenotype of the Gcn5E333st/sex204 heteroallelic combination. As expected from the disruption of the catalytic acetyltransferase domain, the acetylation of polytene chromosomes at the H3 K9 and K14 residues was not restored by the Gcn5ΔHAT variant in the mutant animals compared to the rescue of acetylation by the wild-type dGcn5 protein (Fig. 6A and C). However, it should be noted that the Gcn5ΔHAT variant still localized to the interbands of the polytene chromosomes in a manner similar to that of the wild-type dGcn5 protein expressed under the control of the da-GAL4 driver (Fig. 6B and D).
FIG. 6.
Complementation of histone acetylation and dGcn5 chromosome binding in dGcn5 mutants. Polytene chromosomes from Gcn5E333st/sex204 da-GAL4 third-instar mutant larvae heterozygous for the indicated transgenes were stained with an antibody directed against acetylated H3 K9 and H3 K14 residues (A, C, E, G, and I) or an antibody against the bromodomain of the dGcn5 protein (B, D, F, H, and J). A Western blot of da-GAL4 late-third-instar larvae heterozygous for the indicated dGcn5 variant transgenes (K) was probed with an antibody raised against the dGcn5 bromodomain. The asterisk indicates a degradation product of the Gcn5ΔHAT protein.
It was recently shown that the dAda2b component of the Drosophila SAGA-like complex directly interacts with dGcn5 and is required for the acetylation of histone H3. This result suggested that the interaction with dAda2b is essential either for the HAT activity of dGcn5 or for its targeting to chromatin (49). To our surprise, the Gcn5ΔAda protein appeared normally distributed on polytene chromosomes and restored histone H3 acetylation in dGcn5 mutants (Fig. 6E and F). However, dGcn5 mutants were not rescued by Gcn5ΔAda and still arrested at puparium formation (not shown).
The Gcn5ΔPcaf variant protein also appeared to be normally distributed at the interbands of polytene chromosomes and to restore the acetylation of histone H3 (Fig. 6G and H). In addition, about half of the dGcn5 mutant animals expressing Gcn5ΔPcaf formed their puparium and completed metamorphosis but died before eclosion as pharate adults, while the other half gave rise to adult flies (Fig. 7). However, rescued adults displayed held-out, misshapen wings, legs with a severe femur kink, and rough eyes. They died a few hours after eclosion, indicating that the UAS-Gcn5ΔPCAF construct only partially rescues dGcn5 mutants.
FIG. 7.
Gcn5ΔPcaf variant restores the viability of dGcn5 mutants. (A) Gcn5E333st/sex204 da-GAL4 adult fly heterozygous for the UAS-Gcn5 transgene. Rescued animals are indistinguishable from wild-type animals as well as from fully rescued UAS-Gcn5ΔBromo/+ Gcn5E333st/sex204 da-GAL4 animals (not shown). (B) Gcn5E333st/sex204 da-GAL4 adult fly heterozygous for the UAS-Gcn5ΔPcaf transgene. Rescued adults displayed held-out, notched wings (white arrow), rough eyes, and crooked legs (black arrow).
Strikingly, the Gcn5ΔBromo variant protein not only restored histone H3 acetylation (Fig. 6I) but also restored complete viability of the dGcn5 mutants. Adult flies were indistinguishable from dGcn5 mutant flies rescued by the full-length dGcn5 protein (not shown). The only observed defect was female sterility, a result expected from the absence of activity of the UAS promoter in the female germ line. We were not able to examine the chromosomal localization of the Gcn5ΔBromo protein because our dGcn5 antibody was prepared against the dGcn5 bromodomain (Fig. 6J). Nevertheless, the full phenotypic rescue by this variant strongly suggests that its localization to chromosomes was restored.
DISCUSSION
GNAT complexes are essential actors of transcriptional gene regulation in eukaryotes. Their structure and composition have been conserved throughout evolution from yeast to humans, and they are thought to exert their function through the catalytic activity of an HAT component belonging to the GNAT family. Vertebrate Gcn5 and Pcaf proteins have been involved in many biological processes, including the response to steroid/retinoid hormones, cell proliferation, and cell differentiation. However, how the function of these HATs is integrated in higher organisms to control development has still to be defined.
In an extensive EMS-based genetic screen, we isolated and characterized mutations of dGcn5, the unique Drosophila member of the GNAT family. dGcn5 mutants have no discernible phenotype before the onset of metamorphosis. The considerable dGcn5 maternal stock (37; our unpublished data) may be sufficient to fulfill dGcn5 functions in mutant embryos. However, the dGcn5 protein is undetectable in dGcn5 mutant mid-third-instar larvae, and the proliferation of imaginal disks is already strongly impaired in such mutants at this stage. In addition, mutant larvae keep wandering when control animals have already formed a puparium, dramatically extending the duration of their third larval instar by 4 to 5 days before they die. Ada2b mutants die during pupal development (49), while Ada2a mutants fail to form a puparium in a manner very similar to that of dGcn5 mutant larvae (47a). Together with these observations, our results strongly suggest that the function of the GNAT complexes is not required for Drosophila larval life.
The absence of normal induction of the three early puffs at 2B, 74EF, and 75B in dGcn5 mutants is indicative of a major failure in the regulatory hierarchy controlled by the steroid hormone ecdysone at the end of the third larval instar. The extended duration of this stage in dGcn5 null mutants is also characteristic of defects in pathways controlled by ecdysone. For instance, it is observed with EcR-B1-specific alleles of the ecdysone receptor gene (6, 53) and with mutant alleles of the broad gene (5). The Pcaf homology domain has been involved in interactions of human Pcaf with various nuclear receptors (9, 41, 54). Two dGcn5 hypomorphic alleles isolated in this work change amino acids in the Pcaf homology domain, resulting in a partial loss of function of the protein. In heteroallelic combination with the null Gcn5E333st allele, they led to impaired metamorphosis, with strong defects in adult appendage formation. dGcn5 adult mutant escapers rescued by the Gcn5ΔPcaf variant consistently display very similar defects of leg elongation and blistered wings. These defects are also highly suggestive of an impaired ecdysone regulatory cascade during metamorphosis (17, 18, 19). From these observations, we propose that dGcn5 acts as a nuclear receptor coactivator at metamorphosis. The finding that the EcR protein is coimmunoprecipitated from embryonic nuclear extracts with dGcn5 in an ecdysone-dependent manner (A. Mazo, personal communication) points to the ecdysone receptor itself as a dGcn5 target. However, we did not detect interactions between our dGcn5 alleles and EcR mutant alleles in a trans-heterozygous genetic test. Although these results do not rule out a functional interaction between dGcn5 and the ecdysone receptor, they may also indicate that dGcn5 acts in the metamorphic ecdysone regulatory hierarchy through interactions with other Drosophila nuclear receptors.
We have shown that the lack of dGcn5 in the female germ line arrests oogenesis at an early stage. In addition, somatic dGcn5 mutant clones in ovarian follicles induce the formation of compound egg chambers indicative of an oocyte packaging defect (J. R. Huynh, personal communication). Together, these results indicate a strict requirement for dGcn5 during oogenesis, in both the germ line and the somatic line. In light of the role of dGcn5 during ecdysone-triggered metamorphosis, it is noteworthy that ecdysone regulatory hierarchies have also been shown to regulate Drosophila oogenesis (33).
dGcn5 mutant imaginal disks, as well as imaginal tissues silenced by dGcn5 RNAi, display slower cell proliferation. The role of CBP in the cell cycle has been documented for vertebrates (39), and a domain of interaction of CBP with the Gcn5 homologue Pcaf is targeted by viral and cellular factors to regulate cell cycle progression (51). The acetylation of E2F1 by Pcaf also results in cell cycle modulation (43). In yeast, Gcn5 regulates genes required for mitotic exit (34). Together, these data suggest that functions of Gcn5 in cell cycle regulation have been conserved throughout evolution. Interestingly, a mouse knock-in mutant for Trrap, a conserved component of GNAT complexes, results in aberrant mitosis (29, 40). Similarly, the higher mitotic index, together with the increased BrdU incorporation, of dGcn5-silenced imaginal cells suggests that they undergo an aberrant cell cycle. We have shown that apoptosis induced in imaginal tissues by the loss of dGcn5 function may contribute to the net reduction in cell number. The Drosophila SAGA-like complex interacts in vitro with Dmp53 (37), and several authors reported a role of dAda2b in DNA damage-induced Dmp53-dependent apoptosis (49, 47a). However, this is the first time that apoptosis in a mutant with a deletion of a component of the Drosophila SAGA-like complex has been observed in the absence of X-ray-induced DNA damage. Strikingly, Gcn5 mouse mutants also display apoptosis in the absence of any exogenous inducers (66). Although further analysis is required to understand the complex roles of dGcn5 in cell proliferation, we propose that apoptosis in the dGcn5 mutant might be a consequence of cell cycle defects.
Our data demonstrate that dGcn5 is the major acetylase for two distinct histone residues, H3 K9 and H3 K14, in Drosophila. In contrast, its contribution to histone H4 acetylation could not be detected in our global analysis. The loss of Ada2b results in a partial loss of H3 K9 and H3 K14 acetylation on polytene chromosomes, while the loss of Ada2a has no detectable effect on chromosome acetylation (49, 47a). In light of our results, these data suggest that the Drosophila GNAT complexes may retain partial dGcn5 HAT activity in the absence of the Ada2 components. Alternatively, dGcn5 could exert its HAT activity in other complexes which remain to be characterized. The substrate specificities of Drosophila and yeast Gcn5 proteins appear to be identical (26). However, it is interesting that in contrast to what we found for Drosophila, both the Gcn5 and Elp3 HATs must be invalidated in yeast to significantly impair histone H3 K9 and K14 acetylation (62). Recent studies have extensively documented the relationship between histone H3 acetylation and gene transcription (7, 36, 52), but whether or not patterns of histone modification constitute a true code for the control of gene activity in eukaryotes is still a matter of debate. In either case, our results strongly suggest that specific histone acetylation profiles may be established in vivo through the activity of a very limited set of substrate-specific enzymes. The observation that dGcn5 mutant larvae survive for several days without detectable acetylation of H3 K9 and H3 K14 residues is striking and suggests that transcriptional regulation in larval versus embryonic or adult insect tissues involves distinct mechanisms.
Numerous data indicate that histone modifications can influence each other (24). The loss of H3 K9 and H3 K14 acetylation in imaginal disks or salivary glands had no detectable effect either on the levels of H3 S10 phosphorylation and H3 K4 methylation, both of which have been associated with transcriptional activation, or on the level of H3 K9 methylation, which marks HP1 recruitment and silencing. These results suggest that histone H3 acetylation is either a terminal or independent process in the cascade of histone H3 modifications.
Using transgenic dGcn5 variants, we investigated the functions of the conserved regions in the dGcn5 protein. The deletion of the HAT domain completely abolished the ability of dGcn5 to acetylate histones and to rescue dGcn5 mutant larvae. This result provides the first demonstration that Gcn5 in metazoans exerts its regulatory function during development through its histone acetylase activity. As discussed above, defects displayed by dGcn5 mutant adults rescued by the Gcn5ΔPcaf variant suggest a role of dGcn5 in the ecdysone regulatory hierarchy during metamorphosis, possibly through interactions with nuclear receptors. However, it is noteworthy that Gcn5ΔPcaf retains important functions. The variant protein was distributed along polytene chromosomes similarly to the dGcn5 wild-type protein and provided apparently normal H3 K9 and K14 acetylation. This suggests that the core functions of chromatin binding and substrate acetylation may be performed by the ancestral portion of Gcn5, which has been conserved throughout evolution from yeast to humans, and that the Pcaf homology domain has evolved to perform more specific functions in metazoans. The Ada2 interaction domain has been mapped in Gcn5 to an evolutionarily conserved region between the HAT domain and the bromodomain (15, 16, 42). A direct interaction in vitro between dGcn5 and Ada2b was recently shown for Drosophila (49). The finding that the Gcn5ΔAda variant was completely unable to rescue the lethality of the dGcn5 mutants at the time of puparium formation provides strong evidence that the interaction of dGcn5 with Ada2 proteins is crucial for the function of the Drosophila GNAT complexes. Strikingly, however, in dGcn5 mutants Gcn5ΔAda could bind to polytene chromosomes and restore H3 K9 and H3 K14 acetylation patterns that were indistinguishable from those provided by the wild-type dGcn5 protein. This result is at odds with the finding that histone H3 acetylation is strongly reduced in Ada2b null mutant polytene chromosomes (49). It is possible that the loss of interaction between Ada2b and dGcn5 is deleterious for a specific function of the multiprotein SAGA-like complex but is not sufficient to disrupt the architecture of this complex as well as its ability to acetylate histones.
The bromodomain is conserved in a large number of transcription factors and has been shown to bind to acetylated lysines on histone tails. The yeast Gcn5 bromodomain has been shown to be involved in the stabilization of the SWI/SNF remodeling complex through interactions with acetylated nucleosomes at the PHO5 promoter (59). The bromodomain-containing complexes SWI/SNF and TFIID have been shown to be targeted to acetylated histone H3 and H4 tails on the beta interferon promoter in vitro (1, 2). Collectively, these findings suggest that the bromodomain is essential for targeting a large set of transcription factors to acetylated nucleosomes. In this context, the apparently normal chromosome acetylation by the Gcn5ΔBromo variant, and most significantly, the complete rescue of dGcn5 mutant viability by this protein, raises a number of questions. We cannot exclude the possibility that the overexpression of the Gcn5ΔBromo variant may overcome the lack of an essential dGcn5 function or that Gcn5ΔBromo may be unable to supply an essential function under particular stress conditions. However, our observations clearly indicate that the dGcn5 bromodomain is not as strictly required as the other domains of the protein, since variants with truncations of these domains were expressed under the same conditions. Interestingly, others have reported a minor contribution of the bromodomain to the functions of various transcription factors. Notably, in yeast the deletion of the bromodomain of dGcn5 has little consequence on its transcriptional activity (16, 57), while deletion of the bromodomain of Swi2/Snf2 (38) or Spt7 (25) has no phenotypic effect. Similarly, the Brahma protein with a deletion of its bromodomain binds normally to chromosomes in Drosophila and fully rescues brahma null mutants (22). The apparent lack of requirement for the bromodomain in dGcn5 may be due to a functional redundancy of various components of dGcn5 complexes in targeting chromatin. This role has been proposed for the Spt7 factor and the large Tra1 protein, which are both components of the SAGA complex (12, 28). The recent demonstration that a chromodomain of the yeast SAGA component Chd1 interacts with methylated lysine 4 of histone H3 (48) also raises the interesting possibility that different factors in GNAT complexes interact with different histone modifications.
In summary, the deletion of the HAT domain in dGcn5 abolished both its HAT activity and its ability to rescue dGcn5 mutants. dGcn5 variants deleted in either the Pcaf homology domain or the Ada2 interaction domain acetylated chromosomes. However, Gcn5ΔPcaf was able to partially rescue the dGcn5 mutants, while Gcn5ΔAda was not. Finally, the deletion of the dGcn5 bromodomain had no noticeable consequences. This remarkable variety of effects revealed in our complementation experiments strongly suggests that dGcn5 integrates multiple functions during development.
Acknowledgments
We are grateful to P. Maroy for the gift of the 1479/10 line, to N. Dos-Santos, E. Scola, A. Ikmi, D. Cohen, and J.-R. Huynh for their technical help, and to the Institut Pasteur Dynamic Imaging Platform for assistance and advice. We thank I. Boros, L. Tora, and A. Mazo for sharing data before publication, T. Grange and J. A. Lepesant for helpful discussions during the course of this work, and M. Buckingham, D. Fagegaltier, H. Thomassin, and L. Théodore for critical readings of the manuscript.
This work was supported by the Pasteur Institute and the C.N.R.S. (URA 2578) and by grants from the A.R.C. (7742/4383/3202). D.S. and C.C. were supported by the University of Paris VI and the Ministère de la Recherche.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner, M., C. Chihara, P. Meltzer, and G. Richards. 1974. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harbor Symp. Quant. Biol. 38:655-662. [DOI] [PubMed] [Google Scholar]
- 4.Bellaiche, Y., M. Gho, J. A. Kaltschmidt, A. H. Brand, and F. Schweisguth. 2001. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 3:50-57. [DOI] [PubMed] [Google Scholar]
- 5.Belyaeva, E. S., M. G. Aizenzon, V. F. Semeshin, I. I. Kiss, K. Koczka, E. M. Baritcheva, T. D. Gorelova, and I. F. Zhimulev. 1980. Cytogenetic analysis of the 2B3-4-2B11 region of the X-chromosome of Drosophila melanogaster. I. Cytology of the region and mutant complementation groups. Chromosoma 81:281-306. [DOI] [PubMed] [Google Scholar]
- 6.Bender, M., F. B. Imam, W. S. Talbot, B. Ganetzky, and D. S. Hogness. 1997. Drosophila ecdysone receptor mutations reveal functional differences among receptor isoforms. Cell 91:777-788. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand, C., C. Bergounioux, S. Domenichini, M. Delarue, and D. X. Zhou. 2003. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278:28246-28251. [DOI] [PubMed] [Google Scholar]
- 9.Blanco, J. C., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, Y. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 11.Brodu, V., B. Mugat, J. Y. Roignant, J. A. Lepesant, and C. Antoniewski. 1999. Dual requirement for the EcR/USP nuclear receptor and the dGATAb factor in an ecdysone response in Drosophila melanogaster. Mol. Cell. Biol. 19:5732-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 13.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 14.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 15.Candau, R., and S. L. Berger. 1996. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J. Biol. Chem. 271:5237-5245. [DOI] [PubMed] [Google Scholar]
- 16.Candau, R., J. X. Zhou, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Avino, P. P., and C. S. Thummel. 1998. Crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125:1733-1745. [DOI] [PubMed] [Google Scholar]
- 18.D'Avino, P. P., and C. S. Thummel. 2000. The ecdysone regulatory pathway controls wing morphogenesis and integrin expression during Drosophila metamorphosis. Dev. Biol. 220:211-224. [DOI] [PubMed] [Google Scholar]
- 19.Davis, M. B., G. E. Carney, A. E. Robertson, and M. Bender. 2005. Phenotypic analysis of EcR-A mutants suggests that EcR isoforms have unique functions during Drosophila development. Dev. Biol. 282:385-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dequier, E., S. Souid, M. Pal, P. Maroy, J. A. Lepesant, and C. Yanicostas. 2001. Top-DER- and Dpp-dependent requirements for the Drosophila fos/kayak gene in follicular epithelium morphogenesis. Mech. Dev. 106:47-60. [DOI] [PubMed] [Google Scholar]
- 21.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Elfring, L. K., C. Daniel, O. Papoulas, R. Deuring, M. Sarte, S. Moseley, S. J. Beek, W. R. Waldrip, G. Daubresse, A. DePace, J. A. Kennison, and J. W. Tamkun. 1998. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 24.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 25.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 27.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 28.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 29.Herceg, Z., W. Hulla, D. Gell, C. Cuenin, M. Lleonart, S. Jackson, and Z. Q. Wang. 2001. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 29:206-211. [DOI] [PubMed] [Google Scholar]
- 30.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 32.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33-43. [DOI] [PubMed] [Google Scholar]
- 33.Kozlova, T., and C. S. Thummel. 2000. Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrinol. Metab. 11:276-280. [DOI] [PubMed] [Google Scholar]
- 34.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 35.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 36.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 37.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurent, B. C., I. Treich, and M. Carlson. 1993. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 7:583-591. [DOI] [PubMed] [Google Scholar]
- 39.Lehrmann, H., L. L. Pritchard, and A. Harel-Bellan. 2002. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 86:41-65. [DOI] [PubMed] [Google Scholar]
- 40.Li, H., C. Cuenin, R. Murr, Z. Q. Wang, and Z. Herceg. 2004. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J. 23:4824-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X., J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 45.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ornaghi, P., P. Ballario, A. M. Lena, A. Gonzalez, and P. Filetici. 1999. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287:1-7. [DOI] [PubMed] [Google Scholar]
- 47.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Pankota, T., O. Komonyi, L. Bodai, Z. Újfaludi, S. Muratoglu, A. Ciurciu, L. Tora, J. Szabad, and I. Boros. 2005. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. 25:•••-•••. [DOI] [PMC free article] [PubMed]
- 48.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 49.Qi, D., J. Larsson, and M. Mannervik. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24:8080-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roignant, J. Y., C. Carre, B. Mugat, D. Szymczak, J. A. Lepesant, and C. Antoniewski. 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiltz, R. L., and Y. Nakatani. 2000. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta 1470:M37-M53. [DOI] [PubMed] [Google Scholar]
- 52.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schubiger, M., A. A. Wade, G. E. Carney, J. W. Truman, and M. Bender. 1998. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125:2053-2062. [DOI] [PubMed] [Google Scholar]
- 54.Sharma, D., and J. D. Fondell. 2000. Temporal formation of distinct thyroid hormone receptor coactivator complexes in HeLa cells. Mol. Endocrinol. 14:2001-2009. [DOI] [PubMed] [Google Scholar]
- 55.Simon, A. F., I. Boquet, M. Synguelakis, and T. Preat. 1998. The Drosophila putative kinase linotte (derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mech. Dev. 76:45-55. [DOI] [PubMed] [Google Scholar]
- 56.Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 59.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 60.Turner, B. M., A. J. Birley, and J. Lavender. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69:375-384. [DOI] [PubMed] [Google Scholar]
- 61.Vlachonasios, K. E., M. F. Thomashow, and S. J. Triezenberg. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittschieben, B. O., J. Fellows, W. Du, D. J. Stillman, and J. Q. Svejstrup. 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 19:3060-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]
- 64.Wodarz, A., U. Hinz, M. Engelbert, and E. Knust. 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82:67-76. [DOI] [PubMed] [Google Scholar]
- 65.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 66.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]
- 67.Zink, D., and R. Paro. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14:5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]