Abstract
SRC-3/AIB1/ACTR/pCIP/RAC3/TRAM1 is a primary transcriptional coregulator for estrogen receptor (ER). Six SRC-3 phosphorylation sites have been identified, and these can be induced by steroids, cytokines, and growth factors, involving multiple kinase signaling pathways. Using phosphospecific antibodies for six phosphorylation sites, we investigated the mechanisms involved in estradiol (E2)-induced SRC-3 phosphorylation and found that this occurs only when either activated estrogen receptor α (ERα) or activated ERβ is present. Both the activation function 1 and the ligand binding domains of ERα are required for maximal induction. Mutations in the coactivator binding groove of the ERα ligand binding domain inhibit E2-stimulated SRC-3 phosphorylation, as do mutations in the nuclear receptor-interacting domain of SRC-3, suggesting that ERα must directly contact SRC-3 for this posttranslational modification to take place. A transcriptionally inactive ERα mutant which localizes to the cytoplasm supports E2-induced SRC-3 phosphorylation. Mutations of the ERα DNA binding domain did not block this rapid E2-dependent SRC-3 phosphorylation. Together these data demonstrate that E2-induced SRC-3 phosphorylation is dependent on a direct interaction between SRC-3 and ERα and can occur outside of the nucleus. Our results provide evidence for an early nongenomic action of ER on SRC-3 that supports the well-established downstream genomic roles of estrogen and coactivators.
Steroid receptor coactivator 3 (SRC-3, also known as AIB1/ACTR/RAC3/pCIP/TRAM-1) is a member of the SRC/p160 family of transcription coactivators for nuclear receptors and other transcription factors (2, 9, 32, 36, 42, 58, 64, 65, 67, 77). Loss of SRC-3 expression in cells severely impairs the transcriptional output from nuclear receptors (71, 76). SRC-3 is overexpressed in a significant percentage of breast cancers, and in its role as an oncogene, it is involved in the development and maintenance of breast and prostate cancers (2, 19, 27, 68, 79).
The activities of the SRC coactivators are affected by posttranslational modifications such as phosphorylation, acetylation, sumoylation, and ubiquitination (6, 10, 17, 22, 34, 74, 75). We and others have shown that SRC-3 activity is regulated by phosphorylation, which dramatically affects its association with nuclear receptors and other coregulators and transcription factors and its coactivator functions, subcellular localization, and oncogenic activities (53, 68, 74, 75). This phosphorylation can be induced by different stimuli including steroid hormones, growth factors, and cytokines and involves a wide range of kinases including p42/p44 mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase, p38 MAPK, and IκB kinases (IKKs) (17, 50, 75). Six in vivo SRC-3 phosphorylation sites have been identified (Fig. 1A), and phosphorylation state-specific antibodies against each site have been generated and validated (75). Different stimuli induce distinct patterns of SRC-3 phosphorylation, and mutations at different phosphorylation sites have different downstream effects. For example, 17β-estradiol (E2) induces SRC-3 phosphorylation at all six sites, while tumor necrosis factor alpha (TNF-α) induces phosphorylation of all but the serine-860 (S860) site (75). Consistent with these data, mutation of any of the six phosphorylation sites to an alanine residue impairs the ability of the mutant SRC-3 to coactivate E2-induced estrogen receptor α (ERα) target gene expression in reporter assays, while all but S860 mutations affect TNF-α-induced NF-κB target gene activation. Likewise, mutation of the threonine-24 (T24), S543, S857, and S867 sites, but not the S505 and S860 sites, negatively influences the expression of the SRC-3 target gene interleukin-6, while the tumorigenic activity of SRC-3, demonstrated via its ability to potentiate cellular transformation by RasV12, is affected by mutation of all the phosphorylation sites except S505 (75). These observations likely result from the different affinities of various SRC-3 phosphorylation site mutants for other coregulators and transcription factors such as CBP, CARM1, ERα, and NF-κB (75).
FIG. 1.
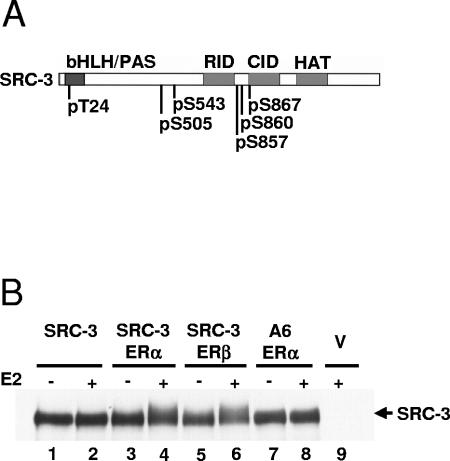
Estradiol induction of SRC-3 phosphorylation. (A) Schematic diagram of SRC-3. The known functional domains of SRC-3 are shown on top: bHLH/PAS, basic helix-loop-helix/Per-Arnt-Sim domain; RID, receptor-interacting domain; CID, CBP/p300-interacting domain; HAT, histone acetyltransferase domain. The identity and position of the six identified phosphorylated amino acids of SRC-3 are shown below. (B) Estradiol induced an ER-dependent change in electrophoretic mobility of SRC-3. HEK293T cells were transfected with plasmids for either Flag-tagged wild-type SRC-3 (SRC-3) or a Flag-SRC-3 mutant (A6) in which alanines were substituted for all six phosphorylation sites (S/T to A), and either ERα or ERβ, or empty vectors (V). Forty-eight hours posttransfection, 0.01% ethanol (−) or 10 nM estradiol (E2; +) was added for 1 h. Cells were then lysed and analyzed by Western blotting using an anti-SRC-3 antibody.
How E2 induces phosphorylation of SRC-3 is poorly understood. The major biological functions of E2 are mediated through ERα and ERβ, which are members of the nuclear receptor superfamily of ligand-dependent transcription factors (46). Estradiol can induce cellular responses through direct “genomic” ER-dependent activation of gene transcription at target promoters as well as by “nongenomic” actions, the latter including rapid activation of various protein kinase cascades independent of prior gene transcription (3). Nongenomic E2-activated pathways may be ER dependent. For example, E2 can induce a rapid and transient activation of the Src/Erk phosphorylation cascade through the association between cytoplasm-localized ERα and MNAR (modulator of nongenomic activity of ER) (73). Alternatively, in an ER-independent fashion, E2 has been shown to activate the MAPK pathway through the membrane-bound G protein-coupled receptor GPR30 (16, 37). Whether E2-induced SRC-3 phosphorylation is mediated through a “genomic” or “nongenomic” activity of E2 and its dependence on ER is unclear at present.
Estrogen receptors regulate the differentiation and maintenance of many tissues including reproductive, neural, skeletal, and cardiovascular tissues (26) and possess several domains essential for their functions (46). The transcriptional activity of ER is dependent on two activation function (AF) domains, AF-1 at the N terminus and AF-2 in the C-terminal ligand binding domain (LBD). The AF-1 domain can exert its activity in a ligand-independent manner (15, 44, 69), while AF-2 activity is dependent on agonist binding to the LBD (28). Both AF-1 and AF-2 activities are associated with their abilities to bind to coregulators. The DNA binding domain (DBD) contains two zinc finger motifs, which bind to specific estrogen response elements (EREs) within the promoters of ER target genes (25). The first zinc finger contains a P box directly contacting DNA, and the latter contains a D box responsible for ER dimerization (57). The hinge region which is located between DBD and LBD contains most of the nuclear localization signal (NLS) (51). Estrogen receptor shuttles between nucleus and cytoplasm, although under steady-state conditions it is mainly detected in the nucleus (14, 39). In the absence of ligand, 5 to 10% of green fluorescent protein (GFP)-ERα is localized in the cytoplasm (39, 78). Estradiol increases ER nuclear localization. It also has been shown that E2 induces a translocalization of SRC-3 from the cytoplasm to the nucleus (53, 71, 74). Up to now, the “classical” mode of activation of coactivators and their interaction with transcription factors were believed to occur on nuclear promoters in response to different stimuli (77).
The ligand-dependent interaction between nuclear receptors and SRC-3 is direct and is mediated through the LBD domains and the LXXLL motifs, respectively (9, 21, 67). Upon binding to an agonist, ERα undergoes a conformational change and presents a coactivator-binding groove on the surface of its LBD capable of binding to the LXXLL motifs (43, 59). This coactivator-binding groove involves residues (e.g., K362, V376, and L539) from several helices, H3 to H5 and H12 (38). After coactivator binding, ERα LBD undergoes a further conformational change, resulting in a more stable receptor (66). There are three LXXLL motifs located in the receptor-interacting domain of SRC-3 (9, 21, 67). Mutations that disrupt the formation of the coactivator-binding groove of ERα or mutations in the LXXLL motifs of SRC-3 abolish interaction between SRC-3 and ERα and thus coactivation of ERα (38).
Here we show that E2-induced SRC-3 phosphorylation requires a direct interaction between SRC-3 and ERα but does not require DNA binding and nuclear localization of ERα, indicating a “nongenomic” action of this receptor. Both ERα AF-1 and AF-2 domains are required for a maximal induction of coactivator phosphorylation. These results indicate that, in addition to “genomic” and transcription-dependent activities of ERα, “nongenomic” activities of ERα can significantly influence SRC-3 functions, suggesting a multilevel functional cooperation between coactivators and nuclear receptors on and off their target genes.
MATERIALS AND METHODS
Chemicals and reagents.
17β-Estradiol (E2), progesterone (P), 4-hydroxytamoxifen (4HT), and raloxifene (Ral) were obtained from Sigma-Aldrich (St. Louis, MO). ICI 182,780 was from Tocris Cookson (Ellisville, MO). Phosphorylation state-specific antibodies against SRC-3 were characterized previously (75). Antibody against SRC-3 was from BD Biosciences (San Jose, CA) when used for Western blotting or from Santa Cruz Biotechnology (Santa Cruz, CA) when used for immunoprecipitation. The EZview red anti-Flag M2 affinity gel used for immunoprecipitating Flag-tagged proteins was from Sigma-Aldrich. Anti-ERα mouse monoclonal antibody was from Cell Signaling Technology (Beverly, MA), and anti-ERα rabbit polyclonal antibody was from Santa Cruz Biotechnology. Anti-ERβ antibody was from GeneTex, Inc. (San Antonio, TX). Antibody against progesterone receptor B (PR-B) was described previously (1). The small interfering RNA (siRNA) against human ERα (siGENOME SMARTpool reagent) was purchased from Dharmacon (Lafayette, CO). The siRNA against luciferase was purchased from Ambion (Austin, TX). Transfection reagents (TransIT-LT1 and TransIT-TKO) were from Mirus Bio Corporation (Madison, WI).
Plasmids.
The expression plasmid for human Flag-tagged SRC-3 (pCMV-Flag-SRC-3) was generated as described previously (75). The wild-type full-length cDNA of SRC-3 was subcloned from the plasmid pCMX-F.RAC3 (32). pCMV-Flag-SRC3-A6 is an expression plasmid for human Flag-tagged SRC-3 mutant (A6) in which all six identified phosphorylation sites (Fig. 1A) have been mutated to alanine (75). The expression plasmid pCMV-Flag-SRC3-AAA encodes a Flag-tagged SRC-3 mutant (AAA) in which all three LXXLL motifs have been mutated to LXXAA and was constructed by replacing the AflII-XbaI fragment of pCMV-Flag-SRC3 with the corresponding region of pCMX-ACTRAAA (36) containing the desired mutations. The expression plasmids for human ERα (pCR3.1-hERα [45], pCMV5-hERα, pCMV5-hERα phosphorylation mutant [S104A/S106A/S118A as 3A] [30], pRST7hERα, pRST7hERα-N282G [amino acids (aa) 1 to 282], and pRST7hERα-179C [aa 179 to 595] [70]), human ERβ (pCXN2-hERβ [47] and pCR3.1hERβ-143C [aa 143 to 530] [11]), and human PR-B (pCR3.1-hPRB [1]) were described previously.
The expression plasmids (pCR3.1-hERα-K362D, pCR3.1-hERα-V376D, or pCR3.1-hERα-L539A) for full-length hERα with single point mutations (K362D, V376D, or L539A) were described previously (34). The expression plasmid (pCR3.1-hERαKV) for the hERα double mutant (K362D/V376D) was constructed as follows. PCR amplification was carried out using the forward primer 5′-TACGGCCCCGGGTCTGAGGCT-3′ containing an XmaI site (underlined) and the reverse primer 5′-ACATTCTAGAAGGTGGTCCTGATCATGGAGGGTCAAATCCACAAAGCCTGGCACCCTATCCGCCCA-3′ containing the K362D and V376D mutations (italics) and an XbaI site (underlined) to amplify a region of the hERα cDNA (pCR3.1-hERα). The PCR product was then digested with XmaI and XbaI and used to replace the corresponding region of pCR3.1-hERα. The expression plasmid (pCR3.1-hERαKVL) for the hERα triple mutant (K362D/V376D/L539A as KVL) was constructed by replacing a BglII-ApaI fragment of pCR3.1-hERαKV with the corresponding region of pCR3.1-hERαL539A containing the L539A mutation. All the mutant plasmid constructs described above were sequenced to ensure that only the intended mutations were introduced.
The expression plasmids HEG0 and HE241G were described previously (78). The plasmid HEG0 encodes a wild-type human ERα, and HE241G encodes a human ERα mutant (ERαΔNLS) with a deletion of aa 250 to 303, which encodes an NLS. The expression plasmid encoding the GFP-tagged human ERα, pEGFP-C1-hERα (GFP-ERαWT), was a gift from M. A. Mancini (Baylor College of Medicine) (63). The expression plasmid encoding the GFP-tagged ERαΔNLS, pEGFP-C1-hERαΔNLS (GFP-ERαΔNLS), was constructed by replacing the XmaI-BlpI fragment of pEGFP-C1-hERα with the corresponding region of the plasmid HE241G containing the ΔNLS mutation.
The expression plasmid pCR3.1-hERαCC, which encodes an hERα mutant containing the double mutation (C202H/C205H as CC) within the DBD, was described previously (34). The expression plasmid pCR3.1-hERαEG encodes an hERα mutant containing the double mutation (E203A/G204A as EG) within the DBD. It was constructed by replacing the XmaI-BglII fragment of pCR3.1-hERα with the corresponding region of pHAhERαE203AG204A (a gift of P. Brown, Baylor College of Medicine).
Cell cultures and transfections.
HEK293T and MCF-7 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Twenty-four hours prior to transfection, cells were plated in phenol red-free Dulbecco's modified Eagle's medium with 5% charcoal-stripped fetal bovine serum. For plasmid DNA transfection, cells were transfected using TransIT-LT1 (3 μl LT1/μg plasmid DNA), while for siRNA transfection, cells were transfected using TransIT-TKO (30 μl TKO/0.1 nmol siRNA) according to the manufacturer's protocol. For immunoprecipitating SRC-3 and its associated proteins, HEK293T cells (6 × 106 cells/10-cm dish) were transfected with a total of 10 μg of plasmid DNA (4 μg of the plasmid DNA encoding Flag-tagged SRC3 or the empty vector pCMVTag2B [Stratagene, La Jolla, CA]; 2 μg of the plasmid DNA encoding hERα, hERβ, hPR-B, or the empty vector pCR3.1 [Invitrogen, Carlsbad, CA]; and 4 μg of the plasmid pBluescript [Stratagene]) for 48 h. Cells were then treated with various nuclear receptor agonists or antagonists (10 nM) or ethanol (0.01%) for different periods of time. For luciferase reporter assays, HEK293T cells (2 × 105 cells/well in six-well plates) were transfected with 1 μg of plasmid DNA (5 ng of the plasmid DNA encoding hERα or the empty vector pCR3.1, 500 ng of pERE-E1b-Luc reporter plasmid [45], and 495 ng of pBluescript) for 24 h and treated with E2 (10 nM) or ethanol (0.01%) for an additional 20 h. For immunofluorescence, HEK293T cells (6 × 105 cells/well in six-well plates with coverslips) were transfected with 1 μg of plasmid DNA (400 ng of the plasmid DNA encoding Flag-tagged SRC-3, 200 ng of the plasmid DNA encoding hERα, and 400 ng of the pBluescript DNA) for 48 h. Cells were then treated with E2 (10 nM) or ethanol (0.01%) for 1 h.
Immunoprecipitation and Western blotting.
Cell lysates were prepared by incubating cells in the lysis buffer (20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 5% glycerol, supplemented with phosphatase inhibitors [1 mM NaF, 1 mM Na3VO4, 1 mM β-glycerophosphate, and 1 mM sodium pyrophosphate], and protease inhibitor cocktail [Roche, Indianapolis, IN]) for 20 min at 4°C, followed by centrifugation at 14,000 × g for 15 min at 4°C. The protein concentration of the lysates was determined using the BCA protein assay kit according to the manufacturer's protocol (Pierce, Rockford, IL). For immunoprecipitating Flag-tagged SRC-3 and its associated proteins, cell lysates containing 500 μg of protein were incubated with EZview red anti-Flag M2 affinity gel (30 μl of packed beads) for 2 h at 4°C with constant rotation. For immunoprecipitating endogenous SRC-3, MCF-7 cells were lysed and 500 μg of protein was incubated with an anti-SRC-3 antibody (1 μg) for 2 h at 4°C with constant rotation; protein G PLUS-Agarose beads (30 μl of packed beads; Santa Cruz Biotechnology) were then added, and the incubation was continued for an additional 2 h. Beads were then washed three times using the lysis buffer. Between washes, beads were collected by centrifugation at 8,000 × g for 30 s at 4°C. The precipitated proteins were eluted from beads by resuspending the beads in NuPage sample buffer (Invitrogen) and heating them at 75°C for 10 min. The resulting materials from immunoprecipitation or total cell lysates for detecting total protein levels were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 3 to 8% NuPage Novex gels (Invitrogen) and transferred onto nitrocellulose membranes. For Western blot analysis, membranes were first blocked using 5% nonfat dry milk in TBS-T buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature (RT) and then blotted using indicated antibodies which were diluted in TBS-T buffer for 1 h at RT or overnight at 4°C followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. Immunoreactive bands were visualized using ECL Plus reagents as recommended by the manufacturer (Amersham Biosciences, Piscataway, NJ). The experiments were repeated at least two times, and representative results are shown in the figures.
Luciferase reporter assay.
After transfection and treatment, HEK293T cells were harvested, and extracts were assayed for luciferase activity using the Luciferase Assay Systems kit (Promega, Madison, WI), which was measured using a Luminoskan Ascent Thermo Labsystems instrument (Thermo Electron Corporation, Vantaa, Finland). Relative luciferase units were normalized to total cellular protein and standardized to a negative control (cells transfected with empty vector and treated with ethanol) which is set as 1. Experiments were done in duplicate, and values represent averages ± standard errors of the means of at least three individual experiments.
Immunofluorescence microscopy.
HEK293T cells were prepared according to the protocol described previously (63). Briefly, after transfection and treatment, cells on the coverslips were fixed in 4% formaldehyde (Polysciences Inc., Warrington, PA) in PEM buffer {80 mM potassium PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 7.5], 5 mM EGTA, and 2 mM MgCl2 } for 30 min on ice. Cells were then washed three times with PEM buffer and incubated for 10 min in 0.1 M ammonium chloride to quench autofluorescence. The cells were then washed two times in PEM buffer and incubated in 0.5% Triton X-100 in PEM buffer for 5 min to permeabilize the cells. After being washed three times with PEM buffer, cells were counterstained for 1 min with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in TBS-T and mounted in Slow Fade reagent (Molecular Probes/Invitrogen). Cells on the coverslips were examined on a Carl Zeiss AxioPhot microscope, and GFP and DAPI signals were recorded using a color charge-coupled device camera with fluorescein and DAPI filters. Images were processed using Adobe Photoshop CS (Adobe Systems Incorporated, San Jose, CA).
RESULTS
Estradiol induces a mobility change of SRC-3 on SDS-PAGE gels in an ER-dependent manner.
It is well known that posttranslational modifications of proteins (e.g., phosphorylation) can change their electrophoretic mobility on SDS-PAGE gels. Protein modification pathways can be activated by various stimuli including hormones and growth factors. To investigate the effect of estradiol (E2) on the migration pattern of SRC-3 on an SDS-PAGE gel, E2 induction (10 nM, 1 h) was performed in HEK293T cells transiently transfected with Flag-tagged wild-type SRC-3 (SRC-3) with or without ERs. As shown in Fig. 1B, E2 treatment caused an upward smearing migration pattern of SRC-3 in the presence of ERα or ERβ (lanes 4 and 6). The mobility shift was not observed in the absence of ER (Fig. 1B, lane 2). Treatment with E2 did not change the mobility pattern of an A6 SRC-3 mutated at all six phosphorylation sites (Fig. 1B, lanes 7 and 8). These results indicated that E2 could alter the electrophoretic mobility of the wild-type SRC-3 but not its phosphorylation-site A6 mutant, implying that the E2-induced mobility change of SRC-3 is the result of SRC-3 phosphorylation. Since this was observed only in the presence of an ER, the E2-induced SRC-3 mobility shift depends on expression of either ERα or ERβ.
Estradiol-induced SRC-3 phosphorylation requires ER.
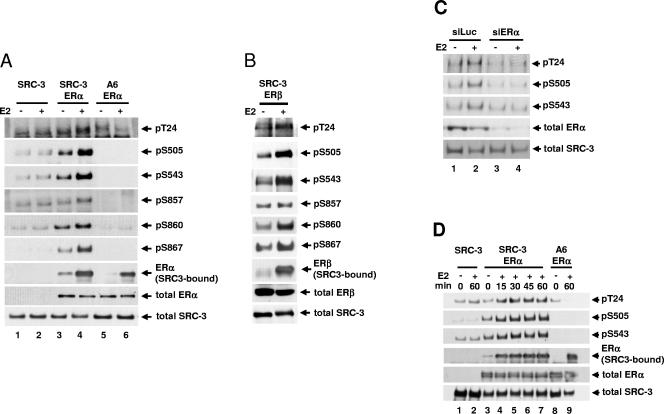
In cells that express ERα, E2 induces SRC-3 phosphorylation at all six phosphorylation sites identified (75). To determine whether E2-induced SRC-3 phosphorylation is dependent on the presence of ER, HEK293T cells transfected with SRC-3 together with or without either ERα or ERβ were treated with E2. SRC-3 phosphorylation state-specific antibodies were used to determine the level of phosphorylation of immunoprecipitated Flag-tagged SRC-3. As shown in Fig. 2A and B, E2 induced SRC-3 phosphorylation at all six sites (T24, S505, S543, S857, S860, and S867) in cells transfected with an expression vector for wild-type ERα (Fig. 2A, lanes 3 and 4, rows 1 to 6) or ERβ (Fig. 2B, rows 1 to 6), but not in cells transfected with an empty expression vector (Fig. 2A, lanes 1 and 2, rows 1 to 6). As a negative control, E2-induced SRC-3 phosphorylation was not observed when the SRC-3 phosphorylation mutant A6 was used (Fig. 2A, lanes 5 and 6, rows 1 to 6), even though the A6 form of SRC-3, like wild-type SRC-3, could still bind to ERα in response to E2 (Fig. 2A, lane 6, row 7). These results indicated that E2-induced SRC-3 phosphorylation requires either form of ER.
FIG. 2.
Estradiol-induced SRC-3 phosphorylation requires the presence of ER. (A) HEK293T cells were transfected with Flag-tagged SRC-3 (SRC-3) or its phosphorylation mutant (A6), in the absence or presence of ERα. Forty-eight hours later, 10 nM E2 (+) or 0.01% ethanol vehicle (−) was added for 1 h. Cells were then lysed, and Flag-SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. Immunoblots were probed with antibodies against each of the six SRC-3 phosphorylation sites (rows 1 to 6). The amount of SRC-3-bound ERα was detected by probing with an anti-ERα antibody (row 7). The total amounts of ERα and SRC-3 in the lysates were determined by immunoblotting with an anti-ERα and an anti-SRC-3 antibody (rows 8 and 9, respectively). (B) Cells were transfected with plasmids for wild-type SRC-3 and ERβ, and the level of SRC-3 phosphorylated at each of the six sites was assessed as described above. (C) siRNA against ERα (siERα) or against luciferase (siLuc) was transfected into MCF-7 cells. Cells were treated as described above. Cells were then lysed, and endogenous SRC-3 was immunoprecipitated using an anti-SRC-3 antibody and separated by SDS-PAGE. The levels of SRC-3 phosphorylated at T24, S505, and S543 were assessed. (D) Cells were transfected as in panel A and treated with 10 nM E2 (+) or 0.01% ethanol (−) for various periods of time (min). The level of SRC-3 phosphorylated at T24, S505, and S543 and the amount of SRC-3-bound ERα were assessed. Shown in the bottom two rows of all the panels are total cell lysates separated by SDS-PAGE and probed with the anti-ERα, anti-ERβ, or anti-SRC-3 antibodies to assess the total cellular levels of ERα, ERβ, and SRC-3, respectively.
In order to confirm the ER dependence with endogenous proteins, siRNA against ERα (siERα) was transfected into MCF-7 cells to reduce the expression of endogenous ERα. Forty-eight hours posttransfection, cells were treated with E2 (10 nM, 1 h), and phosphorylation of immunoprecipitated endogenous SRC-3 was determined using SRC-3 phosphorylation state-specific antibodies as described above. As shown in Fig. 2C, in the presence of siERα, expression of endogenous ERα was greatly reduced (lanes 3 and 4, row 4), and E2-induced phosphorylation of endogenous SRC-3 was abolished (lanes 3 and 4, rows 1 to 3). In contrast, SRC-3 phosphorylation was still induced when a control siRNA against luciferase (siLuc) was used (lanes 1 and 2, rows 1 to 3). This confirmed the data from transient-transfection experiments indicating that E2-induced phosphorylation of SRC-3 requires ER.
The kinetics of E2-induced SRC-3 phosphorylation also were assessed in HEK293T cells cotransfected with Flag-tagged SRC-3 and ERα. As shown in Fig. 2D, SRC-3 phosphorylation could be observed by 15 min after E2 treatment, represented by phosphorylation at sites T24, S505, and S543 (lanes 3 to 7, rows 1 to 3).
Selectivity of hormones and nuclear receptors for induction of SRC-3 phosphorylation.
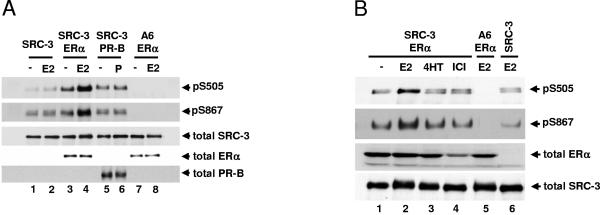
It has been shown that E2 and the synthetic androgen R1881 can induce SRC-3 phosphorylation in the presence of ER and androgen receptor, respectively (Fig. 2) (75). In order to investigate whether other hormones and their cognate nuclear receptors can induce SRC-3 phosphorylation, HEK293T cells transfected with an SRC-3 expression vector in the absence or presence of progesterone receptor B (PR-B) plasmid were treated with progesterone (P; 10 nM, 1 h). As shown in Fig. 3A, P did not induce SRC-3 phosphorylation in the presence of PR-B (lanes 5 and 6), although in parallel experiments E2 induced ER-dependent SRC-3 phosphorylation (lanes 3 and 4). Representative data for SRC-3 phosphorylation at S505 and S867 were shown, and other sites had a similar pattern (data not shown). As in the case of PR-B, dexamethasone did not induce SRC-3 phosphorylation in cells expressing the glucocorticoid receptor (data not shown). These data indicated that different steroids and their receptors have differential effects on SRC-3 phosphorylation.
FIG. 3.
Effects of different agonists and antagonists and their receptors on SRC-3 phosphorylation. HEK293T cells were transfected with Flag-tagged wild-type SRC-3 (SRC-3) or its phosphorylation-site mutant (A6) together with either ERα (A and B), ERβ (C), or progesterone receptor B (PR-B) (A). Forty-eight hours posttransfection, vehicle (−), 10 nM E2, 10 nM progesterone (P), 10 nM 4HT, 10 nM ICI 182,780 (ICI), or 10 nM Ral was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. The levels of SRC-3 phosphorylated at S505 and S867 were assessed by Western blotting using SRC-3 phosphorylation state-specific antibodies. Total cellular levels of ERα, ERβ, SRC-3, and PR-B were also determined by immunoblotting total cell lysates separated by SDS-PAGE with the appropriate antibodies.
SERMs do not induce SRC-3 phosphorylation.
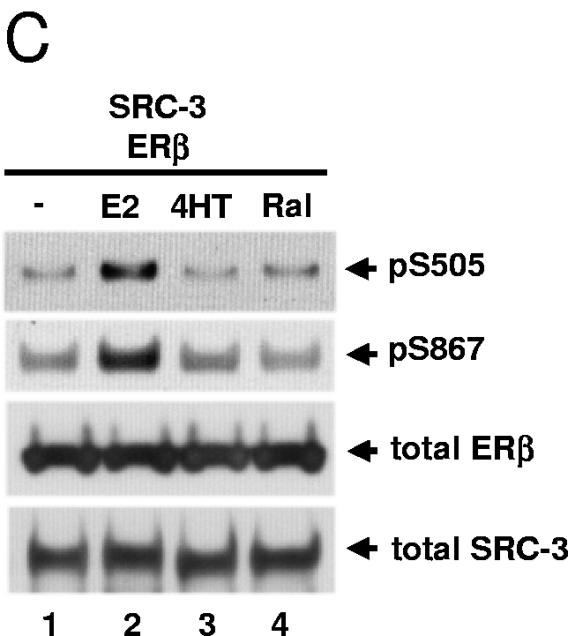
In order to investigate whether selective estrogen receptor modulators (SERMs) such as 4HT and Ral and pure antagonists such as ICI 182,780 (ICI) can induce SRC-3 phosphorylation, drug treatment (10 nM, 1 h) was performed in HEK293T cells cotransfected with plasmids for Flag-tagged SRC-3 and ERα (Fig. 3B) or ERβ (Fig. 3C). Tamoxifen and ICI (Fig. 3B) and 4HT and Ral (Fig. 3C), unlike E2, did not noticeably induce SRC-3 phosphorylation at sites S505 and S867 in the presence of ERα and ERβ, respectively (lanes 3 and 4, rows 1 and 2). ICI treatment significantly reduced the level of total ERα (Fig. 3B, lane 4, row 3) as reported previously (13, 18, 54). Since SERMs do not promote interaction between SRC coactivators and ER in HEK293T (data not shown) and other cell lines (23, 48), the data are consistent with a requirement for a direct binding between ER and SRC-3 for induction of coactivator phosphorylation.
AF-1 and LBD/AF-2 of ERα are required for maximal induction of SRC-3 phosphorylation.
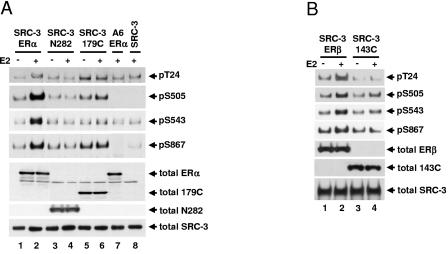
The results above demonstrated that ER is required for E2-induced SRC-3 phosphorylation and implied that an interaction, either direct or indirect, between ER and SRC-3 is required. In order to investigate the contribution of the AF-1 and AF-2 domains of ERα for E2-induced SRC-3 phosphorylation, wild-type full-length ERα and an ERα truncation mutant, ERα1-282 (N282) lacking AF-2/LBD or ERα179-595 (179C) lacking AF-1, together with Flag-tagged SRC-3, were transfected into HEK293T cells. Cells were then treated, and SRC-3 phosphorylation was examined. As shown in Fig. 4A, N282 did not support E2-induced SRC-3 phosphorylation (lanes 3 and 4), which is not surprising since N282 lacks an LBD and is not responsive to E2. The 179C mutant supported only minimal E2-induced SRC-3 phosphorylation at S505 but not at other sites as represented by T24, S543, and S867 (Fig. 4A, lanes 5 and 6). Similarly, an ERβ truncation mutant, 143C (aa 143 to 530), lacking AF-1, supported only minimal E2-induced SRC-3 phosphorylation at S505 and S543 but not at other sites as represented by T24 and S867 (Fig. 4B). These data reveal that the AF-1 or LBD/AF-2 domains are required for maximal induction of E2-induced coactivator phosphorylation.
FIG. 4.
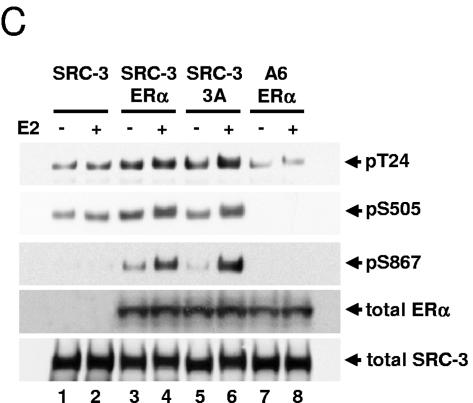
Both AF-1 and LBD/AF-2 domains of ERα are required for maximal induction of SRC-3 phosphorylation in response to E2. HEK293T cells were cotransfected with either Flag-SRC-3 (SRC-3) or Flag-SRC-3 phosphorylation mutant (A6) together with either the full-length wild-type ERα (ERα) (A and C), or ERα1-282 (N282) or ERα179-595 (179C) (A), or the full-length wild-type ERβ (ERβ) or ERβ143-530 (143C) (B), or ERα AF-1 phosphorylation mutant 3A (S104/106/118A) (C). Forty-eight hours posttransfection, vehicle or E2 (10 nM) was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. Immunoblots were probed with SRC-3 phosphorylation state-specific antibodies for T24, S505, S543, and S867. Total cellular levels of wild-type ER and its mutants and SRC-3 were also determined by immunoblotting total cell lysates separated by SDS-PAGE with appropriate antibodies.
To further test the role of AF-1 in SRC-3 phosphorylation, an ERα phosphorylation mutant (S104A/S106A/S118A as 3A) was used to determine whether phosphorylation of AF-1 is required. HEK293T cells were transfected with either wild-type ERα or its mutant 3A together with Flag-tagged SRC-3 and treated as described above. Interestingly, this ERα AF-1 phosphorylation mutant could still induce SRC-3 phosphorylation, at a level comparable to the wild-type ERα (Fig. 4C), suggesting that phosphorylation of ERα is not a prerequisite for promoting SRC-3 phosphorylation.
The coactivator-binding groove of ERα is required for E2-induced SRC-3 phosphorylation.
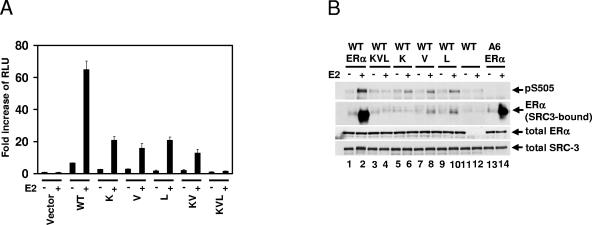
It has been shown that the amino acids K362, V376, and L539 of the ERα LBD are among the residues critical for forming a coactivator-binding groove in response to E2 (38). In order to investigate whether this surface is essential for E2-induced SRC-3 phosphorylation, several mutations which disrupt the direct interaction of ERα with coactivator were examined for their ability to impact E2 induction of SRC-3 phosphorylation in HEK293T cells. The mutants (the single mutants K362D [K], V376D [V], and L539A [L]; the double mutant K362D/V376D [KV]; and the triple mutant K362D/V376D/L539A [KVL]) were transcriptionally impaired as demonstrated in a luciferase reporter assay (Fig. 5A), consistent with previous reports on the effect of the single mutations (34). As shown in Fig. 5B, the ERα single mutants (K, V, or L) were severely compromised in their ability to support E2-induced SRC-3 phosphorylation (lanes 5 to 10, row 1), while the triple mutant (KVL) was unable to induce any phosphorylation of SRC-3 (lanes 3 and 4, row 1). As predicted, the E2-induced interaction between these ERα mutants and SRC-3 was greatly reduced or totally abolished (Fig. 5B, lanes 3 to 10, row 2) in comparison to the positive control (lanes 1 and 2, row 2). These data indicated that an intact coactivator-binding groove within the LBD is essential for E2-induced SRC-3 phosphorylation and imply that a direct interaction between SRC-3 and ERα is necessary.
FIG. 5.
The coactivator-binding groove of ERα is required for estradiol-induced and ER-dependent SRC-3 phosphorylation. (A) HEK293T cells were cotransfected with pERE-E1b-Luc and either the empty vector (Vector), the ERα wild type (WT), the ERα single mutants (K362D as K, V376D as V, or L539A as L), the ERα double mutant (K362D/V376D as KV), or the ERα triple mutant (K362D/V376D/L539A as KVL). Twenty-four hours posttransfection, vehicle or E2 (10 nM) was added for an additional 20 h before cells were harvested for luciferase assays. The error bars represent the standard errors of the means of three individual experiments. (B) HEK293T cells were transfected with either Flag-SRC-3 or Flag-SRC-3 phosphorylation mutant (A6) together with full-length wild-type, single mutant, or triple mutant forms of ERα. Forty-eight hours posttransfection, vehicle or E2 (10 nM) was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. The level of SRC-3 phosphorylated at S505 was assessed by Western blotting using an SRC-3 phosphorylation state-specific antibody. The level of SRC-3-bound ERα was determined by immunoblotting with an anti-ERα antibody. Total cellular levels of wild-type ERα and its mutants and SRC-3 were also determined by immunoblotting total cell lysates separated by SDS-PAGE with appropriate antibodies.
The LXXLL motifs of SRC-3 are required for E2-induced SRC-3 phosphorylation.
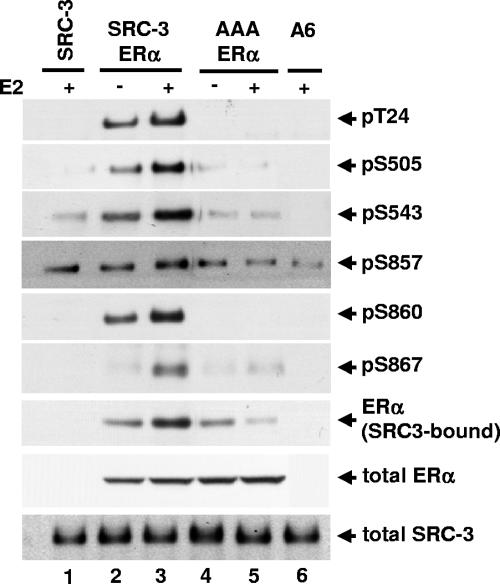
SRC family members contain several LXXLL motifs located in their receptor-interacting domains that mediate their direct binding to the LBD of nuclear receptors such as ERα. The SRC-3-AAA mutant has two amino acid substitutions within each of its three NR boxes, such that the LXXLL motifs are mutated to LXXAA (36); this abolishes the ligand-dependent binding between SRC-3 and the ERα LBD (10, 12, 21, 40). To investigate whether the SRC-3-AAA mutant can be phosphorylated in response to E2, plasmids for the SRC-3 mutant and ERα were transfected into HEK293T cells. As shown in Fig. 6, unlike the wild-type SRC-3 (lanes 2 and 3), the AAA mutant could not be phosphorylated in response to E2 treatment (lanes 4 and 5), substantiating the notion that a direct ligand-induced interaction between coactivator and ERα is necessary for E2-dependent SRC-3 phosphorylation. The interaction observed between the AAA mutant and ERα in the absence of E2 (Fig. 6, lane 4, row 7) likely reflects a hormone- and LXXLL-independent interaction of the ERα AF-1 domain with SRC-3 (72), which is inhibited by E2 as a result of ligand-induced interactions of the N and C termini of the receptor (41).
FIG. 6.
The receptor-interacting domain of SRC-3 is required for estradiol-induced SRC-3 phosphorylation. HEK293T cells were transfected with either wild-type Flag-SRC-3 (SRC-3) or SRC-3 mutant (AAA) having all three LXXLL motifs in the receptor-interacting domain mutated to LXXAA, together with wild-type ERα. Forty-eight hours posttransfection, vehicle or E2 (10 nM) was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. The level of SRC-3 phosphorylated at each of the six phosphorylation sites was assessed by immunoblotting with each SRC-3 phosphorylation state-specific antibody (rows 1 to 6). The level of SRC-3-bound ERα was determined by immunoblotting with an anti-ERα antibody (row 7). Total cellular levels of wild-type SRC-3 and its mutant and ERα were also determined by immunoblotting total cell lysates separated by SDS-PAGE with appropriate antibodies (rows 8 and 9).
Nuclear localization of ERα is not required for E2-induced SRC-3 phosphorylation.
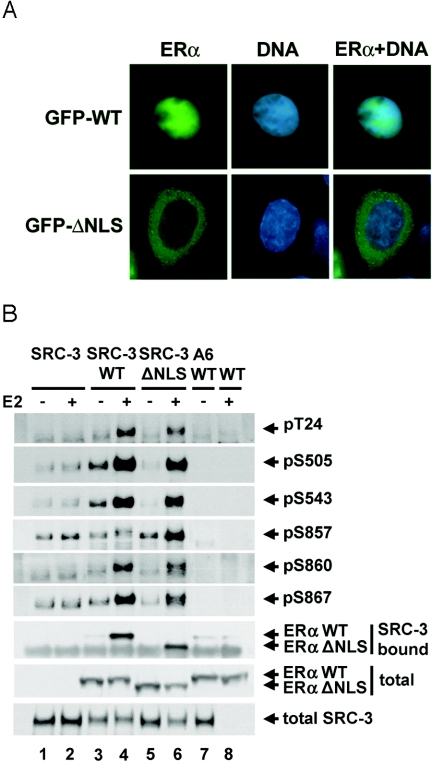
To determine whether phosphorylation of SRC-3 can take place in the cytoplasm, an ERα mutant (ERαΔNLS) lacking amino acids 250 to 303, which encompass the ERα NLS, was used. This mutant was transcriptionally impaired in a luciferase reporter assay and is localized predominantly in the cytoplasm in the absence of ligand (78). In order to determine whether SRC-3 overexpression or ligand would affect the mutant vs subcellular localization, cells were transfected with vectors for Flag-tagged SRC-3 and either wild-type or mutant ERα and then treated with either E2 or vehicle. In the presence of ligand, GFP-ERαΔNLS was localized in the cytoplasm, as opposed to the wild-type ERα (GFP-hERαWT), which was localized primarily in the nucleus (Fig. 7A), regardless of the presence or absence (data not shown) of ligand. Next, the ability of the mutant to promote coactivator phosphorylation was tested. As for wild-type ERα, the mutant was able to induce SRC-3 phosphorylation at all six sites (Fig. 7B), indicating that nuclear localization of ERα may not be required for inducing SRC-3 phosphorylation in response to E2. Furthermore, the mutant could still bind to SRC-3 in the presence of E2 (Fig. 7B, lanes 5 and 6, row 7). It is possible that there is a small amount of the ΔNLS mutant receptor localized in the nucleus (78) which may contribute to SRC-3 phosphorylation. However, it should be noted that, under the conditions used for these assays, there is insufficient nuclear ERαΔNLS to induce the transcription of an ERE-containing reporter gene (data not shown) consistent with a previous report (78).
FIG. 7.
Nuclear localization of ERα is not required for estradiol-induced SRC-3 phosphorylation. (A) HEK293T cells were cotransfected with GFP-tagged wild-type ERα (GFP-WT) or ERα NLS deletion mutant lacking aa 250 to 303 (GFP-ΔNLS), together with Flag-SRC-3. Forty-eight hours posttransfection, vehicle or E2 (10 nM) was added for 1 h. Cell fixing and preparation were performed, and GFP fluorescence signals were determined microscopically (column 1). DNA was counterstained with DAPI (column 2), and the signals for GFP and DAPI were merged (column 3). (B) HEK293T cells were transfected with either Flag-SRC-3 wild type (SRC-3) or its phosphorylation mutant (A6), together with either wild-type ERα (WT), or ERα mutant (ΔNLS). Forty-eight hours posttransfection, E2 (10 nM) was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. The level of SRC-3 phosphorylated at each of the six phosphorylation sites was assessed by immunoblotting with each SRC-3 phosphorylation state-specific antibody (rows 1 to 6). The level of SRC-3-bound ERα was determined by immunoblotting with an anti-ERα antibody (row 7). Total cellular levels of ERα and SRC-3 were determined by immunoblotting total cell lysates separated by SDS-PAGE with appropriate antibodies (rows 8 and 9).
ERα DNA binding activity is not required for E2-induced SRC-3 phosphorylation.
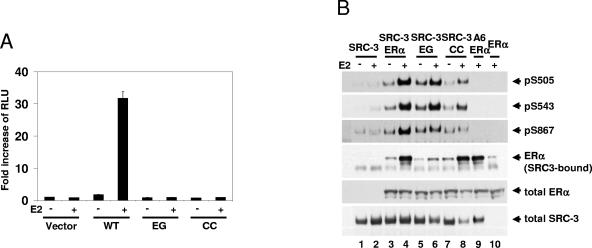
Previous data indicated that E2-induced SRC-3 phosphorylation requires the formation of a complex containing SRC-3 and ER. Assembly of ERα, coactivators, and general transcription factors at target gene promoters is associated with recruitment of protein kinases and receptor phosphorylation (8, 50). Whether E2-dependent SRC-3 phosphorylation requires ERα to be bound to DNA was investigated. The receptor binds directly to estrogen response elements mainly through the P box in the first of the two functionally distinct cysteine-cysteine zinc fingers located within the DBD. Two ERα DBD mutants were tested, ERαCC and ERαEG, each containing a double mutation in different regions of the P box (C202H/C205H as CC and E203A/G204A as EG, respectively). Neither of the two ERα DBD mutants could activate ERE-dependent transcription in a luciferase reporter assay (Fig. 8A). However, both of the CC and EG ERα DBD mutants induced SRC-3 phosphorylation, represented by sites S505, S543, and S867 (Fig. 8B, lanes 5 and 6 and lanes 7 and 8, respectively). The CC mutant appeared to be less effective at supporting SRC-3 phosphorylation than wild-type ERα was (lanes 3 and 4) or the EG mutant, even though the CC mutant binds to SRC-3 as well as does the wild-type receptor. In contrast, a weaker interaction between the EG mutant and SRC-3 was noted (Fig. 8B, row 4). It was observed that E2 treatment resulted in decreased SRC-3 levels in cells transfected with either ERα DBD mutant (Fig. 8B, lanes 5 to 8, row 6); the reason for this is unknown, but it does suggest that ERα can exert control over SRC-3 levels in some contexts.
FIG. 8.
Estradiol-induced and ER-dependent SRC-3 phosphorylation does not require ER DNA binding. (A) HEK293T cells were cotransfected with pERE-E1b-Luc, and either the empty vector (Vector), or vectors for wild-type ERα (WT), or the EG (E203A/G204A) or CC (C202H/C205H) double ERα DNA-binding domain mutant. Twenty-four hours posttransfection, vehicle or E2 (10 nM) was added for an additional 20 h. The luciferase activity was assayed as described in Materials and Methods. (B) HEK293T cells were cotransfected with either Flag-SRC-3 wild type (SRC-3) or its phosphorylation mutant (A6), together with either the wild-type ERα (ERα), or the ERα double mutant EG or CC. Forty-eight hours posttransfection, vehicle or E2 (10 nM) was added for 1 h. Cells were then lysed, and Flag-tagged SRC-3 was immunoprecipitated by anti-Flag antibody and separated by SDS-PAGE. The level of SRC-3 phosphorylated at S505, S543, and S867 was assessed by Western blotting using the SRC-3 phosphorylation state-specific antibody against each site (rows 1 to 3). The level of SRC-3-bound ERα was determined by immunoblotting with an anti-ERα antibody (row 4). Total cellular levels of wild-type ERα and its mutants and SRC-3 were also determined by immunoblotting total cell lysates separated by SDS-PAGE with appropriate antibodies (rows 5 and 6).
DISCUSSION
The phosphorylation status of SRC family members has been shown to be important for their biological functions including their coactivator activities and tumorigenic potentials (17, 20, 55, 74, 75). Hormones and growth factors induce SRC phosphorylation, but the mechanisms are largely unknown. Estrogen can induce posttranslational modifications through either “genomic” or “nongenomic” pathways (3). The “genomic” effects of estradiol are mainly mediated by ERs over the time course of hours to days and relate to an activated receptor's ability to bind to promoters of estrogen-responsive target genes involved in cellular signaling (3). The “nongenomic” effects of estrogen are mediated through activation of cellular signaling pathways independent of transcription and can be either ER dependent or independent, occurring over a time course of seconds to minutes (3). Here we report that E2-induced SRC-3 phosphorylation occurs within minutes and requires a direct interaction between SRC-3 and ERα but does not require nuclear localization or the DNA binding activity of ERα. However, both the AF-1 and LBD/AF-2 domains of ERα are essential for maximal E2 induction of SRC-3 phosphorylation. These results lead to a model in which the posttranslational modification of SRC-3 is the result of a transcription-independent and “nongenomic” action of ER that occurs in an extranuclear complex containing SRC-3 and ERα.
We found that both the ERα AF-1 and AF-2 domains are necessary for robust induction of SRC-3 phosphorylation by E2 and that a direct interaction between the ERα LBD and coactivator is required. The inability of the SERMs, 4HT and raloxifene, to induce SRC-3 phosphorylation is consistent with both of these points. Even though SERMs allow AF-1 to exert its ligand-independent function, they block AF-2 activity and inhibit interaction between coactivators and the LBD of ERs (5, 61). Moreover, steroid-induced phosphorylation of SRC-3 is nuclear receptor specific. Previously, we showed that androgen can induce SRC-3 phosphorylation (75), but in this report we demonstrate that neither PR-B nor GR is able to induce phosphorylation of any of the six known SRC-3 phosphorylation sites in response to their cognate ligands. This dichotomy suggests that the extensive phosphorylation of SRC-3 induced by estrogen and androgen may relate to the well-known growth-promoting actions of their cognate receptors. However, failure to induce extensive phosphorylation was surprising in view of the ability of SRC-3 to coactivate and participate in nuclear PR-B- and GR-dependent gene expression, albeit in conjunction with SRC-1 and SRC-2, respectively (33). Conceivably, PR or GR and their cognate ligands may induce phosphorylation of SRC-3 at sites distinct from those known to be phosphorylated in response to ER or AR ligands. Nonetheless, the differences between ER/AR and GR/PR and their cognate ligands, relative to their abilities to induce SRC-3 phosphorylation, indicate that the relative importance of SRC-3 phosphorylation at the six sites tested can be receptor specific.
Consistent with the model that E2-induced coactivator phosphorylation can be mediated through the transcription-independent and “nongenomic” signaling of ER, we showed that ERα nuclear localization is not required for promoting SRC-3 phosphorylation. It has been shown that both ER and SRC-3 shuttle between the cytoplasm and the nucleus (14, 39, 53), and the collective data indicate that SRC-3 phosphorylation can occur in both cytoplasm and nucleus and that it generally promotes nuclear localization of SRC-3 (53, 68, 71, 74). For example, TNF-α induces phosphorylation and nuclear translocation of SRC-3, and nuclear SRC-3 is hyperphosphorylated compared to the cytoplasmic fraction (74), indicating that SRC-3 may be partially phosphorylated in the cytoplasm and further phosphorylated after translocation to the nucleus and recruitment to the promoter. However, the precise extent to which each of the six known sites of SRC-3 is phosphorylated in each component is unknown.
Our model, in which E2-induced SRC-3 phosphorylation occurs in a complex with either ERα or ERβ, suggests that the high proximity of receptor and coactivator may enable the same kinase to efficiently phosphorylate SRC-3 and ER in response to E2. Indeed, the kinases ERK1/2, p38 MAPK, protein kinase A, and IKKα can phosphorylate both ER and SRC family members (7, 24, 29, 35, 50, 56, 75). However, the mechanism of kinase recruitment to the complex is not clear. It is possible that E2-bound ERα can first recruit kinases via its AF-1 or AF-2 domain, allowing subsequent phosphorylation of SRC-3, or vice versa. Alternatively, a kinase could be recruited by either ER or SRC-3 to an E2-induced preexisting complex of SRC-3 and ERα, followed by phosphorylation of both molecules. Although our current data cannot distinguish between these models, or whether the ER is attached to the membrane (31, 52, 62), our results indicate that phosphorylation of three of the major phosphorylation sites of the ERα AF-1 domain is not a prerequisite for SRC-3 phosphorylation.
In addition to its phosphorylation by an E2/ER-dependent pathway, SRC-3 phosphorylation also can be induced by growth factors and cytokines (17, 75). Previously, we reported that TNF-α induces phosphorylation of SRC-3 in an ER-independent fashion and that the patterns of phosphorylated residues induced by TNF-α and E2 overlapped but were not identical (75). Moreover, these distinct phosphorylation patterns lead to distinct effects on SRC-3 association with nuclear receptors, other transcription factors (e.g., NF-κB), and coregulators (e.g., CBP and CARM1), leading to distinct potentials for maximal transcriptional output (75). It is highly probable that the stimuli encountered by SRC-3, and hence its phosphorylation pattern and coactivator activity, will be cell and tissue specific. For example, in HER2-overexpressing breast cancers, SRC-3 phosphorylation induced by HER2 signaling may facilitate coactivator function under conditions of low E2 or tamoxifen exposure and thereby contribute to endocrine resistance (4, 49, 60). Indeed, tamoxifen induces SRC-3 phosphorylation as shown by its mobility shift and phosphatase treatment and serves as an agonist of ER/SRC-3-dependent transcription in HER2-overexpressing breast cancer cells (60). However, the status of SRC-3 phosphorylation at each of the six sites in the presence of high levels of HER2 has not been investigated yet. In contrast, estrogen may attenuate cellular responses to inflammatory stimuli, such as the cytokine TNF-α, because of E2-induced phosphorylation of SRC-3 that renders the coactivator less effective at cytokine target genes.
Taken together, our results expand the established model of action of coactivators relative to their activation of gene expression. This “classical” model indicates that E2-bound ER is first recruited to the promoter of its target gene, which in turn recruits coactivators and other transcription factors. Our data provide the first indication of an E2-induced assembly of an extranuclear SRC-3- and ERα-containing complex prior to promoter binding, which is essential for E2-induced SRC-3 phosphorylation and thus for its biological functions. The complex could serve as a convergence point for cross talk between cell signaling and ER pathways, resulting in fine tuning of coactivator activation to support receptor nuclear functions. Since this step serves as a middle step in the multistep process of target gene activation by nuclear receptor and coactivator from their upstream “nongenomic” to downstream “genomic” responses leading to activation of transcription, it also is an intervention point for drug development for diseases sensitive to SRC-3 action.
Acknowledgments
We are grateful to N. L. Weigel, P. H. Brown, R. X. Song, and H.-W. Chen for reagents and to M. A. Mancini for reagents and microscopy assistance.
This work was supported by grants from the National Institutes of Health to B.W.O. (HD08818 and NIDDK NURSA) and to C.L.S. (DK53002).
REFERENCES
- 1.Agoulnik, I. U., X. W. Tong, D. C. Fischer, K. Korner, N. E. Atkinson, D. P. Edwards, D. R. Headon, N. L. Weigel, and D. G. Kieback. 2004. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J. Clin. Endocrinol. Metab. 89:6340-6347. [DOI] [PubMed] [Google Scholar]
- 2.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 3.Bjornstrom, L., and M. Sjoberg. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19:833-842. [DOI] [PubMed] [Google Scholar]
- 4.Bouras, T., M. C. Southey, and D. J. Venter. 2001. Overexpression of the steroid receptor coactivator AIB1 in breast cancer correlates with the absence of estrogen and progesterone receptors and positivity for p53 and HER2/neu1. Cancer Res. 61:903-907. [PubMed] [Google Scholar]
- 5.Bramlett, K. S., and T. P. Burris. 2002. Effects of selective estrogen receptor modulators (SERMs) on coactivator nuclear receptor (NR) box binding to estrogen receptors. Mol. Genet. Metab. 76:225-233. [DOI] [PubMed] [Google Scholar]
- 6.Chauchereau, A., L. Amazit, M. Quesne, A. Guiochon-Mantel, and E. Milgrom. 2003. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J. Biol. Chem. 278:12335-12343. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., P. E. Pace, R. C. Coombes, and S. Ali. 1999. Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol. Cell. Biol. 19:1002-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, D., T. Riedl, E. Washbrook, P. E. Pace, R. C. Coombes, J.-M. Egly, and S. Ali. 2000. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol. Cell 6:127-137. [PubMed] [Google Scholar]
- 9.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, K. M., M. Dutertre, A. El-Gharbawy, B. G. Rowan, N. L. Weigel, and C. L. Smith. 2003. Mechanistic differences in the activation of estrogen receptor-alpha (ER alpha)- and ER beta-dependent gene expression by cAMP signaling pathway(s). J. Biol. Chem. 278:12834-12845. [DOI] [PubMed] [Google Scholar]
- 12.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauvois, S., S. Danielian, R. White, and M. G. Parker. 1992. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. USA 89:4037-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauvois, S., R. White, and M. G. Parker. 1993. The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J. Cell Sci. 106:1377-1388. [DOI] [PubMed] [Google Scholar]
- 15.Deblois, G., and V. Giguere. 2003. Ligand-independent coactivation of ERalpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J. Steroid Biochem. Mol. Biol. 85:123-131. [DOI] [PubMed] [Google Scholar]
- 16.Filardo, E. J. 2002. Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. J. Steroid Biochem. Mol. Biol. 80:231-238. [DOI] [PubMed] [Google Scholar]
- 17.Font de Mora, J. F., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, M. K., L. A. Nemmers, Jr., W. C. Beckman, V. L. Davis, S. W. Curtis, and K. S. Korach. 1991. The mechanism of ICI 164,384 antiestrogenicity involves rapid loss of estrogen receptor in uterine tissue. Endocrinology 129:2000-2010. [DOI] [PubMed] [Google Scholar]
- 19.Gnanapragasam, V. J., H. Y. Leung, A. S. Pulimood, D. E. Neal, and C. N. Robson. 2001. Expression of RAC 3, a steroid hormone receptor co-activator in prostate cancer. Br. J. Cancer 85:1928-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory, C. W., X. Fei, L. A. Ponguta, B. He, H. M. Bill, F. S. French, and E. M. Wilson. 2004. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J. Biol. Chem. 279:7119-7130. [DOI] [PubMed] [Google Scholar]
- 21.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 22.Hoang, T., I. S. Fenne, C. Cook, B. Borud, M. Bakke, E. A. Lien, and G. Mellgren. 2004. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J. Biol. Chem. 279:49120-49130. [DOI] [PubMed] [Google Scholar]
- 23.Jaber, B. M., R. Mukopadhyay, and C. L. Smith. 2004. Estrogen receptor-alpha interaction with the CREB binding protein coactivator is regulated by the cellular environment. J. Mol. Endocrinol. 32:307-323. [DOI] [PubMed] [Google Scholar]
- 24.Kato, S., H. Endoh, Y. Masuhiro, T. Kitamoto, S. Uchiyama, H. Sasaki, S. Masushige, Y. Gotoh, E. Nishida, H. Kawashima, D. Metzger, and P. Chambon. 1995. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491-1494. [DOI] [PubMed] [Google Scholar]
- 25.Klein-Hitpass, L., G. U. Ryffel, E. Heitlinger, and A. C. B. Cato. 1988. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 16:647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korach, K. S. 1994. Insights from the study of animals lacking functional estrogen receptor. Science 266:1524-1527. [DOI] [PubMed] [Google Scholar]
- 27.Kuang, S.-Q., L. Liao, H. Zhang, A. V. Lee, B. W. O'Malley, and J. Xu. 2004. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 64:1875-1885. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, V., S. Green, G. Stack, M. Berry, J.-R. Jin, and P. Chambon. 1987. Functional domains of the human estrogen receptor. Cell 51:941-951. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H., and W. Bai. 2002. Regulation of estrogen receptor nuclear export by ligand-induced and p38-mediated receptor phosphorylation. Mol. Cell. Biol. 22:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeGoff, P., M. M. Montano, D. J. Schodin, and B. S. Katzenellenbogen. 1994. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J. Biol. Chem. 269:4458-4466. [PubMed] [Google Scholar]
- 31.Levin, E. R. 2003. Bidirectional signaling between the estrogen receptor and the epidermal growth factor receptor. Mol. Endocrinol. 17:309-317. [DOI] [PubMed] [Google Scholar]
- 32.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, X., J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-α and coactivator turn-over and for efficient estrogen receptor-α transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 35.Lopez, G. N., C. W. Turck, F. Schaufele, M. R. Stallcup, and P. J. Kushner. 2001. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 co-activator activity. J. Biol. Chem. 276:22177-22182. [DOI] [PubMed] [Google Scholar]
- 36.Louie, M. C., J. X. Zou, A. Rabinovich, and H. W. Chen. 2004. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol. Cell. Biol. 24:5157-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maggiolini, M., A. Vivacqua, G. Fasanella, A. G. Recchia, D. Sisci, V. Pezzi, D. Montanaro, A. M. Musti, D. Picard, and S. Ando. 2004. The G protein-coupled receptor GPR30 mediates c-fos up-regulation by 17beta-estradiol and phytoestrogens in breast cancer cells. J. Biol. Chem. 279:27008-27016. [DOI] [PubMed] [Google Scholar]
- 38.Mak, H. Y., S. Hoare, P. M. A. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruvada, P., C. T. Baumann, G. L. Hager, and P. M. Yen. 2003. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J. Biol. Chem. 278:12425-12432. [DOI] [PubMed] [Google Scholar]
- 40.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T.-M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInerney, E. M., M.-J. Tsai, B. W. O'Malley, and B. S. Katzenellenbogen. 1996. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc. Natl. Acad. Sci. USA 93:10069-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 43.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 44.Metzger, D., S. Ali, J. M. Bornert, and P. Chambon. 1995. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J. Biol. Chem. 270:9535-9542. [DOI] [PubMed] [Google Scholar]
- 45.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 96:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson, S., S. Mäkelä, E. Treuter, M. Tujague, J. Thomsen, G. Andersson, E. Enmark, K. Pettersson, M. Warner, and J.-Å. Gustafsson. 2001. Mechanisms of estrogen action. Physiol. Rev. 81:1535-1565. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa, S., S. Inoue, T. Watanabe, H. Hiroi, A. Orimo, T. Hosoi, Y. Ouchi, and M. Muramatsu. 1998. The complete primary structure of human estrogen receptor β (hERβ) and its heterodimerization with ERα in vivo and in vitro. Biochem. Biophys. Res. Commun. 243:122-126. [DOI] [PubMed] [Google Scholar]
- 48.Onate, S. A., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 49.Osborne, C. K., V. Bardou, T. A. Hopp, G. C. Chamness, S. G. Hilsenbeck, S. A. W. Fuqua, J. Wong, D. C. Allred, G. M. Clark, and R. Schiff. 2003. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 95:353-361. [DOI] [PubMed] [Google Scholar]
- 50.Park, K. J., V. Krishnan, B. W. O'Malley, Y. Yamamoto, and R. B. Gaynor. 2005. Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol. Cell 18:71-82. [DOI] [PubMed] [Google Scholar]
- 51.Picard, D., V. Kumar, P. Chambon, and K. R. Yamamoto. 1990. Signal transduction by steroid hormones: nuclear localization is differentially regulated in estrogen and glucocorticoid receptors. Cell Regul. 1:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietras, R. J., I. Nemere, and C. M. Szego. 2001. Steroid hormone receptors in target cell membranes. Endocrine 14:417-427. [DOI] [PubMed] [Google Scholar]
- 53.Qutob, M. S., R. N. Bhattacharjee, E. Pollari, S. P. Yee, and J. Torchia. 2002. Microtubule-dependent subcellular redistribution of the transcriptional coactivator p/CIP. Mol. Cell. Biol. 22:6611-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reese, J. C., and B. S. Katzenellenbogen. 1992. Examination of the DNA-binding ability of estrogen receptor in whole cells: implications for hormone-independent transactivation and the actions of antiestrogens. Mol. Cell. Biol. 12:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowan, B. G., N. Garrison, N. L. Weigel, and B. W. O'Malley. 2000. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol. Cell. Biol. 20:8720-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowan, B. G., N. L. Weigel, and B. W. O'Malley. 2000. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J. Biol. Chem. 275:4475-4483. [DOI] [PubMed] [Google Scholar]
- 57.Schwabe, J. W. R., L. Chapman, J. T. Finch, and D. Rhodes. 1993. The crystal structure of the complex between the oestrogen receptor DNA-binding domain and DNA at 2.4A: how receptors discriminate between their response elements. Cell 75:567-578. [DOI] [PubMed] [Google Scholar]
- 58.Sheppard, K. A., D. W. Rose, Z. K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M. G. Rosenfeld, C. K. Glass, and T. Collins. 1999. Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol. 19:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 60.Shou, J., S. Massarweh, C. K. Osborne, A. E. Wakeling, S. Ali, H. Weiss, and R. Schiff. 2004. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 96:926-935. [DOI] [PubMed] [Google Scholar]
- 61.Smith, C. L., and B. W. O'Malley. 2004. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 25:45-71. [DOI] [PubMed] [Google Scholar]
- 62.Song, R. X., R. A. McPherson, L. Adam, Y. Bao, M. Shupnik, R. Kumar, and R. J. Santen. 2002. Linkage of rapid estrogen action to MAPK activation by ERα-Shc association and Shc pathway activation. Mol. Endocrinol. 16:116-127. [DOI] [PubMed] [Google Scholar]
- 63.Stenoien, D. L., M. G. Mancini, K. Patel, E. A. Allegretto, C. L. Smith, and M. A. Mancini. 2000. Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol. Endocrinol. 14:518-534. [DOI] [PubMed] [Google Scholar]
- 64.Suen, C.-S., T. J. Berrodin, R. Mastroeni, B. J. Cheskis, C. R. Lyttle, and D. E. Frail. 1998. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J. Biol. Chem. 273:27645-27653. [DOI] [PubMed] [Google Scholar]
- 65.Takeshita, A., G. R. Cardonna, N. Koibuchi, C.-S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 66.Tamrazi, A., K. E. Carlson, A. L. Rodriguez, and J. A. Katzenellenbogen. 2005. Coactivator proteins as determinants of estrogen receptor structure and function: spectroscopic evidence for a novel coactivator-stabilized receptor conformation. Mol. Endocrinol. 19:1516-1528. [DOI] [PubMed] [Google Scholar]
- 67.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 68.Torres-Arzayus, M. I., J. F. de Mora, J. Yuan, F. Vazquez, R. Bronson, M. Rue, W. R. Sellers, and M. Brown. 2004. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 6:263-274. [DOI] [PubMed] [Google Scholar]
- 69.Tremblay, A., G. B. Tremblay, F. Labrie, and V. Giguère. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activation function AF-1. Mol. Cell 3:513-519. [DOI] [PubMed] [Google Scholar]
- 70.Tzukerman, M. T., A. Esty, D. Santiso-Mere, P. Danielian, M. G. Parker, R. B. Stein, J. W. Pike, and D. P. McDonnell. 1994. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol. Endocrinol. 8:21-30. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Z., D. W. Rose, O. Hermanson, F. Liu, T. Herman, W. Wu, D. Szeto, A. Gleiberman, A. Krones, K. Pratt, R. Rosenfeld, C. K. Glass, and M. G. Rosenfeld. 2000. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl. Acad. Sci. USA 97:13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Webb, P., P. Nguyen, J. Shinsako, C. Anderson, W. Feng, M. P. Nguyen, D. Chen, S.-M. Huang, S. Subramanian, E. McKinerney, B. S. Katzenellenbogen, M. R. Stallcup, and P. J. Kushner. 1998. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol. 12:1605-1618. [DOI] [PubMed] [Google Scholar]
- 73.Wong, C. W., C. McNally, E. Nickbarg, B. S. Komm, and B. J. Cheskis. 2002. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc. Natl. Acad. Sci. USA 99:14783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Wu, R.-C., J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2002. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by IκB kinase. Mol. Cell. Biol. 22:3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIBI coactivator integrate genomic responses to multiple cellular signaling pathways. Mol. Cell 15:937-949. [DOI] [PubMed] [Google Scholar]
- 76.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu, J., and B. W. O'Malley. 2002. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Rev. Endocr. Metab. Disord. 3:185-192. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, Z., B. Maier, R. J. Santen, and R. X. Song. 2002. Membrane association of estrogen receptor alpha mediates estrogen effect on MAPK activation. Biochem. Biophys. Res. Commun. 294:926-933. [DOI] [PubMed] [Google Scholar]
- 79.Zhou, G., Y. Hashimoto, I. Kwak, S. Y. Tsai, and M.-J. Tsai. 2003. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol. Cell. Biol. 23:7742-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]