Abstract
While small Maf proteins have been suggested to be essential for the Nrf2-mediated activation of antioxidant response element (ARE)-dependent genes, the extent of their requirement remains to be fully documented. To address this issue, we generated mafG::mafF double-mutant mice possessing MafK as the single available small Maf. Induction of the NAD(P)H:quinone oxidoreductase 1 (NQO1) gene was significantly impaired in double-mutant mice treated with butylated hydroxyanisole, while other ARE-dependent genes were less affected. Similarly, in a keap1-null background, where many of the ARE-dependent genes are constitutively activated in an Nrf2-dependent manner, only a subset of ARE-dependent genes, including NQO1, were sensitive to a simultaneous deficiency in MafG and MafF. Examination of single and double small maf mutant cells revealed that MafK also contributes to the induction of ARE-dependent genes. To obtain decisive evidence, we established mafG::mafK::mafF triple-mutant fibroblasts that completely lack small Mafs and turned out to be highly susceptible to oxidative stress. We found that induction in response to diethyl maleate was abolished in a wider range of ARE-dependent genes in the triple-mutant cells. These data explicitly demonstrate that small Mafs play critical roles in the inducible expression of a significant portion of ARE-dependent genes.
Antioxidant or electrophile response elements (AREs or EpREs) are cis-regulatory elements vital for the transcription of genes encoding antioxidant and phase II drug-metabolizing enzymes (reviewed in reference 9). In response to oxidative stress, numerous cytoprotective genes are induced in an ARE-dependent manner. The ARE consensus sequence TGA(G/C)nnnGC resembles the Maf recognition element (MARE) TGCTGA(G/C)TCAGCA, which was originally identified as the binding site for the v-maf oncoprotein (8, 15). This similarity originally suggested that small Maf proteins might also be essential coeffectors for transcriptional regulation mediated through the ARE. At present, a consensus of evidence suggests that small Maf proteins form homodimers, various combinations of small Maf heterodimers, heterodimers with transcriptional activator proteins of the CNC family (Nrf1, Nrf2, Nrf3, and p45 NF-E2), and heterodimers with the transcriptional repressor Bach proteins Bach1 and Bach2. Any of these dimeric combinations can then bind to MAREs, as well as to other related sequences, including AREs (for reviews, see references 29 and 30).
The GC residues of the ARE (underlined above), which flank the TRE (TPA-response element) core sequence [TGA(G/C)TCA] of MARE, are critical for the induction of genes in response to oxidative stimuli (38). Furthermore, a recent nuclear magnetic resonance structural analysis suggested that the small Maf proteins contact the GC sequence in the MARE (or ARE) using a unique DNA binding motif termed the extended homology region, which is located N terminal to the basic region-leucine zipper (bZIP) motif (18, 20). Hence, the small Maf proteins are generally regarded as important components of the cellular stress response mediated through the cis-acting ARE elements. Since typical MAREs contain a central TRE, bZIP-type transcription factors, including Jun, Fos, and ATF family proteins, may also bind to MAREs and their related sequences and may affect transcriptional responses through these elements (15).
The biological significance of ARE-binding transcription factors has also been examined and confirmed by germ line genetic manipulation. Analysis of nrf2-null mutant mice showed that Nrf2 is a central regulator of the induction of many antioxidant-responsive genes and phase II detoxification enzyme genes and that Nrf2 is a key transcriptional activator of AREs (6, 8). Under normal physiological conditions, Nrf2 is captured in the cell cytoplasm by a molecule called Keap1 and turned over rapidly by proteasomal degradation (10, 11, 24, 33, 41). However, in response to oxidative stress, Nrf2 is stabilized, relocates to the nucleus, and binds to and activates target response genes (10, 11). As part of the proof elucidating this pathway, it was shown that Nrf2 accumulates in the nuclei of keap1-null mutant mice even in the absence of exogenous stress stimuli, leading to constitutive activation of ARE-dependent genes (43). Analysis of single nrf1-null mice and nrf1::nrf2 compound mutant mice suggests that Nrf1 also contributes to the induction of ARE-dependent genes (22).
Many studies have attempted to clarify the contribution of the small Maf proteins to ARE-dependent gene regulation. To date, no clear functional differences have been reported among the three small Maf proteins MafG, MafK, and MafF (29). Unlike their CNC and Bach partner molecules, canonical functional domains other than the DNA binding and dimerization motifs have not been identified in these small Maf proteins (29). Published studies have led to the conclusion that the CNC and Bach proteins require small Maf proteins as obligatory partners to promote site-specific ARE/MARE association (1, 2, 7, 19, 25, 37). One interpretation of these data is that the small Maf proteins contribute to transcriptional activation and repression as a consequence of the intrinsic activities of their heterodimeric partners (the CNC and Bach proteins). However, it is well documented that the small Maf proteins form homodimers, which can act as repressive competitors for MAREs with large Maf homodimers, as well as with small Maf-CNC heterodimers (28). In contrast to this view of small Maf contributions to MARE-mediated gene activation, several reports describe evidence that led their authors to conclude that the small Maf proteins contribute exclusively to the repression of ARE-dependent genes and not to their activation (3, 14, 32). In summary, according to divergent interpretations of biochemical, transfection, and in vivo data, it is clear that the contribution of the small Maf proteins to transcriptional activation and repression through ARE elements has not been definitively resolved.
To address the function of the small Maf proteins in vivo, we originally reported their individual germ line disruptions in mice (35, 36, 39). Although mafF−/− (F0) and mafK−/− (K0) mice displayed no apparent mutant phenotypes, mafG-null mutant mice displayed mild thrombocytopenia accompanied by a late-onset neurological disorder (39). The neurological abnormality was exacerbated in mafG−/−::mafK+/− (G0K1) compound mutant mice, suggesting that MafG and MafK share compensatory functions in the central nervous system (16). Since several neurodegenerative disorders are suspected to result from a dysregulated oxidative stress response, we analyzed the expression of ARE-dependent genes in the central nervous systems of these symptomatic mutant mice. These studies showed that the heme oxygenase 1 gene (HO-1) was induced in G0K1 compound mutant animals, whereas many other ARE-dependent genes were not significantly affected (16). We also found that Bach proteins failed to accumulate in neuronal nuclei in the G0K1 central nervous system, which could explain the observed HO-1 transcriptional derepression (16, 42). However, these studies did not resolve whether or not the small Maf proteins collaboratively, with a CNC partner, contribute to the activation of ARE-dependent genes. Here, we provide direct and decisive genetic evidence that the small Maf proteins are required for the activation of ARE-dependent gene expression collaboratively with Nrf2 in vivo, thereby excluding the notion that the small Maf proteins contribute merely to ARE-mediated repression.
MATERIALS AND METHODS
Generation of compound mutant mice.
Germ line mutagenesis of the mouse mafG, mafK, mafF, nrf2, and keap1 genes has been described previously (8, 35, 36, 39, 43). All mice examined in this study were of a mixed genetic background, with contributions from the 129Sv/J, C57BL/6J, and ICR strains. Genotypes were determined by PCR as described previously (8, 35, 36, 39, 43).
Mouse embryonic fibroblasts (MEFs).
MEFs were prepared from individual embryos at embryonic day 11.0 (E11.0) or E13.5. The head and internal organs were removed, and the torso was minced and dispersed in 0.25% trypsin-EDTA. MEFs were maintained in Dulbecco's modified Eagle's medium (Sigma Chemical Co., St. Louis, Mo.) containing 10% fetal bovine serum and antibiotics.
Cell viability analysis.
MEFs prepared from mafG+/+::mafK−/−::mafF−/− (G2K0F0) and mafG−/−::mafK−/−::mafF−/− (G0K0F0) embryos were seeded at an initial density of 5 × 103 cells/well in 96-microwell plates. Cells were initially cultured in 25 μM diethyl maleate (DEM). After 24 h, the medium was replaced with medium containing 0, 5, or 10 μM 1-chloro-2,4-dinitrobenzene (CDNB; Wako Pure Chemicals, Osaka, Japan). After 12 h of culturing, a cell viability assay (Cell Counting Kit-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to measure the activities of dehydrogenase enzymes as an indicator of cell viability according to the manufacturer's protocol.
RNA extraction.
Total RNA was prepared from mouse liver or mouse embryonic fibroblasts using an Isogen RNA extraction kit (Nippon Gene, Toyama, Japan) and following the manufacturer's protocol.
RNA blot analysis.
Purified total RNA was electrophoresed on formaldehyde-agarose gels and transferred onto a nylon membrane (Zeta Probe membrane; Promega). 32P-labeled probes were prepared from the cDNA by random primer labeling. cDNAs encoding HO-1 and NQO1 were previously described (6, 8). cDNA that can detect both glutathione S-transferase P1 (GSTP1) and GSTP2 mRNAs was a gift from Kimihiko Satoh (Hirosaki University School of Medicine, Hirosaki, Japan) and designated GSTP1/2. To generate the probes, cDNA fragments were amplified by PCR using various primer pairs as follows: 5′-CAG TGT TTG AAC GGA ACA GA-3′ and 5′-TAC TTG GTC CAA GAC TTG AC-3′ for the ferritin light chain (FTL) gene, 5′-ATG TAT GCA GAT GGC ACC CAG GAC CTG-3′ and 5′-GGA CAA TCC TGA CCA CCT CAA CAT AG-3′ for the GSTA4 gene, and 5′-GAA GAC CCT AGT AGT TGG TGC ATC-3′ and 5′-AAG GAG TAA ATA CAG TCG TTG GGA C-3′ for the thioredoxin reductase 1 gene (TXNRD1). The PCR products were subcloned into the pGEM-T easy vector (Promega).
Administration of butylated hydroxyanisole (BHA) to mice.
BHA (Sigma Chemicals) was administered by oral gavage at a dose of 0.6 g/kg dissolved in corn oil. Animals were sacrificed 24 h after treatment, and livers were processed for the purification of RNA. All mice used in this analysis were 10- to 15-week-old females.
cDNA microarray analysis.
Isolated total RNA was further purified by RNeasy RNA isolation kit (QIAGEN) by following the manufacturer's protocol. The purified RNA was processed and hybridized to a mouse expression array 430A gene chip (Affymetrix, Santa Clara, CA). Experimental procedures for GeneChip were performed according to the Affymetrix Gene Chip Expression Analysis technical manual.
Quantitative real-time PCR.
cDNA was synthesized from the isolated RNAs by reverse transcriptase using random hexamer oligonucleotide primers, and real-time PCR was performed using an ABI Prism 7700 (Applied Biosystems, Foster City, CA) as previously described (35). The primer and probe sequences used for detecting NQO1, GSTA4, TXNRD1, glutamate-cysteine ligase catalytic subunit (GCLC), and HO-1 mRNAs are shown in Table 1. The rRNA primers and probes were purchased from Applied Biosystems.
TABLE 1.
Oligonucleotide primers and TaqMan probes used in this study
| Gene | Primers (5′ to 3′) | TaqMan probe (5′ to 3′)a |
|---|---|---|
| NQO1 | Sense: AGC TGG AAG CTG CAG ACC TG | FAM-ATT TCA GTT CCC ATT GCA GTG GTT TGG G-TAMRA |
| Antisense: CCT TTC AGA ATG GCT GGC A | ||
| GSTA4 | Sense: GGG AAC AGT ATG AGA AGA TGC AAA A | FAM-TGG ACA CCT GCT TTT CGG CCA AG-TAMRA |
| Antisense: CCC ATC GAT TTC AAC CAA GG | ||
| TXNRD1 | Sense: AGA AAG TGC TGG TCT TGG ATT TTG | FAM-TCT GGT CCC AAG AGG AGT CGG TGT G-TAMRA |
| Antisense: ACA CGT TCC TCC GAG ACC C | ||
| GCLC | Sense: ATC TGC AAA GGC GGC AAC | FAM-ACG GGT GCA GCA AGG CCC A-TAMRA |
| Antisense: ACT CCT CTG CAG CTG GCT C | ||
| HO-1 | Sense: GTG ATG GAG CGT CCA CAG C | FAM-CGA CAG CAT GCC CCA GGA TTT GTC-TAMRA |
| Antisense: TTG GTG GCC TCC TTC AAG G |
FAM, reporter fluorescent dye; TAMRA, quencher fluorescent dye.
RESULTS
Impaired induction of the NQO1 gene in mafG::mafF double-mutant mice.
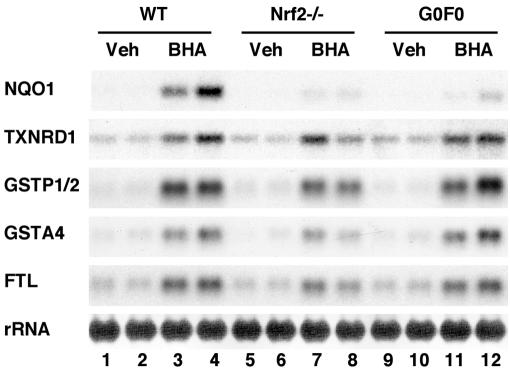
In order to prove how small Maf proteins contribute to Nrf2-dependent gene regulation, we first exploited mafG−/−::mafF−/− double-mutant (G0F0) mice. BHA was administered to wild-type (WT), G0F0, and nrf2-null mutant mice, and the expression of ARE-dependent genes (4, 21, 34, 43) was analyzed by RNA blot analysis. Induction of the NQO1 gene was severely compromised in the livers of both nrf2-null and G0F0 mice compared to that of WT mice (Fig. 1). In contrast, the induction of the other ARE-dependent genes, including TXNRD1, GSTP1/2, GSTA4, and FTL, was affected only marginally by the simultaneous deficiency of MafG and MafF, while the expression of these ARE-dependent genes was affected differentially, but more substantially, by the lack of Nrf2 (Fig. 1). The induction of GSTP1/2, GSTA4, and FTL mRNAs by BHA was decreased in the nrf2-null mouse liver, while the level of TXNRD1 was not changed much. These results suggest that, in MafG and MafF double-mutant mice, the remaining small Maf protein, MafK, compensated for lack of the small Maf activity required for Nrf2-mediated induction of ARE-dependent genes except NQO1. In the case of the NQO1 gene, the requirement level of small Maf activity may be relatively high, so that MafK could not compensate for lack of MafG and MafF. As for the residual induction by BHA of the TXNRD1, GSTP1/2, GSTA4, and FTL genes in Nrf2-null mouse liver, we surmise that there may be important contributions from certain other pathways that are activated by BHA but are independent of Nrf2. Thus, based on these analyses, we conclude that development of a much improved assay system is necessary to evaluate individual contributions of small Maf proteins and Nrf2 to the induction of ARE-dependent genes.
FIG. 1.
ARE-dependent gene expression in livers of adult small maf mutant mice. RNA blot analysis was performed to examine the expression of NQO1, TXNRD1, GSTP1/2, GSTA4, and FTL. RNA samples were prepared from the livers of adult female WT controls (lanes 1 to 4), nrf2-null mutants (Nrf2−/−; lanes 5 to 8), and mafG−/−::mafF−/− mutants (G0F0; lanes 9 to 12). BHA (lanes 3, 4, 7, 8, 11, and 12) or corn oil (Veh; lanes 1, 2, 5, 6, 9, and 10) was administered to the mice.
Requirement of small Maf proteins for ARE-dependent gene activation in keap1-null mutant mice.
To examine the contributions of small Maf proteins and Nrf2 to the regulation mediated by the ARE pathway without interference from other (real or postulated) influences, we investigated ARE-dependent activity in keap1-null mutant mice. The rationale for this approach is that, in the absence of Keap1, Nrf2 constitutively accumulates in the nucleus and activates ARE-dependent genes (43). Therefore, the keap1-null mutant mouse would serve as a powerful means for analyzing Nrf2-dependent gene regulation (27).
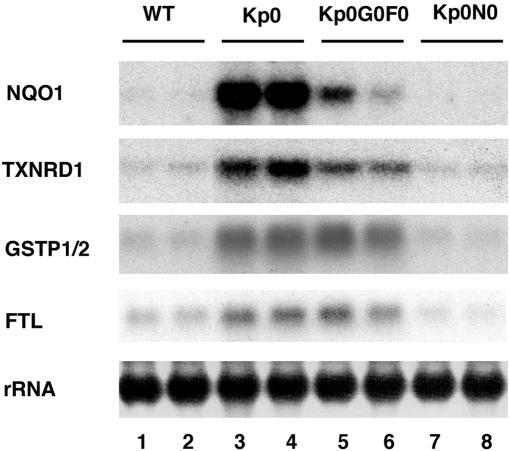
We generated two lines of compound keap1-null mutant mice, keap1−/−::mafG−/−::mafF−/− (Kp0G0F0) and keap1−/−::nrf2−/− (Kp0N0). Since keap1-null mutant pups die before weaning, the expression of ARE-dependent genes was examined in the livers of mice at postnatal day 10. As previously documented (43), multiple ARE-dependent genes were constitutively induced in the keap1-null mutant mice (Kp0) and the induction was fully abrogated in Kp0N0 compound mutant mice (Fig. 2), confirming that ARE-dependent gene activation in the absence of Keap1 is completely dependent on Nrf2. However, in the Kp0G0F0 mutant mice, only NQO1 and TXNRD1 induction was partially decreased, while GSTP1/2 and FTL were essentially unaffected by the absence of the small Maf proteins (Fig. 2). Taken together, these results demonstrate that the expression of MafG and/or MafF is indispensable for the induction of the NQO1 and TXNRD1 genes through the Nrf2-ARE pathway, while the GSTP1/2 and FTL genes are not sensitive to the loss of both MafG and MafF. Two possibilities were raised regarding the regulation of the GSTP1/2 and FTL genes. One is that the remaining small Maf protein, MafK, is sufficient for the Nrf2-mediated activation of the latter genes. The other possibility is that bZIP proteins apart from small Maf proteins cooperate with Nrf2 and activate these genes.
FIG. 2.
Induction of NQO1 and TXNRD1 in keap1 mutants is reversed by simultaneous disruption of nrf2 or mafG plus mafF. RNA blot analysis was performed to examine the expression of NQO1, GSTP1/2, FTL, and TXNRD1. RNA samples were prepared from the postnatal livers of 10-day-old female WT control mice (lanes 1 and 2) and keap1−/− (Kp0, lanes 3 and 4), keap1−/−::mafG−/−::mafF−/− (Kp0G0F0, lanes 5 and 6), and keap1−/−::nrf2−/− (Kp0N0, lanes 7 and 8) mutant mice.
All three small Maf proteins are functional in the regulation of ARE-dependent gene expression.
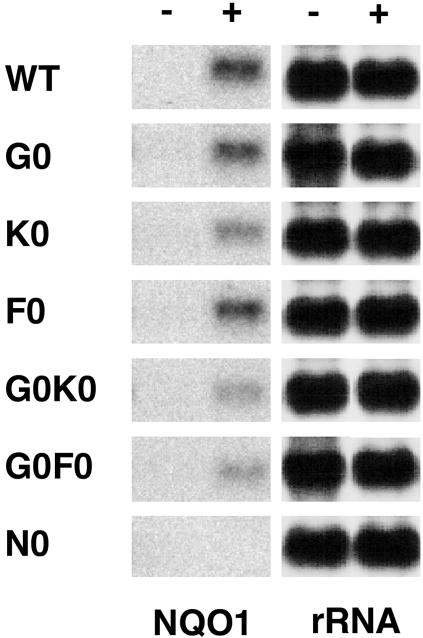
We next examined the contribution of individual small Maf proteins to the expression of ARE-dependent genes. To this end, we established MEF lines from various mutant mouse embryos, including WT, mafG−/− (G0), mafF−/− (F0), mafK−/− (K0), mafG−/−::mafF−/− (G0F0), mafG−/−::mafK−/− (G0K0), and nrf2−/− (N0) mutant embryos. These lines of mutant mice have been described in previous studies (27). We treated the fibroblasts with DEM and examined the NQO1 mRNA level, as the NQO1 gene is one of the representative Maf-dependent genes (above). Induction of NQO1 gene expression by DEM was slightly diminished in G0 and F0 MEFs in comparison to WT MEFs (Fig. 3). In contrast, induction of the NQO1 gene in K0, G0F0, and G0K0 MEFs was significantly impaired (Fig. 3), and the induction was almost abolished in nrf2-null mutant MEFs (Fig. 3). These results suggest that all three small Maf proteins are functional in the regulation of ARE-dependent gene expression in collaboration with Nrf2.
FIG. 3.
ARE-dependent gene expression in small maf single- and double-mutant embryonic fibroblasts. RNA blot analysis was performed to examine the expression levels of NQO1. RNA samples were prepared from the MEFs of WT mice and mafG−/− (G0), mafK−/− (K0), mafF−/− (F0), mafG−/−::mafK−/− (G0K0), mafG−/−::mafF−/− (G0F0), and nrf2-null (N0) mutant mice. MEFs were treated with DEM (+) or vehicle (−).
Loss of all small Maf proteins renders embryonic fibroblasts insensitive to electrophilic priming against oxidative stress.
The above results suggest that the remaining MafK makes a substantial contribution to the induction of ARE-dependent genes in G0F0 mice. So, we finally went on to generate mafG−/−::mafK−/−::mafF−/− (G0K0F0) triple-mutant mice to see the effect of a complete loss of small Maf proteins. However, the simultaneous disruption of all three small maf genes was found to lead mice to embryonic lethality before E11.5 (precise analyses will be published elsewhere). Therefore, we prepared MEFs from the triple small maf gene mutant embryos at E11.0. These embryos were reproducibly recovered from mafG+/−::mafK−/−::mafF−/− (G1K0F0) mouse intercrossing.
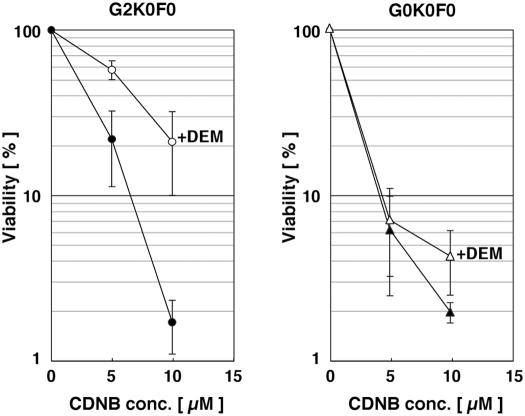
We selected a pair of fibroblasts with the G2K0F0 (mafG+/+::mafK−/−::mafF−/−) and G0K0F0 genotypes. To determine whether the G2K0F0 fibroblasts represent an appropriate reference for the gene expression analysis of G0K0F0 cells, we analyzed and compared the sensitivities of these cells to oxidative stress. These cells were incubated for 12 h with 0, 5, and 10 μM CDNB, which is known to act as a strong oxidative stress agent (6). When we recorded an indirect measure of cell viability, the differences in cell viability that represented sensitivity to CDNB toxicity were insignificant in MEFs with the G2K0F0 or G0K0F0 genotype (Fig. 4).
FIG. 4.
Small maf triple-mutant MEFs are sensitive to an oxidative stress reagent. The viability of MEFs established from mafG+/+::mafK−/−::mafF−/− (G2K0F0; left graph) and mafG−/−::mafK−/−::mafF−/− (G0K0F0; right graph) embryos is shown. MEFs were exposed to increasing concentrations (conc.) of CDNB (0, 5, and 10 μM) with or without DEM pretreatment. Cell viability was examined as described in the text, and the viability of the 0 μM CDNB sample was set to 100%. Closed circles, open circles, closed triangles, and open triangles indicate G2K0F0 MEFs without DEM, G2K0F0 MEFs with DEM, G0K0F0 MEFs without DEM, and G0K0F0 MEFs with DEM, respectively. Error bars indicate standard deviations.
Therefore, as a second test for the oxidative stress response, we pretreated MEFs for 24 h with 25 μM DEM, a mildly cytotoxic electrophile that has the potential to induce antioxidative stress enzymes. Following DEM treatment, we further treated the cells with CDNB for an additional 12 h. We found that G2K0F0 cells pretreated with DEM were much less sensitive to CDNB compared to cells without DEM pretreatment (Fig. 4). In contrast, there was no significant difference in sensitivity to CDNB between DEM-treated and untreated G0K0F0 cells (Fig. 4). These results allowed us to predict that MafG in G2K0F0 MEFs significantly contributes to the induction of ARE-dependent antioxidant and drug-metabolizing enzyme genes, while these genes were induced only weakly in G0K0F0 cells. Thereby, we concluded that G2K0F0 MEFs could be used as a reference for the gene expression analysis of G0K0F0 MEFs.
Microarray analysis documents the broad contribution of small Maf proteins to ARE-dependent gene transcription.
To comprehensively examine the effect of a complete loss of small Maf proteins on gene regulation, we exploited oligonucleotide microarrays and analyzed the expression of genes that are inducible by DEM in MEFs with the genotype G2K0F0 or G0K0F0. These fibroblasts were treated with DEM or a vehicle control, followed by RNA extraction for probe processing. In this analysis, we focused on the genes encoding drug-metabolizing enzymes and antioxidant enzymes, as these genes seem to play important roles in the acquisition of cellular resistance to oxidative stress. We found that in G2K0F0 cells, DEM treatment induced a number of genes coding for chaperone-related proteins, drug-metabolizing enzymes, and antioxidant enzymes (Table 2). A detailed comparison of the results obtained with the previous microarray analyses utilizing nrf2-null mice and nrf2::keap1 compound mutant mice (6, 21) revealed that many of the DEM-inducible genes in G2K0F0 mice were identified as the genes whose expression is heavily dependent on Nrf2 (Table 2).
TABLE 2.
Effect of small Maf protein deficiency on the regulation of DEM-induced genes in fibroblastsa
| Gene category and description | G2K0F0-DEM/G2K0F0b | G0K0F0-DEM/G0K0F0c | G0K0F0/G2K0F0d | G0K0F0-DEM/G2K0F0-DEMe |
|---|---|---|---|---|
| Chaperone system/stress response | ||||
| Heat shock protein 1A | 7.46 | 8.00 | 0.71 | 0.71 |
| Heat shock protein 1B | 6.99 | 7.20 | 0.79 | 0.83 |
| HSP40 homolog, subfamily B, member 1 | 2.56 | 2.46 | 0.97 | 0.90 |
| HSP40 homolog, subfamily B, member 9 | 2.46 | 1.32 | 1.23 | 0.71 |
| Crystallin, alpha C | 2.15 | 1.28 | 0.93 | 0.55 |
| Heat shock protein 1 | 1.58 | 1.47 | 0.80 | 0.77 |
| Crystallin, alpha B | 1.56 | 1.19 | 1.04 | 0.84 |
| DnaJ (HSP40) homolog, subfamily B, member 10 | 1.52 | 1.15 | 0.81 | 0.62 |
| DnaJ (HSP40) homolog, subfamily A, member 1 | 1.52 | 1.32 | 0.93 | 0.87 |
| Xenobiotic metabolism | ||||
| Metallothionein 2 | 12.13 | 10.56 | 1.07 | 0.93 |
| Metallothionein 1 | 9.66 | 7.84 | 1.04 | 1.00 |
| Glutamate-cysteine ligase, modifier subunit | 6.06 | 1.00 | 0.87 | 0.16 |
| Glutamate-cysteine ligase, catalytic subunit | 3.43 | 1.00 | 0.97 | 0.28 |
| NAD(P)H:quinone oxidoreductase 1 | 3.25 | 1.41 | 0.47 | 0.18 |
| UDP-glycosyltransferase 1 family, polypeptide A1 | 3.16 | 1.52 | 0.68 | 0.35 |
| Glutathione S-transferase, alpha 4 | 2.46 | 1.15 | 0.13 | 0.06 |
| Carbonyl reductase 3 | 2.30 | 0.93 | 1.00 | 0.38 |
| Microsomal glutathione S-transferase 1 | 2.14 | 1.15 | 0.66 | 0.35 |
| GlutathioneS-transferase, alpha 2 | 1.87 | 1.52 | 1.23 | 0.62 |
| GlutathioneS-transferase, mu 1 | 1.52 | 1.23 | 0.81 | 0.57 |
| P450 (cytochrome) oxidoreductase | 1.52 | 1.32 | 1.00 | 0.93 |
| Antioxidant enzymes | ||||
| Heme oxygenase 1 | 10.56 | 1.23 | 11.31 | 1.23 |
| Thioredoxin reductase 1 | 2.32 | 1.22 | 0.77 | 0.41 |
| Glutathione reductase 1 | 2.00 | 1.07 | 0.71 | 0.35 |
| Ferritin heavy chain | 1.94 | 1.11 | 1.11 | 0.67 |
| Glutaredoxin 1 | 1.87 | 1.07 | 1.00 | 0.50 |
| Catalase | 1.74 | 0.93 | 1.11 | 0.56 |
| Peroxiredoxin 1 | 1.67 | 1.09 | 0.87 | 0.58 |
| Peroxiredoxin 4 | 1.62 | 1.52 | 0.81 | 0.54 |
Each value was calculated from raw data obtained from microarray analysis. The genes categorized into the four classes shown were listed if they were induced by over 1.5-fold after DEM treatment in mafG+/+::mafK−/−::mafF−/− (G2K0F0-DEM/G2K0F0 > 1.5) cells. Gene products in boldface were previously reported to be encoded by Nrf2-dependent genes (4, 6, 21, 34, 43).
Each value indicates the n-fold change in the gene expression of G2K0F0-DEM cells relative to that of control G2K0F0 cells.
Each value indicates the n-fold change in the gene expression of DEM-treated mafG−/−::mafK−/−::mafF−/− (G0K0F0-DEM) cells relative to that of control mafG−/−::mafK−/−::mafF−/− (G0K0F0) cells.
Each value indicates the n-fold change in the gene expression of G0K0F0 cells relative to that of G2K0F0 cells.
Each value indicates the n-fold change in the gene expression of G0K0F0-DEM cells relative to that of G2K0F0-DEM cells.
Several genes that are not regarded as ARE-dependent genes, such as heat shock protein genes and metallothionein genes, were also induced by DEM but less affected by the loss of MafG (Table 2). On the other hand, we found that the induction of ARE-dependent genes was severely impaired in G0K0F0 cells in comparison to that in G2K0F0 fibroblasts (Table 2). The contribution of MafG to basal gene expression was diverse. The NQO1 and GSTA4 genes seemed to be strongly dependent on MafG for their basal expression, while the basal expression levels of the other genes were not significantly different between the two genotypes (Table 2). While regulation of GSTA4 has not been well characterized, accumulating lines of evidence suggest that GSTA4 is a typical ARE-dependent gene (4, 21). HO-1 was the only gene whose basal level of expression was increased in the absence of MafG (Table 2). This observation is in very good agreement with our previous analysis showing that the small Maf proteins collaborate with Bach1 and that the Bach1-small Maf heterodimer acts to repress HO-1 gene transcription in murine neuronal cells (16, 42).
Deficiency of small Maf proteins affects the electrophile-mediated induction of gene expression.
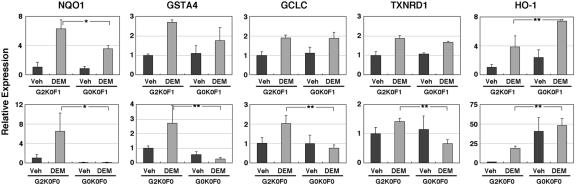
To verify the microarray results and to gain further insights into the small Maf contribution to the expression of genes induced by electrophiles, we examined the expression of several pertinent genes by quantitative reverse transcription-PCR analyses. In the analyses, in addition to the triple-null mutant MEFs (G0K0F0), we newly adopted G0K0F1 (mafG−/−::mafK−/−::mafF+/−) cells that contain only one copy of MafF, expecting the lowest expression of small Maf activity, and compared the gene expression profile of the cells with that of G2K0F1 (mafG+/+::mafK−/−::mafF+/−) cells. Consistent with the microarray data, G0K0F0 cells showed no apparent induction of the NQO1, GSTA4, TXNRD1, or GCLC gene (Fig. 5, lower panels). The HO-1 gene was similarly derepressed in both the basal and induced states, and no induction was observed anymore in G0K0F0 fibroblasts (Fig. 5, lower right panel). In the presence of one mafF allele (G0K0F1 cells versus G2K0F1 cells), the loss of MafG did not affect significantly the induction of the GSTA4, GCLC, and TXNRD1 genes and decreased and increased moderately the induction of the NQO1 and HO-1 genes, respectively (Fig. 5, upper panels). These results strongly support our contention that MafF functionally compensates the lack of MafG and MafK. In summary, we conclude that the small Maf proteins are essential for the induction of multiple ARE-dependent genes encoding xenobiotic-metabolizing enzymes and antioxidant enzymes.
FIG. 5.
Deficiency of small Maf proteins affects the electrophile-mediated induction of gene expression. Expression profiles of xenobiotic-metabolizing enzyme and antioxidant enzyme genes were examined by quantitative real-time reverse transcription-PCR. Based on the result of the microarray analysis, five genes were selected. The expression level of each mRNA was normalized to the rRNA abundance. The average values of three independent experiments (upper panels, comparison between G2K0F1 and G0K0F1 cells) or five independent experiments (lower panels, comparison between G2K0F0 and G0K0F0 cells) are shown, with error bars indicating standard deviations. The gene expression levels in vehicle (Veh)-treated G2K0F1 or G2K0F0 cells were set to 1. Asterisks and double asterisks indicate statistically significant differences by unpaired t test (*, P < 0.05; **, P < 0.01).
DISCUSSION
The small Maf proteins are unique transcription factors with no recognizable transcriptional regulatory domains other than DNA binding and dimerization motifs. The small Maf proteins have been shown to form heterodimers in combination with CNC and Bach family proteins and can also form homodimers, as well as heterodimers, among themselves, depending on the abundance or activity of each molecule. The physiological concentration of each participant molecule is known to be crucial for the final output of transcriptional activity (28). Therefore, it would be ideal to elucidate the function of the small Maf proteins by a loss-of-function strategy in order to evaluate the contribution of each of these molecules to specific transcriptional responses under physiological conditions. However, since the three small Mafs are expressed in broadly overlapping patterns (35, 39) and since no functional differences among them have been documented, analysis of the single-gene loss-of-function mutants has been limited.
Our group and others have documented that the small Maf proteins act cooperatively with p45 NF-E2 to activate target genes in megakaryocytes (28, 36, 40). Therefore, heterodimers of the other CNC family (Nrf1, Nrf2, and Nrf3) and small Maf proteins were anticipated to act as transcriptional activators as well. However, cell culture studies have shown that small Maf proteins can behave as negative regulators of Nrf2-dependent reporter gene activation (3, 32). To definitively address whether or not the Nrf2 and small Maf proteins act as cooperative transcriptional activators in vivo, we examined the expression of ARE-dependent genes in small Maf and other (keap1 and nrf2) mutant mice whose biochemical pathways hypothetically intersect. The prediction of these experiments was that if CNC-small Maf heterodimers positively regulate transcription, the induction of Nrf2-dependent genes should be diminished in small maf mutant mice, as well as in nrf2 mutant mice. In contrast, if the small Mafs contribute only to negative regulation through AREs, a loss of small Maf activity would be expected to confer an activation phenotype.
In this study, we first examined G0F0 mice to prove the contribution of small Maf proteins in vivo. We failed to observe any significant effects of small Maf reduction on the abundance of ARE-dependent genes other than NQO1 and TXNRD1. When G0K0F0 fibroblasts lacking all three small Maf proteins were established and examined, we finally observed a complete loss of induction in almost all of the Nrf2-dependent genes induced in this analysis. Intriguingly, many of the ARE-dependent genes, with a few exceptions, responded to electrophilic reagents in G2K0F0 cells and G0K0F1 cells, as well as in G0F0 (G0K2F0) mouse livers. This implies that each single small Maf protein is more or less capable of supporting Nrf2-mediated transcriptional activation. This is the first report that genetically demonstrates the ability of each small Maf protein to contribute to ARE-dependent gene activation and the essential roles of the family of small Maf proteins in the activation.
Strictly speaking, in this study, we could evaluate the contribution of small Maf proteins only in relation to inducible ARE-dependent genes in fibroblasts. Since GSTP1/2 and FTL were not significantly induced in fibroblasts, it was still possible that in the liver the GSTP1/2 and FTL genes are regulated in an Nrf2-dependent and Maf-independent manner. Indeed, it has been reported that Nrf2 heterodimerizes with c-Jun (13) and ATF4 (5). In contrast, a comprehensive analysis of all possible protein-protein interactions was recently reported using a protein array that was fabricated with an almost complete set of coiled-coil domains of all the human bZIP transcription factors. The data revealed that the binding affinities between the small Maf and CNC proteins were much stronger than those between CNC proteins and c-Jun or ATF4 (31). This result leads us to believe that small Maf proteins are substantial partners of Nrf2 and that induction of GSTP1/2 and FTL in a G0F0 background was maintained by Nrf2-MafK heterodimer. Nonetheless, it remains an open question whether or not Nrf2-c-Jun or Nrf2-ATF4 heterodimers might be able to functionally compensate for Nrf2-Maf loss in vivo. Simultaneous disruption of keap1 and c-jun or ATF4 may provide unique insights into this issue. In any case, our present data rejected the notion that the small Maf proteins contribute exclusively to ARE-mediated repression.
Since we here provide genetic evidence that small Maf proteins are essential for the ARE-dependent genes, it might be possible that small Maf proteins are essential for DNA binding of Nrf2 and/or for other processes, including the localization and stabilization of Nrf2. We previously reported that the nuclear localization of Bach proteins is dependent on small Maf proteins (16). Since Bach proteins possess a nuclear export signal (NES), we surmised that small Maf proteins prevent Bach proteins from being exported from nuclei by the NES binding protein CRM1 (exportin 1). Recently, a NES has been identified in the leucine zipper motif of Nrf2 (12, 23), which suggests the possibility that the nuclear accumulation of Nrf2 is also influenced by small Maf proteins.
In conclusion, we have provided evidence that a genetic reduction in the abundance of small Maf proteins has a great influence on the expression of a wide range of ARE-dependent genes. Our in vivo experiments utilizing small maf double-mutant mice also implied the differential contribution of small Maf proteins to the expression of ARE-dependent genes. For instance, the NQO1 gene seems to be more sensitive to the reduction of small Maf proteins compared to several other ARE-dependent genes. We envisage that the differential requirement of small Maf proteins might be attributed to the variation in the ARE and its flanking sequence of each gene, which might result in the differential affinity for the activator complex containing Nrf2-small Maf heterodimer (34). However, we could not find a clear correlation between the sensitivity to the reduction in small Mafs and the ARE sequence variations. We also surmise that such a differential threshold might enable small Maf proteins, by virtue of a change in their abundance, to differentially regulate specific ARE-dependent genes. Since the abundance of small Maf proteins has been reported to fluctuate in response to oxidative stress (17, 26), the intriguing possibility emerges that the small Maf proteins may generate diversity in Nrf2-mediated gene regulation by changing their abundance.
Acknowledgments
We greatly appreciate the assistance of Y. Meguro in the microarray analysis. We are grateful to K. Itoh, M. Kobayashi, and T. Suzuki for advice and helpful comments. We thank T. O'Connor, Y. Tamagawa, R. Kawai, and A. Sakurai for help.
This work was supported by grants from the NIH (CA80088 and GM28896 to F.K., H.M., and J.D.E.), ERATO-JST (M.Y.), the Ministry of Education, Science, Sports and Culture (H.M. and M.Y.), the Atherosclerosis Foundation (M.Y.), the Yamanouchi Foundation for Research on Metabolic Disorders (H.M.), and the Uehara Memorial Foundation (H.M.). F.K. is a JSPS Research Fellow.
REFERENCES
- 1.Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. [DOI] [PubMed] [Google Scholar]
- 2.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 90:11371-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhakshinamoorthy, S., and A. K. Jaiswal. 2000. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:quinone oxidoreductase1 gene. J. Biol. Chem. 275:40134-40141. [DOI] [PubMed] [Google Scholar]
- 4.Hayes, J. D., J. U. Flanagan, and I. R. Jowsey. 2005. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45:51-88. [DOI] [PubMed] [Google Scholar]
- 5.He, C. H., P. Gong, B. Hu, D. Stewart, M. E. Choi, A. M. Choi, and J. Alam. 2001. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 276:20858-20865. [DOI] [PubMed] [Google Scholar]
- 6.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 7.Itoh, K., K. Igarashi, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1995. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 158:4184-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, K., T. Ishii, N. Wakabayashi, and M. Yamamoto. 1999. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 31:319-324. [DOI] [PubMed] [Google Scholar]
- 10.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 12.Jain A. K., D. A. Bloom, and A. K. Jaiswal. 17 May 2005. Nuclear import and export signals in control of NRF2. J. Biol. Chem. doi: 10.1074/jbc.M502083200. [DOI] [PubMed]
- 13.Jeyapaul, J., and A. K. Jaiswal. 2000. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of γ-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 59:1433-1439. [DOI] [PubMed] [Google Scholar]
- 14.Johnsen, O., P. Murphy, H. Prydz, and A. B. Kolsto. 1998. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 26:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka, K., M. Noda, and M. Nishizawa. 1994. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14:700-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsuoka, F., H. Motohashi, Y. Tamagawa, S. Kure, K. Igarashi, J. D. Engel, and M. Yamamoto. 2003. Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Mol. Cell. Biol. 23:1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsuoka, F., H. Motohashi, J. D. Engel, and M. Yamamoto. 2005. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J. Biol. Chem. 280:4483-4490. [DOI] [PubMed] [Google Scholar]
- 18.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 19.Kobayashi, A., E. Ito, T. Toki, K. Kogame, S. Takahashi, K. Igarashi, N. Hayashi, and M. Yamamoto. 1999. Molecular cloning and functional characterization of a new Cap′n' collar family transcription factor Nrf3. J. Biol. Chem. 274:6443-6452. [DOI] [PubMed] [Google Scholar]
- 20.Kusunoki, H., H. Motohashi, F. Katsuoka, A. Morohashi, M. Yamamoto, and T. Tanaka. 2002. Solution structure of the DNA-binding domain of MafG. Nat. Struct. Biol. 9:252-256. [DOI] [PubMed] [Google Scholar]
- 21.Kwak, M. K., N. Wakabayashi, K. Itoh, H. Motohashi, M. Yamamoto, and T. W. Kensler. 2003. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 278:8135-8145. [DOI] [PubMed] [Google Scholar]
- 22.Leung, L., M. Kwong, S. Hou, C. Lee, and J. Y. Chan. 2003. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 278:48021-48029. [DOI] [PubMed] [Google Scholar]
- 23.Li, W., M. R. Jain, C. Chen, X. Yue, V. Hebbar, R. Zhou, and A. N. Kong. 23 May 2005. NRF2 possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J. Biol. Chem. doi: 10.1074/jbc.M410601200. [DOI] [PubMed]
- 24.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 25.Moi, P., K. I. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, J. A., E. L. Dahl, and R. T. Mulcahy. 2002. Differential induction of mafF, mafG and mafK expression by electrophile-response-element activators. Biochem. J. 361:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motohashi, H., F. Katsuoka, J. D. Engel, and M. Yamamoto. 2004. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 101:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motohashi, H., F. Katsuoka, J. A. Shavit, J. D. Engel, and M. Yamamoto. 2000. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103:865-875. [DOI] [PubMed] [Google Scholar]
- 29.Motohashi, H., T. O'Connor, F. Katsuoka, J. D. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Motohashi, H., J. A. Shavit, K. Igarashi, M. Yamamoto, and J. D. Engel. 1997. The world according to Maf. Nucleic Acids Res. 25:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, T., H. C. Huang, and C. B. Pickett. 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 275:15466-15473. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 34.Nioi, P., M. McMahon, K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onodera, K., J. A. Shavit, H. Motohashi, F. Katsuoka, J. E. Akasaka, J. D. Engel, and M. Yamamoto. 1999. Characterization of the murine mafF gene. J. Biol. Chem. 274:21162-21169. [DOI] [PubMed] [Google Scholar]
- 36.Onodera, K., J. A. Shavit, H. Motohashi, M. Yamamoto, and J. D. Engel. 2000. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 19:1335-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oyake, T., K. Itoh, H. Motohashi, N. Hayashi, H. Hoshino, M. Nishizawa, M. Yamamoto, and K. Igarashi. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16:6083-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rushmore, T. H., M. R. Morton, and C. B. Pickett. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632-11639. [PubMed] [Google Scholar]
- 39.Shavit, J. A., H. Motohashi, K. Onodera, J. Akasaka, M. Yamamoto, and J. D. Engel. 1998. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 12:2164-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shivdasani, R. A., M. F. Rosenblatt, D. Zucker-Franklin, C. W. Jackson, P. Hunt, C. J. Saris, and S. H. Orkin. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81:695-704. [DOI] [PubMed] [Google Scholar]
- 41.Stewart, D., E. Killeen, R. Naquin, S. Alam, and J. Alam. 2003. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278:2396-2402. [DOI] [PubMed] [Google Scholar]
- 42.Sun, J., H. Hoshino, K. Takaku, O. Nakajima, A. Muto, H. Suzuki, S. Tashiro, S. Takahashi, S. Shibahara, J. Alam, M. M. Taketo, M. Yamamoto, and K. Igarashi. 2002. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 19:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]