Abstract
Archaeal RNA polymerases (RNAPs) are recruited to promoters through the joint action of three basal transcription factors: TATA-binding protein, TFB (archaeal homolog of TFIIB), and TFE (archaeal homolog of TFIIE). Our results demonstrate several new insights into the mechanisms of TFB and TFE during the transcription cycle. (i) The N-terminal Zn ribbon of TFB displays a surprising degree of redundancy for the recruitment of RNAP during transcription initiation in the archaeal system. (ii) The B-finger domain of TFB participates in transcription initiation events by stimulating abortive and productive transcription in a recruitment-independent function. TFB thus combines physical recruitment of the RNAP with an active role in influencing the catalytic properties of RNAP during transcription initiation. (iii) TFB mutations are complemented by TFE, thereby demonstrating that both factors act synergistically during transcription initiation. (iv) An additional function of TFE is to dynamically alter the nucleic acid-binding properties of RNAP by stabilizing the initiation complex and destabilizing elongation complexes.
A comprehensive understanding of the molecular mechanisms operating within pro- and eukaryotic basal transcriptional machineries is a formidable intellectual goal. The combination of biochemical, genetic, and structural approaches to study the functions of basal factors and RNA polymerases (RNAPs) has started to provide very detailed insights into some of these events (reviewed in references 6 and 33). These efforts have more recently been boosted by the development of unique experimental tools based on archaeal model systems. Archaea are prokaryotes, but they contain a simplified transcriptional apparatus that is very similar to the eukaryotic core RNAPII system (reviewed in references 4 and 37). This machinery consists of TATA-binding protein (TBP), TFB (a homolog of eukaryotic TFIIB), TFE (a homolog of the eukaryotic TFIIEα subunit), and RNAP (homologous to eukaryotic RNAPII). Archaeal TBPs, like their eukaryotic counterparts, are responsible for the recognition of TATA elements that are located approximately 20 to 30 nucleotides upstream of the transcription start site. The DNA-bound TBP is subsequently recognized by TFB, which stabilizes the TBP/DNA complex and often enhances the sequence specificity of promoter recognition by making additional contacts with the B-recognition element (32, 40) located next to the TATA element. This TBP/TFB complex is then capable of recruiting RNAPs for promoter-specific transcription. TFE stimulates transcription from suboptimal promoters but is not strictly essential for in vitro transcription (5, 22). Unlike eukaryotic systems, no other basal factors (such as TFIIA, TFIIF, and TFIIH) are required for transcript initiation/promoter escape, and none of these steps are dependent on nucleoside triphosphate (NTP) hydrolysis to induce specific conformational changes.
The entire transcriptional machinery derived from the hyperthermophilic archaeon Methanocaldococcus jannaschii has recently been reconstituted in recombinant form (47). The recombinant system is fully responsive to stimulation by transcriptional activators and thus faithfully mimics all known functions of archaeal transcription systems (36). In this study we sought to investigate the molecular mechanism governing the early steps of RNAP recruitment and transcription initiation, with special emphasis on the functional interplay of TFB and TFE with RNAP. The results reveal previously undocumented interactions between the basal factors TFB and TFE with RNAP that occur mostly at the postrecruitment stage and culminate in a carefully orchestrated modulation of core RNAP functions. Our data also demonstrate a surprising degree of redundancy of the archaeal N-terminal zinc ribbon of TFB, a novel transcription stimulatory role of the archaeal B-finger domain and a role for TFE in promoter melting and template loading. All these processes have the potential to influence key aspects of transcript initiation and promoter escape and therefore have implications for our understanding of core RNAP functions and gene expression mechanisms in archaeal and eukaryotic systems.
MATERIALS AND METHODS
Recombinant RNA polymerases and transcription factors.
The Methanocaldococcus jannaschii RNAP subunits and TBP were cloned, expressed, and purified as described previously (47). The recombinant wild-type (12-subunit) RNAP was assembled in vitro and purified by heat inactivation and size exclusion chromatography on an S100HR column as described previously (47).
The open reading frames encoding M. jannaschii TFB and TFE were cloned from genomic DNA and subcloned into pET21(a)+, eliminating the stop codon using 5′ NdeI and 3′ XhoI restriction sites. This creates in-frame fusions of TFB and TFE to a vector-encoded C-terminal His6 tag. The TFB domain deletion variants were created by a PCR splice by overlap extension (46) strategy and subcloned as described above. A complete list of oligonucleotide sequences used in this study can be requested from the authors. All recombinant expression plasmids were transformed into Escherichia coli BL21(DE3)Rosetta (Novagen). Expression cultures were typically expanded using 50 ml overnight starter culture per liter expression culture at 37°C under vigorous shaking and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an A600 of ∼0.6 to 0.8 in rich medium. Three hours after induction, the expression cultures were harvested and soluble recombinant proteins were extracted in N500 buffer (500 mM sodium chloride, 20 mM Tris-acetate, 10 mM magnesium acetate, 10% glycerol, 100 μM zinc sulfate, 1 mM β-mercaptoethanol) and purified using cobalt affinity chromatography according to the manufacturer's instructions (Talon resin; BD Clontech).
In vitro transcription assays.
Nonspecific and promoter-directed transcription assays were performed as described previously (47). Briefly, the nonspecific transcription assay measures the incorporation of α-32P-labeled UTP ([α-32P]rUTP) into trichloroacetic acid-insoluble material using activated calf thymus DNA template (Fluka). Assays were performed using 0.5 μg RNAP for 30 min at 70°C. The trichloroacetic acid precipitate was collected on glass filters and quantitated by scintillation counting. In the promoter-directed transcription assay, 100 ng TBP and 100 ng TFB were incubated with 0.1 to 1.0 μg RNAP and 100 ng supercoiled template (pGEM-SSV T6) for 30 min at 70°C. Under these conditions both basal factors are in excess to ensure saturation of the promoter. The generated RNA transcripts were subsequently detected by primer extension, and the resulting cDNA was visualized and quantitated by either autoradiography or phosphorimaging (Fuji FLA 5000).
Transcription assays using the 3′-tailed template were performed by incubating 2.0 μg RNAP (200 nM) ± 1.0 μg TFB (1 μM) with 130 nM linear template DNA (see Fig. 3C); 300 μM rGTP, rCTP, and rUTP; 10 μM rATP; 1 μCi [α-32P]rATP in 1× transcription buffer (50 mM Tris-HCl [pH 7.5], 75 mM KCl, 25 mM MgCl2, 10 mM dithiothreitol) for 1 h at 65°C. The products were separated on a 22% denaturing polyacrylamide gel and visualized by autoradiography (Kodak Biomax MR film) or a Fuji BAS phosphorimager device. The abortive transcription assays were performed by incubating 2.0 μg RNAP with or without 1.0 μg TFB or TFB-ΔB with 0.6 μg DNase I-treated calf thymus DNA template, 1 μM dinucleotide ApG (adenine and phosphorylated guanine) or ApA or UpG, 10 μM rATP, 1 μCi [α-32P]rATP in 1× transcription buffer for 1 h at 65°C. The products were separated and visualized as described above.
FIG. 3.
The TFB B-finger is stimulating RNAP in a recruitment-independent manner. (A) M. jannaschii TFB stimulates RNAP activity in a nonspecific transcription assay. This effect is dependent on the B-finger domain but independent of the core domain (reaction mixtures contain 1 μg TFB or TFB variants and 1.0 μg RNAP). (B) Nonspecific abortive initiation assay with RNAP in the presence (+) or absence (−) of TFB using one of the three dinucleotide substrates (ApG, ApA, or UpG) (reaction mixtures contain 1.0 μg TFB or TFB-ΔB and 2.0 μg RNAP). 32P-labeled RNA 9-mer marker was run in one lane. (C) Factor-independent transcription initiation using the 3′-tailed template is stimulated by TFB (reaction mixtures contain 1.0 μg TFB or TFB-ΔB and 2.0 μg RNAP). nt, nucleotides. (D) RNAP forms heparin-stable complexes with the 3′-tailed template in the presence (+) of TFB, and this effect is dependent on the B-finger (reaction mixtures contain 0.5 μg TFB or TFB-ΔB and 2.0 μg RNAP). (E) Sequence of the 3′-tailed template based on the SSV6 promoter; the transcription start site of the double-stranded promoter is indicated by bold type.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed using 32P-labeled oligonucleotides as probes. Either template or nontemplate strand was labeled using [γ-32P]rATP (5,000 Ci/mmol, Amersham) and polynucleotide kinase (Roche) and annealed to the unlabeled complementary strand (in 5:4 molar ratio excess of nonlabeled oligonucleotide) or an RNA 9-mer corresponding to a short transcript by heating to 100°C for 2 min and letting the mixture cool to room temperature for 5 min.
32P-labeled probe (0.1 pmol) was incubated with recombinant TBP (0.5 μg), TFB (0.5 μg), TFE (0.5 μg), and RNAP (2.0 μg) in HNME buffer (40 mM HEPES, pH 7.3, 250 mM sodium chloride, 2.5 mM magnesium chloride, 0.1 mM EDTA, 5% glycerol, and 10 mM dithiothreitol) containing 1 μg bovine serum albumin in a total volume of 15 μl for 20 min at 65°C. One microliter of heparin (0.5 mg/ml; Sigma) was added to suppress nonspecific protein-nucleic acid interactions, and the reaction mixture was incubated for another 20 min at 65°C. Ten microliters of the reaction mixture was transferred into 6 μl of native Tris-glycine loading buffer and loaded onto a native 4 to 20% Tris-glycine gradient gel (Invitrogen). The complexes were separated at room temperature for 1 h at 180 V, and the gel was dried for 2 h at 80°C under vacuum and subjected to autoradiography (Kodak Biomax MR film) and/or phosphorimager analysis (Fuji FLA 5000).
RESULTS
The TFB zinc ribbon plays a redundant role during RNAP recruitment in archaea.
In eukaryotic systems TFIIB-mediated recruitment of RNAPII to promoters has been extensively investigated (e.g., references 8 and 21). The accumulated data, together with recent crystallographic studies of the Saccharomyces cerevisiae RNAPII-TFIIB complex, have provided unprecedented insights into the structural basis of the interaction between TFIIB and RNAPII (9, 11-13). The compact N-terminal zinc ribbon motif of TFIIB (TFIIB-Zn) interacts with the dock domain in the largest RNAPII subunit, RPB1 (9, 15). This interaction has been identified as a major stabilizing factor because mutations in critical residues in TFIIB-Zn interfere with RNAPII recruitment or abolish it altogether (e.g., references 1, 7, and 38).
The high degree of structural and functional similarity between archaeal and eukaryotic RNAPII core transcriptional machineries can be exploited to shed more light on this mechanism. Sequence alignments of eukaryotic RPB1 and euryarchaeal A′ RNAP subunits reveal unambiguously the overall position of the likely dock domain within the M. jannaschii A′ subunit because it lies between the two evolutionarily highly conserved sequence blocks C and D (Fig. 1A). Interestingly, the predicted interaction domain itself displays only a rather low degree of sequence similarity—in fact, M. jannaschii A′ and two other closely related methanogens contain eight amino acids inserted near the N-terminal part of the putative dock domain that are not present in other archaea or eukaryotes. To test whether this region was playing a role similar to that of its eukaryotic counterpart, we used a site-directed mutagenesis approach. In the yeast RNAPII dock domain, a single residue (RPB1-R412) is crucial for determining the interaction between TFIIB and the polymerase (12, 43). Charge reversal mutations of arginine 412 cause lethality in vivo, while a more subtle substitution of the same residue with histidine results in an RNAPII that cannot be recruited to promoters under in vitro conditions. Due to the low degree of sequence conservation between the archaeal and eukaryotic dock domains, the identification of a putative archaeal homolog of RPB1-R412 is not entirely unambiguous, but the M. jannaschii A′-K396 residue emerged as the most likely candidate and M. jannaschii A′-R388 as a weaker, but still feasible alternative (Fig. 1A). We introduced charge reversal mutations of these residues into the M. jannaschii A′ RNAP subunit and produced recombinant RNAP variants harboring the mutated subunits by in vitro assembly (47). In promoter-directed transcription assays, the M. jannaschii A′-R388E mutation displayed a reproducible but relatively minor phenotype, whereas the M. jannaschii A′-K396E RNAP was noticeably impaired (Fig. 1B). Both mutant enzymes displayed the same catalytic activity as wild-type RNAP in nonspecific transcription assays (Fig. 1C), proving that the impairment observed in the promoter-directed transcription assays was not due to damage of the catalytic center but was most likely caused by diminished recruitment of the mutant enzymes to the promoter-bound basal factor platform.
FIG. 1.
The RNA polymerase dock domain is involved in promoter-directed transcription. (A) Sequence alignment of euryarchaeal and eukaryotic dock domains. Euryarchaeal sequences (vertical white bar) and eukaryotic sequences (black bar) are identified to the left of the species. The sequences of Methanocaldococcus jannaschii, Methanococcus maripaludis, Methanococcus vannielii, Methanothermobacter thermautotrophicus (M. thermauto.), Pyrococcus furiosus, Pyrococcus horikoshii, Thermococcus celer, Halobacter salinarum, Methanococcoides burtonii, Methanosarcina barkeri, Methanosarcina acetivorans, Methanosarcina mazei, Archaeoglobus fulgidus, Methanopyrus kandleri, Saccharomyces cerevisiae, Drosophila melanogaster, Mus musculus, and Homo sapiens are shown. The dock domain is located between the conserved sequence blocks C and D (12). The Methanocaldococcus jannaschii A′ residue R388 (mjA′-R388) and K396 residue and the Saccharomyces cerevisiae RPB1 R412 residue (scRPB1-R412) are indicated. The alignment was performed using MultAlin (14). All invariant residues are shown in red, and highly conserved residues are shown in blue. Gaps introduced to maximize alignment are indicated by dashes. (B) The dock mutant M. jannaschii RNAP variants are defective in promoter-directed transcription assays (reaction mixtures contain 0.1 μg TBP; 0.1 μg TFB; and 0.1, 0.3, or 1.0 μg RNAP). (C) The dock mutant M. jannaschii RNAP variants display a comparable amount of activity in nonspecific transcription assays (reaction mixtures contain 0.5 μg RNAP). neg. ctrl., negative control.
These results are generally compatible with the phenotypes previously observed in the yeast system. It was, however, surprising that the archaeal mutants with charge reversal mutations in the gene encoding the dock domain did not display more severe phenotypes in promoter-directed transcription assays. A possible explanation is that the interaction between the RNAP dock domain and the TFB zinc ribbon plays a less critical role in archaea. To test this hypothesis and to clarify the roles of individual TFB domains in RNAP recruitment, we constructed several TFB deletion variants (Fig. 2A). First, we analyzed the roles of the zinc ribbon and linker in archaeal RNAP recruitment by examining whether they were capable of competing with full-length TFB in promoter-directed transcription assays. The addition of either the zinc ribbon (TFB-Zn), or the zinc ribbon fused to the complete linker sequence (TFB-Zn-B-L), competitively inhibited promoter-specific transcription in a concentration-dependent manner (Fig. 2B).
FIG. 2.
Deletion analysis of Methanocaldococcus jannaschii TFB. (A) M. jannaschii TFB is comprised of four distinct regions: N-terminal Zn ribbon (red), B-finger (yellow), flexible linker (green), and C-terminal core domain (blue). The various domain deletion variants used in this study are shown. The B-finger sequences of M. jannaschii TFB (mjTFB) and Saccharomyces cerevisiae TFIIB (scTFIIB) are shown at the bottom of the panel. (B) The N-terminal fragments TFB-Zn and TFB-Zn-B-L are capable of competing for full-length TFB in promoter-directed transcription assays (reaction mixtures contain 0.1 μg TBP, 0.1 μg TFB, 1.0 μg RNAP, and 0.5 or 1.0 μg TFB-Zn or TFB-Zn-B-L). (C) Activity of the TFB deletion variants in promoter-directed transcription assays (reaction mixtures contain 0.1 μg TBP, 0.1 μg TFB or TFB variants, and 1.0 μg RNAP). One lane contains no TFB variant (−). (D) Activity of the TFB B-finger mutants in promoter-directed transcription assays (reaction mixtures contain 0.1 μg TBP, 0.1 μg TFB or TFB variants, 1.0 μg RNAP, and 10 or 200 μM NTP). wt, wild type.
We next examined the abilities of the TFB variants to support promoter-directed transcription. TFB lacking the Zn ribbon altogether (TFB-ΔZn) was able to support promoter-specific transcription (Fig. 2C). This finding is in good agreement with the previously observed absence of any severe phenotypes of the archaea with mutations in the gene encoding the RNAP dock domain (Fig. 1B). Extending the deletion further by removing both the zinc ribbon and the B-finger region of the flexible linker (creating TFB-ΔZn-ΔB) resulted in a substantial reduction of transcription that is comparable to the activity of the TFB core domain (Fig. 2C). This result suggests that the B-finger domain, in conjunction with the zinc ribbon, contributes actively to promoter-directed transcription.
To explore the function of the TFB linker region further, we targeted the B-finger region by deleting it altogether (TFB-ΔB) or by introducing a single amino acid substitution. Two residues, E78 and R92, are thought to form a stabilizing salt bridge within the B-finger (9, 39). We introduced a charge reversal mutation (R92E) into full-length TFB (TFB-R92E) and TFB-ΔZn and tested their ability to support promoter-directed transcription in the presence of TBP and RNAP. Like wild-type TFB, both TFB-ΔB and TFB-R92E were capable of supporting transcription initiation at saturating (200 μM), but not at low recombinant NTP (rNTP), concentrations (10 μM; Fig. 2D) (3). The TFB-ΔZn/R92E construct, combining the zinc ribbon deletion with the salt bridge mutation, was inactive even at saturating rNTP (200 μM) concentrations. Thus, compromising two TFB-RNAP interaction sites by deleting the zinc ribbon and interfering with B-finger stability impairs the ability of TFB to support transcription initiation in the promoter-directed assay to a level comparable to that of the double-deletion variant TFB-ΔZn-ΔB (Fig. 2C).
B-finger mutations in eukaryotic TFIIBs result in altered start site selection on some promoters (e.g., reference 11). We do not observe this phenomenon due to the strong Sulfolobus shibatae virus (SSV) T6 promoter used in this study (see also reference 3).
In summary, compromising the TFB B-finger interferes with transcription in a substrate concentration-dependent manner and therefore suggests a postrecruitment function of this domain.
TFB stimulates transcription.
All studies involving TFB/TFIIB in promoter-specific transcription systems are intrinsically limited by the fact that both factors are essential for RNAP recruitment. This makes it difficult to dissect the recruitment functions from any postrecruitment contributions that TFB or TFIIB might be involved in. We overcame this problem by assaying the effect of TFB on RNAP transcription directly in recruitment-independent nonspecific transcription assays. These assays take advantage of the fact that all RNAPs are capable of initiating transcription from 3′ overhangs and nicks in double-stranded DNA (dsDNA) templates independent of promoter sequences and in the absence of general transcription factors.
We set up a nonspecific in vitro transcription assay using activated (nuclease-treated) DNA as the template and measured the activity of recombinant RNAP in this assay in the presence of full-length TFB and various TFB domain deletion variants. Addition of TFB markedly increased the transcriptional activity of RNAP (up to fourfold; Fig. 3A). This stimulation is not due to any recruitment function of TFB because TFB does not bind DNA in the absence of TBP, even at high concentrations (23). Neither of the two other basal factors (TBP and TFE) had any detectable effect under these assay conditions, either in the presence or absence of TFB (data not shown). The increased transcriptional activity reveals a novel RNAP stimulatory function of TFB that is distinct from its promoter recruitment activity or from other postrecruitment functions documented for archaeal TFBs and eukaryotic TFIIBs (3, 11). The stimulation is most pronounced at low rNTP concentrations and results in the formation of transcripts that can be several hundred nucleotides in length (data not shown).
In subsequent experiments we tested whether the TFB deletion variants described earlier retained the stimulatory activity of full-length TFB. The zinc ribbon fused to the entire linker region (TFB-Zn-B-L) was able to stimulate RNAP activity on its own but only to a lesser extent (approximately twofold; Fig. 3A). None of the remaining TFB variants stimulated RNAP activity (Fig. 3A and data not shown). Moreover, a targeted deletion of the B-finger subdomain from full-length TFB (TFB-ΔB) abolished the stimulatory properties of TFB, indicating that the B-finger was essential for the observed stimulation (Fig. 3A). All these findings show that the B-finger contributes actively to transcription and can be delivered to the RNAP either via the TFB core domain (when bound to the TBP platform in the promoter-directed assays), via the zinc ribbon (in nonspecific transcription assays), or by both (in vivo and in promoter-directed assay with full-length TFB).
The B-finger stimulates abortive transcription.
We next set out to determine at which stage of the transcription cycle the B-finger acts. The nonspecific transcription assays reveal only an overall stimulation of RNAP activity which could be due to increased rates of transcription initiation, elongation, or less frequent termination. Two observations favor transcription initiation as the most likely target for the stimulation: the S. cerevisiae TFIIB-B-finger penetrates deeply into the active site of RNAPII (9, 13), and archaeal TFB can be cross-linked to promoter DNA around the transcription start site (2, 41). It is therefore likely that the B-finger productively interacts with the template DNA, the DNA/RNA hybrid, and/or the substrate rNTPs during transcription initiation.
We carried out abortive transcription initiation assays to gain further insights into the stimulatory effect of TFB. During the initiation stage all known types of RNAPs repeatedly produce a series of short transcripts (usually 2 to 12 nucleotides in length) before they successfully enter the elongation stage (reviewed in references 16 and 26; see also reference 45). Abortive initiation assays typically employ a DNA template and a specific diribonucleotide complementary to the +1/+2 (or −1/+1) position of the promoter in the presence of a single type of radiolabeled rNTP which are added to the dinucleotide precursor through RNAP action. We modified this assay by using nuclease-treated genomic DNA as template. This setup yields a heterogeneous mixture of short abortive RNA products derived from random initiation sites and therefore samples the abortive transcripts produced by RNAPs on all sites capable of transcript initiation. In order to assay a cross-section of different subsets of abortive transcripts, we used one of three different dinucleotide substrates (ApG, UpG, and ApA) in conjunction with [α-32P]rATP. RNAP on its own gave rise to a reproducible signal of 8 to 10 nucleotides under these assay conditions. Under the same conditions the addition of TFB caused a substantial stimulation of the generation of abortive transcription products with any of the three dinucleotide substrates (Fig. 3B). A hallmark of the generation of abortive transcripts is the resistance to a nonspecific competitor such as heparin (10, 20). Heparin precludes reinitiation of RNAP from the DNA template, and therefore, only elongating RNAPs or RNAPs engaged in abortive transcription can synthesize transcripts under such conditions. The continuous accumulation of RNA products under the assay conditions described above is indeed resistant to heparin, thus strongly supporting the notion that they are bona fide abortive initiation products (data not shown). In addition, the substrate limitations of the assay (ribodinucleotide plus rATP), in combination with the high temperature at which the assays are carried out (65°C), may also cause transcript slippage of the newly formed RNA products. We therefore conclude that the TFB-mediated stimulation observed in nonspecific transcription assays is mostly (if not entirely) due to a stimulation of the abortive stage.
In a further attempt to characterize the recruitment-independent stimulatory role of TFB under more defined conditions, we made use of a 3′-tailed template (Fig. 3E). RNAPs are able, in a situation very similar to the nuclease-treated genomic DNA, to initiate transcription approximately three nucleotides downstream of the 3′-tail junction, albeit with low efficiency (18, 28). In the absence of factors, M. jannaschii RNAP utilizes the 3′-tailed template to generate transcripts up to 18 nucleotides long (Fig. 3C). The addition of TFB increases the overall amount of transcripts synthesized and, more importantly, generates a predominant transcript that is 20 nucleotides in length (Fig. 3C). This result is in good agreement with our previous observation that TFB stimulates transcription initiation in a manner that is independent of upstream promoter regions and therefore rules out a conventional, TBP-dependent recruitment of RNAP to the template DNA.
The TFB-mediated stimulation of transcription on a defined 3′-tailed template allowed us to investigate the molecular basis of this mechanism in more detail. During promoter melting, RNAPII undergoes significant conformational changes that largely consist of closing of the clamp domain over the template DNA strand by a hinge movement of 30o (19). In this closed clamp configuration, the RNAP/nucleic acid complex is rendered more resistant to heparin, a polyanion acting as a competitor for DNA binding. M. jannaschii RNAP does not form heparin-stable complexes with 3′-tailed templates in the absence of basal factors (Fig. 3D). However, the addition of TFB, but not TFB-ΔB, did lead to the formation of heparin-stable RNAP/tailed template complexes (Fig. 3D; i.e., template strand loaded and clamp closed). This result supports the interpretation that the B-finger of TFB stabilizes the RNAP/template complex, e.g., by decreasing the rate of dissociation of RNAP from the DNA template.
The observed stimulatory function of TFB on the abortive transcription rate of RNAP has not been previously documented in eukaryotic systems. We therefore expressed and purified recombinant yeast TFIIB and incubated it with highly purified yeast RNAPII (gift from Patrick Cramer) in the presence of nuclease-activated double-stranded DNA and 3′-tailed templates. No stimulation of RNAPII activity was observed under these conditions (data not shown), indicating that this type of functional interaction either does not occur in eukaryotes or that the stimulatory role of TFIIB depends on the presence of other basal factors.
A functional overlap between TFB and TFE: stabilization of the preinitiation complex.
The experiments described in the previous section took advantage of the fact that only two basal transcription factors (TBP and TFB) are necessary and sufficient for promoter-specific archaeal transcription in vitro. This is a useful experimental setup because it emphasizes the functional contributions that these two factors make by themselves. The majority of archaeal genomes sequenced so far also encode TFE, a homolog of one of the two subunits present in the eukaryotic TFIIE heterodimer (TFIIEα) (5, 22, 27). In archaeal in vitro transcription systems, TFE is not strictly necessary, but it stimulates transcription two- to threefold on weak promoters (5, 22). The mechanism by which TFE acts during transcription initiation is currently not known. In the RNAPII system TFIIE has been shown to stimulate promoter melting (probably through a direct interaction of TFIIEα with the upstream end of the partially opened transcription bubble; 16) and through the recruitment of TFIIH (25, 34, 35). Importantly for our investigation, it has been suggested that TFIIE changes the template topology of the transcription initiation complex (24) and thus might be involved in the same process (template loading/DNA strand separation; 17, 20, 42) in which, according to data presented earlier, TFB might also be implicated.
We first asked whether recombinant M. jannaschii TFE could enhance transcription driven by the strong SSV T6 promoter, RNAP, TBP, and wild-type and domain deletion variants of TFB in promoter-directed transcription assays. In agreement with published results (5), transcription facilitated by full-length TFB could not be enhanced any further. Intriguingly, however, TFE was capable of partially rescuing the reduced transcription signals obtained with the weakened TFB variants that lack the Zn ribbon motif (TFB-ΔZn) or contain the additional B-finger salt bridge mutation (TFB-ΔZn-R92E; Fig. 4A). We therefore conclude that there is at least a partial overlap between the functional spectra of TFE and TFB. To study the impact of TFE on RNAP recruitment more directly, we tested different combinations of RNAP, TBP, TFE, and TFB variants in EMSAs (Fig. 4B). In the presence of TBP and TFB, RNAP forms heparin-resistant complexes with the double-stranded SSV T6 promoter template (Fig. 4B). Under the same conditions neither the zinc ribbon nor the B-finger mutants of TFB are capable of recruiting RNAP in a sufficiently stable manner to yield a signal (Fig. 4B). In EMSAs the stability of the initiation complex plays a more important role than in transcription assays because all components need to remain associated throughout the duration of the experiment. The addition of TFE led to stable RNAP recruitment by both TFB-ΔZn and TFB-R92E, whereas TFE was not capable of recruiting RNAP on its own or in combination with TBP (Fig. 4B). The addition of TFE increased the electrophoretic mobility of the initiation complexes in EMSAs only marginally. This is most likely due to the small contribution of TFE (molecular mass of ∼20 kDa) to the overall hydrodynamic radius of the RNAP/TBP/TFB/promoter complex (molecular mass of ∼0.5 MDa).
FIG. 4.
TFE complements TFB mutations in vitro. (A) Promoter-directed transcription assays using RNAP, TBP, full-length TFB, TFB-ΔZn, and TFB-ΔZn/R92E in combination with TFE (reaction mixtures contain 0.1 μg TBP, 0.1 μg TFB, 0.25 or 0.5 μg TFE, and 1.0 μg RNAP). (B) Electrophoretic mobility shift assays using various combinations of RNAP, TBP, and TFE together with full-length and mutant variants of TFB as indicated (reaction mixtures contain 0.5 μg TBP, 0.5 μg TFB or TFB variant, 0.5 μg TFE, and 2.0 μg RNAP). The presence of TFE causes a minor decrease in the mobility of the preinitiation complexes. wt, wild type. (C) In the presence (+) of TFB and TFE, RNAP is capable of forming heparin-stable complexes with the template strand (tDNA) but not with the nontemplate strand (nDNA) of the SSV T6 promoter. This interaction is dependent on the TFB B-finger. Both single-stranded DNA probes tDNA and nDNA form a nonspecific shift (marked with an asterisk in panel C) that is presumably due to a stable secondary structure not to be confused with the TBP/DNA shift in panel B. Reaction mixtures contain 0.5 μg TFB or TFB-ΔB, 0.5 μg TFE, and 2.0 μg RNAP.
The results confirm the conclusions derived earlier from the activity-based assays and demonstrate that TFE increases transcription initiation complex stability. In effect, TFE can compensate for TFB (mutant) defects in promoter-directed transcription assays and in RNAP-recruitment EMSAs through a direct stabilization of the RNAP/TFB/TBP/DNA complex.
TFB and TFE dynamically alter the nucleic acid-binding properties of RNAP.
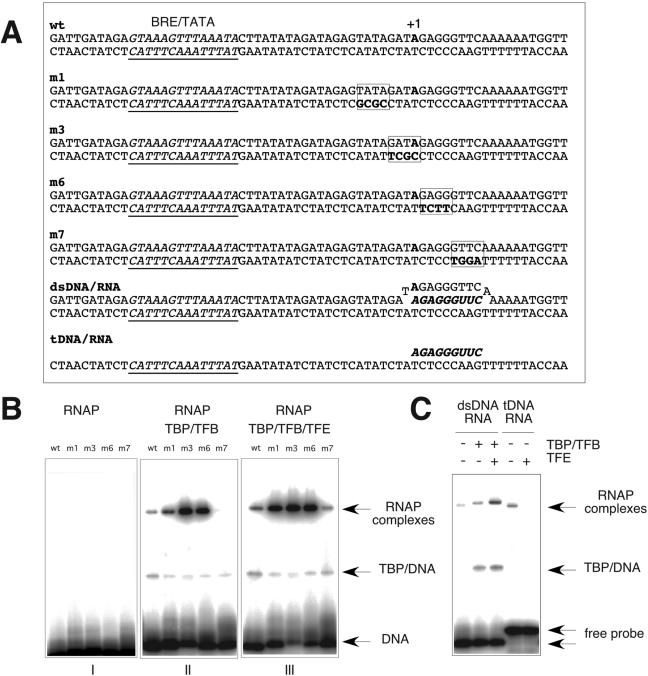
In eukaryotes, TFIIE is thought to be involved in DNA strand separation during transcription initiation by directly influencing the overall template topology (17) and indirectly by influencing the recruitment and helicase activity of TFIIH (25). These effects can be studied experimentally by mimicking different template topologies with premelted DNA templates containing locally mismatched strands (heteroduplexes) (17, 24). Having shown that TFB and TFE influence the loading and/or arrangement of the DNA template within RNAP, we decided to investigate this phenomenon in greater detail by designing a series of heteroduplex variants of the SSV T6 promoter that carry a window of four noncomplementary bases at different positions relative to the TATA box and the transcription start site (Fig. 5A). These promoter variants were used as templates in EMSAs using RNAP, TBP, TFB, and TFE. Figure 5B illustrates that promoters carrying the heteroduplex region surrounding the transcription start site (−3/+1 and + 2/+5; m3 and m6, respectively, in Fig. 5A) displayed a significantly increased affinity for RNAP compared to that for the homoduplex promoter. This is strictly dependent on the presence of TBP and TFB (Fig. 5B, panels I and II), proving that the effect is specific for initiation complexes and not simply due to an increased affinity of RNAP for single-stranded DNA stretches. Moving the heteroduplex window either upstream or downstream decreased the affinity to a level comparable to that of the homoduplex promoter. These experiments demonstrate that premelted sequences in close proximity to the transcription start site increase the stability of initiation complexes, most likely by mimicking open complexes. We next investigated whether TFE could enhance the recruitment of RNAP, TBP, and TFB to wild-type and heteroduplex promoter templates. The addition of TFE increased the RNAP/TFB/TBP shift on the wild-type and on the weak (m1 and m7) heteroduplex promoter templates, but not on the strong (m3 and m6) heteroduplex templates (Fig. 5B, panels II and III). It is therefore tempting to speculate that TFE leads to an increased initiation complex stability by the same means as the heteroduplex promoter variants, e.g., by promoting DNA melting and/or template loading (similar to the properties reported for TFIIEα in reference 17). To investigate the contributions of the template and nontemplate strands during initiation complex formation, we performed EMSAs with both strands separately. Surprisingly, RNAP can form heparin-stable complexes with the template strand in the presence of TFB/TFE, but not TFB-ΔB/TFE (Fig. 4C). This interaction is quasidependent on the sequence, since the SSV T6 nontemplate strand and other nonrelated sequences do not form stable complexes (Fig. 4C and data not shown). More importantly, although the recruitment of the template strand into RNAP requires TFB and TFE, there is no requirement for TBP. This result has two unexpected implications. Firstly, it shows that TFB and TFE can interact with RNAP in the absence of TBP and any double-stranded promoter DNA (reminiscent of the eukaryotic RNAPII holoenzyme complexes) (30). Secondly, it directly demonstrates a degree of sequence specificity in the interaction between RNAP and the template strand.
FIG. 5.
RNAP binding to heteroduplex DNA and DNA/RNA-hybrid templates. (A) Sequences of nucleic acid templates based on the SSV T6 promoter used for EMSAs. The B-recognition element (BRE)/TATA elements are underlined, the transcription start site is indicated by +1 and bold type, the heteroduplex mutations are boxed and in italic type, and RNA sequences are indicated by bold italic type. wt, wild type. (B) RNAP was used in EMSAs with various combinations of TBP, TFB, and TFB on the homoduplex and heteroduplex templates m1, m3, m6, and m7 (reaction mixtures contain 0.5 μg TBP, 0.5 μg TFB, 0.5 μg TFE, and 2.0 μg RNAP). wt, wild type. (C) RNAP forms stable complexes with DNA/RNA-hybrid probes (reaction mixtures contain 0.5 μg TBP, 0.5 μg TFB, 0.5 μg TFE, and 2.0 μg RNAP).
Next we investigated whether TFE altered the interactions of RNAP with RNA/DNA hybrid templates. During transcription elongation, the RNAP active site accommodates the template DNA strand and approximately 9 nucleotides of the RNA transcript annealed to the template DNA strand. During the abortive initiation phase, a similar configuration is likely to occur within the active site, with the important difference that the RNAP still occupies the promoter and is likely to be associated with the basal factors TBP, TFB, and TFE. The elongation/abortive initiation complexes can be mimicked in vitro by the use of double-stranded DNA templates that have incorporated a short RNA oligonucleotide (dsDNA/RNA) (Fig. 5A) (31). We tested the interactions between RNAP and dsDNA/RNA hybrid templates in the presence and absence of basal transcription factors. The formation of heparin-stable complexes on double-stranded DNA templates depends on the presence of both TBP and TFB (Fig. 4B). In contrast, RNAP can stably associate—in the absence of TBP and TFB—with templates consisting of double-stranded DNA containing a short RNA oligonucleotide (Fig. 5A, dsDNA/RNA). The addition of TBP/TFB leads to RNAP complex with reduced mobility that can be further stabilized by the addition of TFE in a manner very similar to double-stranded templates (Fig. 4B and 5C).
Bacterial RNAPs and eukaryotic RNAPIIs form transcription-competent (elongation) complexes with templates consisting of the template DNA strand preannealed to short RNA transcript but lacking the nontemplate strand (tDNA/RNA) (Fig. 5A) (31). The M. jannaschii RNAP also forms heparin-stable elongation complexes of this kind (Fig. 5C). In contrast to the stabilizing effect of TFE on RNAP/TBP/TFB complexes bound to dsDNA and dsDNA/RNA templates, TFE destabilizes RNAP complexes bound to the tDNA/RNA complexes (Fig. 5C). Thus, TFE stabilizes initiation complexes but destabilizes elongation complexes in vitro. A similar conclusion has emerged from work on the RNAPII system where TFIIE has been shown to play a role during the early (promoter melting) and late (promoter escape) stages of transcription initiation (17, 20).
DISCUSSION
A key advantage of the archaeal transcriptional machinery is the substantially reduced number of accessory basal factors compared to that of the RNAPII system (typically TBP, TFB, and TFE versus TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, respectively). The functional interactions observed in such a minimal system are likely to be stronger and more distinct. We therefore believe that at least some of the novel functions of the archaeal basal factors TFB and TFE uncovered in this study are also part of the functional spectra of their eukaryotic counterparts but may be less obvious there because of the increased complexity of the larger eukaryotic nuclear transcription initiation complexes (4).
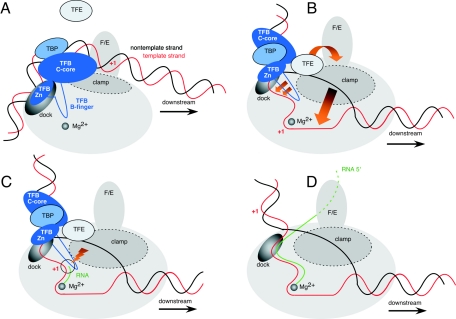
The emerging picture of the archaeal transcription initiation process is summarized in Fig. 6. During the early phase of transcription initiation, RNAP is recruited to the promoter by the TBP/TFB/DNA complex. This interaction is mediated by the TFB Zn ribbon and the RNAP dock domain (Fig. 6A). At this stage, the closed initiation complex-RNAP makes only very weak or no direct contacts with the promoter DNA, partly because (in parallel to the eukaryotic system) the TFB core domain is probably situated between the major DNA-binding cleft of RNAP and the template DNA (Fig. 6A) (13). The conversion of the closed complex into the open complex is accompanied by conformational changes of both the RNAP and promoter that most notably result in the separation of the DNA strands (melting) and loading of the template strand into the active center of the enzyme (Fig. 6B). In the open complex, the template strand is then structurally stabilized in the active site by the B-finger domain of TFB; as a consequence, the rate of abortive initiation product formation is stimulated, presumably by stabilizing a template DNA-rNTP substrate geometry that is conducive to catalysis (Fig. 6C) (9). This situation is reminiscent of the one found in bacteria where many RNAP functions, ranging from transcript initiation to promoter escape, are directed by the regulatory interplay between core RNAP, σ-factors and particular promoter sequence elements. σ-factors control the rate, yield, and length of abortive transcripts, resulting in indirect control of the kinetics of promoter escape (26, 44, 48).
FIG. 6.
Modulation of RNAP core functions by TFB and TFE during transcription initiation. (A) RNAP is recruited to the TBP/TFB/DNA complex through interactions between the TFB Zn ribbon and the RNAP dock domain. In this closed complex, RNAP makes only superficial contacts with promoter DNA. F/E, RNAP subunits F and E. (B) During open complex formation, the DNA strands are melted and the template strand is inserted into the active site where it is stabilized by the TFB B-finger. TFE furthermore stabilizes this complex by influencing the position of the RNAP clamp by subunits F and E and possibly by interacting directly with the nontemplate strand. (C) The abortive initiation phase is characterized by the production of short RNA transcripts. This step is stimulated by TFB presumably because the TFB B-finger facilitates a DNA/rNTP or even DNA/RNA/rNTP configuration that is conducive to catalysis (indicated by the red-orange flash). (D) During the escape of the RNAP from the promoter, basal transcription factors TBP, TFB, and TFE are shed and the RNA transcript is directed towards RNAP subunits F and E in the elongation complex.
The modulation effect of basal factors on RNAP functions is further exemplified by the novel roles of TFE uncovered in our work. This third archaeal basal transcription factor increases the stability of the initiation complex and functionally interacts with RNAP subunits F and E (Fig. 6B) (36). The presence of F/E and the positioning of the RNAP mobile clamp are almost certainly closely related—as are the nucleic acid-binding properties of RNAP and the position of the clamp. We therefore propose a model where TFE stabilizes the transcription initiation complex by closing the clamp via RNAP subunits F and E. TFIIE can be cross-linked to promoter sequences between the TATA box and the transcription start site (17, 29, 42). This places TFE at the upstream face of the initiation complex in proximity to RNAP subunits F and E, the RNA exit channel, and the nontemplate strand (Fig. 6B and C). According to this model, extension of the RNA transcript beyond the size of abortive product length (approximately 10 nucleotides) (45) is concomitant with a downstream movement of RNAP and displacement of both TFB and TFE, resulting in a transcription elongation complex (Fig. 6D).
A remaining challenge is how these observations concerning the workings of archaeal systems can be correlated with eukaryotic transcriptional machineries. The more-complex initiation and promoter escape mechanisms and the involvement of additional basal factors (TFIIF and TFIIH) imply that the activities that we have discovered in the archaeal initiation complex are likely to be more functionally distributed and entangled in the additional RNAPII-specific basal factors. This underscores the importance of the archaeal transcriptional machinery for highlighting the presence of discrete steps that would be difficult to dissect out experimentally in the more-complex eukaryotic systems.
Acknowledgments
We thank the members of Martin Buck's laboratory for advice regarding heteroduplex promoter variants and for many helpful discussions. We are especially grateful to Ramesh Wigneshweraraj, Peter Brick, Silvia Onesti, Kristine Arnvig, and Mohamed Ouhammouch for valuable comments on the manuscript.
This work was supported by a Wellcome Trust Project grant (grant 068764/Z/02/Z) to R.O.J.W.
REFERENCES
- 1.Barberis, A., C. W. Muller, S. C. Harrison, and M. Ptashne. 1993. Delineation of two functional regions of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90:5628-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett, M. S., M. Thomm, and E. P. Geiduschek. 2004. Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 279:894-903. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. D., and S. P. Jackson. 2000. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 275:12934-12940. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. D., A. B. Brinkman, J. van der Oost, and S. P. Jackson. 2001. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeger, H., D. A. Bushnell, R. Davis, J. Griesenbeck, Y. Lorch, J. S. Strattan, K. D. Westover, and R. D. Kornberg. 2005. Structural basis of eukaryotic gene transcription. FEBS Lett. 579:899-903. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski, S., and H. Zhou. 1993. Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90:5633-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski, S., M. Sopta, J. Greenblatt, and P. A. Sharp. 1991. RNA polymerase II-associated proteins are required for a DNA conformation change in the transcription initiation complex. Proc. Natl. Acad. Sci. USA 88:7509-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushnell, D. A., K. D. Westover, R. E. Davis, and R. D. Kornberg. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303:983-988. [DOI] [PubMed] [Google Scholar]
- 10.Cashel, M., L. M. Hsu, and V. J. Hernandez. 2003. Changes in conserved region 3 of Escherichia coli sigma 70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 278:5539-5547. [DOI] [PubMed] [Google Scholar]
- 11.Chen, B. S., and M. Hampsey. 2004. Functional interaction between TFIIB and the Rpb2 subunit of RNA polymerase II: implications for the mechanism of transcription initiation. Mol. Cell. Biol. 24:3983-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H. T., and S. Hahn. 2003. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12:437-447. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H. T., and S. Hahn. 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119:169-180. [DOI] [PubMed] [Google Scholar]
- 14.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876.11313498 [Google Scholar]
- 16.Dvir, A. 2002. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta 1577:208-223. [DOI] [PubMed] [Google Scholar]
- 17.Forget, D., M. F. Langelier, C. Therien, V. Trinh, and B. Coulombe. 2004. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol. Cell. Biol. 24:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnatt, A., J. Fu, and R. D. Kornberg. 1997. Formation and crystallization of yeast RNA polymerase II elongation complexes. J. Biol. Chem. 272:30799-30805. [DOI] [PubMed] [Google Scholar]
- 19.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876-1882.11313499 [Google Scholar]
- 20.Goodrich, J. A., and R. Tjian. 1994. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 77:145-156. [DOI] [PubMed] [Google Scholar]
- 21.Ha, I., S. Roberts, E. Maldonado, X. Sun, L. U. Kim, M. Green, and D. Reinberg. 1993. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7:1021-1032. [DOI] [PubMed] [Google Scholar]
- 22.Hanzelka, B. L., T. J. Darcy, and J. N. Reeve. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hausner, W., and M. Thomm. 2001. Events during initiation of archaeal transcription: open complex formation and DNA-protein interactions. J. Bacteriol. 183:3025-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holstege, F. C., D. Tantin, M. Carey, P. C. van der Vliet, and H. T. Timmers. 1995. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 14:810-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holstege, F. C., P. C. van der Vliet, and H. T. Timmers. 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15:1666-1677. [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, L. M. 2002. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta 1577:191-207. [DOI] [PubMed] [Google Scholar]
- 27.Inostroza, J., O. Flores, and D. Reinberg. 1991. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of general transcription factor IIE. J. Biol. Chem. 266:9304-9308. [PubMed] [Google Scholar]
- 28.Kadesch, T. R., and M. J. Chamberlin. 1982. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem. 257:5286-5295. [PubMed] [Google Scholar]
- 29.Kim, T. K., R. H. Ebright, and D. Reinberg. 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418-1422. [DOI] [PubMed] [Google Scholar]
- 30.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 31.Kireeva, M. L., N. Komissarova, D. S. Waugh, and M. Kashlev. 2000. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275:6530-6536. [DOI] [PubMed] [Google Scholar]
- 32.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 34.Maxon, M. E., J. A. Goodrich, and R. Tjian. 1994. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 8:515-524. [DOI] [PubMed] [Google Scholar]
- 35.Ohkuma, Y. 1997. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J. Biochem. 122:481-489. [DOI] [PubMed] [Google Scholar]
- 36.Ouhammouch, M., F. Werner, R. O. Weinzierl, and E. P. Geiduschek. 2004. A fully recombinant system for activator-dependent archaeal transcription. J. Biol. Chem. 279:51719-51721. [DOI] [PubMed] [Google Scholar]
- 37.Ouhammouch, M., G. E. Langham, W. Hausner, A. J. Simpson, N. M. El-Sayed, and E. P. Geiduschek. 2005. Promoter architecture and response to a positive regulator of archaeal transcription. Mol. Microbiol. 56:625-637. [DOI] [PubMed] [Google Scholar]
- 38.Pardee, T. S., C. S. Bangur, and A. S. Ponticelli. 1998. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 273:17859-17864. [DOI] [PubMed] [Google Scholar]
- 39.Pinto, I., W. H. Wu, J. G. Na, and M. Hampsey. 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269:30569-30573. [PubMed] [Google Scholar]
- 40.Qureshi, S. A., and. S. P. Jackson. 1998. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1:389-400. [DOI] [PubMed] [Google Scholar]
- 41.Renfrow, M. B., N. Naryshkin, L. M. Lewis, H. T. Chen, R. H. Ebright, and R. A. Scott. 2004. Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J. Biol. Chem. 279:2825-2831. [DOI] [PubMed] [Google Scholar]
- 42.Robert, F., D. Forget, J. Li, J. Greenblatt, and B. Coulombe. 1996. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J. Biol. Chem. 271:8517-8520. [DOI] [PubMed] [Google Scholar]
- 43.Scafe, C., C. Martin, M. Nonet, S. Podos, S. Okamura, and R. A. Young. 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severinov, K., D. Fenyö, E. Severinova, A. Mustaev, B. T. Chait, A. Goldfarb, and S. A. Darst. 1994. The σ-subunit conserved region 3 is part of the “5′-face” of the active center of Escherichia coli RNA polymerase. J. Biol. Chem. 269:20826-20828. [PubMed] [Google Scholar]
- 45.Spitalny, P., and M. Thomm. 2003. Analysis of the open region and of DNA-protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 278:30497-30505. [DOI] [PubMed] [Google Scholar]
- 46.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]
- 47.Werner, F., and R. O. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10:635-646. [DOI] [PubMed] [Google Scholar]
- 48.Zenkin, N., and K. Severinov. 2004. The role of RNA polymerase sigma subunit in promoter-independent initiation of transcription. Proc. Natl. Acad. Sci. USA 101:4396-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]