Abstract
Spatial organization of chromatin in the interphase nucleus plays a role in gene expression and inheritance. Although it appears not to be random, the principles of this organization are largely unknown. In this work, we show an explicit relationship between the intranuclear localization of various chromosome segments and the pattern of gene distribution along the genome sequence. Using a 7-megabase-long region of the Drosophila melanogaster chromosome 2 as a model, we observed that the six gene-poor chromosome segments identified in the region interact with components of the nuclear matrix to form a compact stable cluster. The six gene-rich segments form a spatially segregated unstable cluster dependent on nonmatrix nuclear proteins. The resulting composite structure formed by clusters of gene-rich and gene-poor regions is reproducible between the nuclei. We suggest that certain aspects of chromosome folding in interphase are predetermined and can be inferred through in silico analysis of chromosome sequence, using gene density profile as a manifestation of “folding code.”
In the interphase nucleus, individual chromosomes occupy discrete and largely autonomous chromosome territories (CTs) (10, 25). Spatial organization of chromatin within CTs plays a role in gene expression (8) and inheritance (7). However, mechanisms of chromosome folding within CTs are far from clear. The observed remodeling of chromosome configuration during interphase (11) and regulated changes in the intranuclear position of certain genetic loci during cell differentiation (5, 9, 15, 20, 23, 30) exemplify a dynamic nature of organization of CTs. However, studies on the localization of other genes (17, 20) and of entire chromosome segments (12) in interphase nuclei suggest that they occupy fixed positions within the CT, and these positions do not change with alterations in gene activity. This implies that certain aspects of chromatin folding are invariable and perhaps preprogrammed. Irregular localization of RNA polymerase and splicing factors within the interphase nucleus (1, 16, 28) indicates that certain compartments are enriched with these components and thus perhaps are better suited for gene expression than other compartments. Preprogrammed chromosome folding may result in consistent intranuclear positioning of different chromosome regions relative to such compartments. The observation that gene-enriched chromosome bands tend to associate with the splicing factor (SC-35) domains (26) implies that gene-rich chromosome segments are tethered to nuclear subcompartments favorable for large-scale transcription. Alternatively, the presence of a gene-rich chromosome segment may create a nuclear compartment with elevated gene density; such a compartment would recruit transcription and splicing machinery, thus establishing the transcription/splicing hub. This possibility is supported by analysis of repositioning of activated muscle genes to the splicing hubs (the SC-35 domains) (22), which demonstrated that the location of the genes within the CT did not change. This suggests the reorganization of the SC-35 domains rather than gross alterations in chromosome folding during changes in transcription program and implies the dynamic nature of the SC-35 domains. The accumulating evidence for preprogrammed folding of chromosomes in the interphase nucleus and for the association of chromosome regions with intranuclear structures according to gene density led us to suggest that the pattern of chromosome folding within the CT may be predicted from the profile of gene density along the chromosome. Our findings support this hypothesis, indicating that certain aspects of three-dimensional chromatin organization are encoded in the linear genomic sequence.
MATERIALS AND METHODS
Genome sequence analysis.
To compute gene density, we used annotated genome sequences of Drosophila melanogaster (http://www.fruitfly.org) and of Caenorhabditis elegans (http://www.wormbase.org). For each gene, local gene density was calculated by dividing the preset number of genes in a sliding window centered on the query gene by the length of the region occupied by these genes. Presented data are for a sliding window size of 50 genes for D. melanogaster and 25 genes for C. elegans. A moving average of gene density for a window of 150 genes was also generated. Gene-rich regions were identified as the ones with a gene density above the local average (as indicated by a moving average curve) and gene-poor regions as the ones with a density below that. For calculations of the sizes of the regions, the borders between the gene-rich and gene-poor regions were placed at the intersections between the gene density curve and the moving average curve, which in most cases corresponded roughly to half of the gene density peak's height. Putative scaffold/matrix attachment regions (S/MARs) were predicted from the genome sequence with the help of SMARtest software (14). A sliding window of 300 kb was moved along the chromosome sequence in 100-kb increments, and the number of genes and the number of predicted S/MARs were calculated for each window. Correlation between the gene number and the S/MAR number per window (i.e., between the gene density and the S/MAR density) was calculated using Microsoft Excel software.
Cell culture.
Schneider-2 Drosophila cells were grown in Shields and Sang M3 medium supplemented with 10% fetal calf serum and were synchronized in the late G1 phase in the presence of 200 μg/ml l-mimosine. Cells were incubated in fresh medium for 1 h after removal of the l-mimosine and then fixed for fluorescence in situ hybridization (FISH).
FISH.
Cells were washed in phosphate-buffered saline (PBS), then fixed in 4% paraformaldehyde and 0.1% Triton X-100 in PBS, treated with 0.1 N HCl for 20 min, washed with PBS, and treated with 100 μg/ml of RNase A in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 h. Then, cells were incubated with 8 mg/ml salmon sperm DNA and 0.01% psoralen, cross-linked under the UV source, and hybridized with fluorescent probes. Alternatively, the treatment with psoralen and salmon sperm DNA was omitted. To extract nonmatrix proteins, nuclei were incubated with 2 M NaCl in PBS for 20 min, then fixed with paraformaldehyde, treated with 0.1 N HCl for 20 min, washed with PBS, treated with RNase A, and hybridized. Probes were prepared using the Prime-It Fluor labeling kit (Stratagene) and deoxynucleoside triphosphates conjugated with either Alexa Fluor 488 or Alexa Fluor 647 for gene-poor regions and with Alexa Fluor 594 for gene-rich regions (Molecular Probes). The templates used were bacterial artificial chromosomes (BACs) isolated by the alkaline lysis procedure. Exact chromosome coordinates and sizes of individual BACs used (BACR13P06, BACR14B02, BACR10M14, BACR20O22, BACR35F01, BACR33L22, BACR38O04, BACR02C23, BACR30G11, BACR09B12, BACR37I09, BACR36J03, BACR07P02, BACR30O03, BACR07P02, BACR03D24, BACR29I14, BACR36P23, BACR03E17, BACR32P08, BACR21A09, BACR20E20, BACR48A20, BACR01N09, and BACR19I21) are shown in Drosophila genome sequence annotations (www.flybase.net). The BAC clones were obtained from the BACPAC resources center at the Children's Hospital Oakland Research Institute. Hybridizations were performed according to the manufacturer's protocols, and slides were counterstained for total DNA with DAPI (4′,6′-diamidino-2-phenylindole) and mounted in VectaShield medium (Vector).
Microscopic analysis and image processing and analysis.
Nuclei were examined in a Nikon Eclipse E800 microscope equipped with a 60× objective and an Orca-ER digital charge-coupled-device camera (Hamamatsu). Images were captured using OpenLab 3.1.5 software (Improvision) and pseudocolored as follows: Alexa Fluor 488 or Alexa Fluor 647, green; Alexa Fluor 594, red; and DAPI, blue. Each channel was processed in Adobe Photoshop 7.0 (Adobe Systems) to adjust the input levels to fit the actual signal intensity distribution and to apply threshold to eliminate general nonspecific background. Three-dimensional reconstructions were performed from the serial optical sections using raw data with Volocity 2.0.1 software (Improvision).
Images were analyzed visually to determine the mode of distribution of signal. Only the nuclei with both red and green signals clearly visible and in focus were selected. Distribution of the signal was considered “clustered” when one or two closely located and interconnected spots were observed. In nuclei with two spots located more than a nucleus radius apart or with three or more spots, the distribution was considered “dispersed.” Morphological analysis of the clustered signals was performed using Image-Pro Plus 4.1.1.2 software (Media Cybernetics).
RESULTS AND DISCUSSION
Variations of gene density along chromosomes: gene-rich and gene-poor regions.
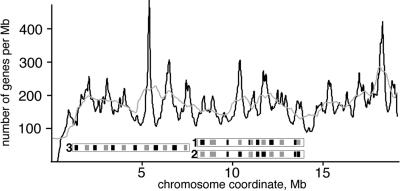
First, we computed the gene density profile along the right arm of chromosome 2 in D. melanogaster and found a cyclic pattern of peaks and troughs of gene density with an average period of 0.9 megabase pairs (Mb) (standard deviation [SD] = 0.3 Mb) (Fig. 1). Overall gene density varied along the chromosome; therefore, we applied a moving average as a reference of local average gene density (Fig. 1, gray line). Chromosome regions with a gene density above the local average were identified as gene rich and those with a gene density below the local average as gene poor. An average gene-rich region spans over 390 kb (SD = 170 kb) and contains 59 genes (SD = 25); a gene-poor region spans 500 kb (SD = 230 kb) and contains 67 genes (SD = 25). A similar pattern was detected for chromosome 1 in C. elegans (data not shown). Variations of gene density along the chromosomes have also been found in humans (29), indicating that this observation reflects a general trend of eukaryotic genome organization.
FIG. 1.
Gene density profile reveals alternating pattern of gene-rich and gene-poor regions on chromosome arm 2R of D. melanogaster. The solid line indicates local gene density; the gray line shows a moving average for 150 genes on the D. melanogaster chromosome. Probe sets used for FISH are shown below the gene density curve, where gray rectangles indicate probes labeled with Alexa Fluor 594 (pseudocolored red on other figures) and black rectangles indicate probes labeled with Alexa Fluor 488 or Alexa Fluor 647 (pseudocolored green).
Clusters of gene-rich and gene-poor chromosome regions in the interphase nucleus.
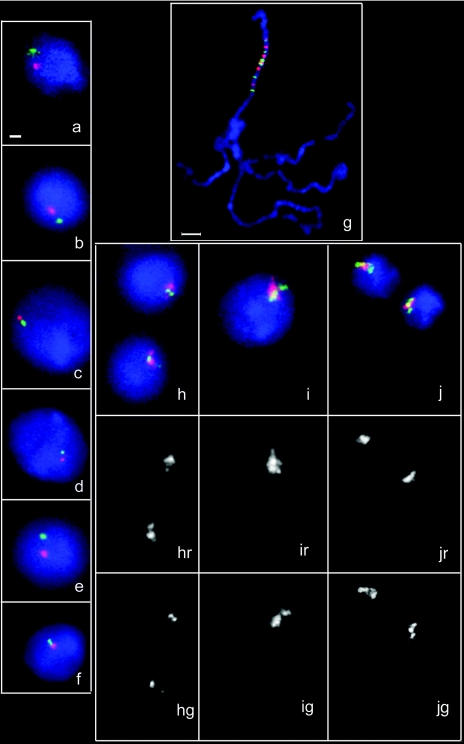
To study the localization of the gene-rich and gene-poor regions in Drosophila interphase nuclei, we employed a mixture of 12 fluorescent hybridization probes that surveyed about one-third of the right arm of chromosome 2 (Fig. 1, probe set 1). Probes corresponding to the gene-rich regions were labeled with Alexa Fluor 594 (red) and probes for gene-poor regions with Alexa Fluor 488 (Fig. 2, 3, and 4) or Alexa 647 (Fig. 5), both pseudocolored green. Each individual probe gave a single signal when hybridized with the interphase nuclei using the FISH procedure. When mixed together and hybridized to the polytene chromosomes, probes hybridized to the expected cytological sites, covering about one-third of the chromosome arm 2R, and showed no significant homology elsewhere in the genome (Fig. 2g). In the interphase nuclei hybridized with the same mixture of probes, we observed consistent spatial segregation of the gene-rich regions from the gene-poor regions. In addition, the gene-poor regions usually clustered together, as did the gene-rich regions, so that individual signals fused together, forming two separate entities. This pattern was observed both in the cultured Schneider-2 cells (Fig. 2h) and in the cells of larval brain (Fig. 2i). Control hybridization of cultured cells with the probes with random assignment of colors to gene-rich and gene-poor regions (Fig. 1, probe set 2) did not show segregation between the red and green signals (Fig. 3h).
FIG. 2.
Characterization of the set 1 of probes for FISH. Individual BACs representing the probes from set 1 (Fig. 1) were labeled with either Alexa Fluor 594 (red) or Alexa Fluor 488 (green) and hybridized pairwise with the interphase nuclei of the Schneider-2 cells (panels a to f). Combinations of probes were as follows: a, probes 1 and 2; b, probes 3 and 4; c, probes 5 and 6; d, probes 7 and 8; e, probes 9 and 10; and f, probes 11 and 12. All probes were mixed and hybridized with the squashed polytene nuclei from the larval salivary glands (g) and with interphase nuclei of Schneider-2 cells (h and i) or of the cells from larval brain (j). Images from the red channel (marked “r”) and from the green channel (marked “g”) are shown below the merged images in panels h to j. The horizontal bar (a) represents 1 μm. All images were taken at the same magnification except for panel g, where the bar represents 10 μm.
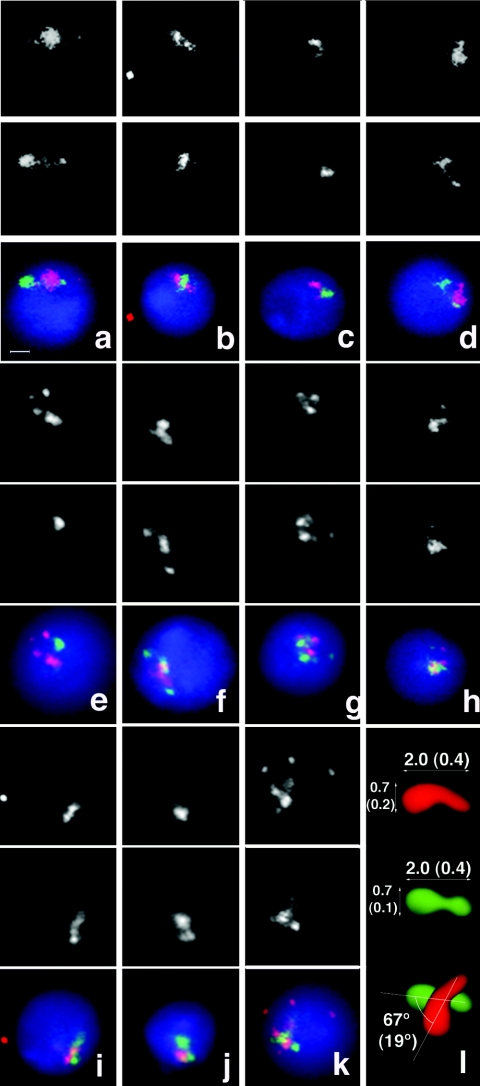
FIG. 3.
Gene-rich and gene-poor regions form distinct clusters in interphase nuclei. FISH probes used are as shown in Fig. 1. Probe set 1 was used for all hybridizations except for panel h, where set 2 was used. Total nuclear DNA is stained with DAPI (blue). (a to g) Images of the formaldehyde/psoralen-fixed nuclei hybridized with probe set 1. The upper panel represents the red channel (probes for gene-rich regions) and the middle panel the green channel (probes for gene-poor regions), and lower panels show red and green merged with the DAPI stain. (h) Nucleus treated as in panels a to g but hybridized with probe set 2. (i to k) Nuclei fixed with formaldehyde alone and hybridized with probe set 1. All images were taken at the same magnification. The horizontal bar (a) represents 1 μm. (l) Schematic representation of thequantitation of images showing the sizes of the gene-rich and the gene-poor clusters in micrometers and the angle between long axes of the clusters (standard deviations shown in parentheses).
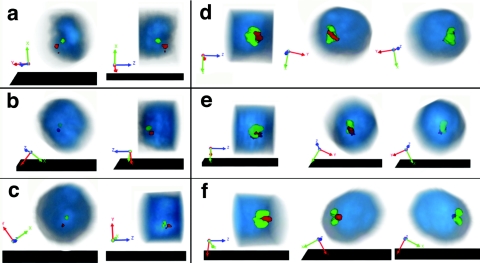
FIG. 4.
Three-dimensional reconstructions show spatial segregation between clusters of gene-rich and gene-poor regions. Three-dimensional models were reconstructed from serial optical sections. Red signal is for gene-rich regions and green signal is for gene-poor regions. Bulk chromatin stained with DAPI is shown in blue. (a to c) The models of three different nuclei hybridized with probe set 1, each shown in two projections. In each row, the left panel represents a frontal view and the right panel a side view of a model. (d to f) The models of three different nuclei hybridized with probe set 3 that covers a different region of chromosome 2 (Fig. 1); each model is shown in three projections.
FIG. 5.
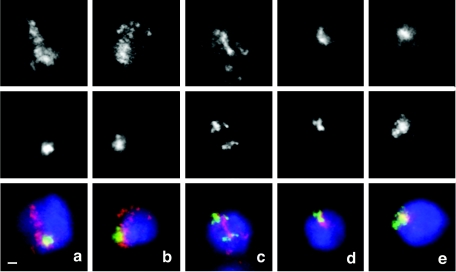
Nuclei preextracted with 2 M NaCl and hybridized with set 1 of fluorescent probes (Fig. 1). (a and b) In a majority of nuclei, extraction of nonmatrix proteins with high salt concentration results in further unfolding of gene-rich chromosomal segments (red), which are seen as dispersed specks and threads, but does not lead to breakdown of the clusters of gene-poor regions (green). (c to e) Representative images of other types of nuclei observed with both the red and green signals dispersed (c) and both signals clustered (d and e). All images were taken at the same magnification. The horizontal bar (a) represents 1 μm.
To study the distribution of signals from the fluorescent probes in interphase nuclei in more detail, we analyzed nuclei of the cultured cells and found that both the gene-rich and the gene-poor regions were clustered in 68% of cells (standard error [SE] = 8%). Flat images (Fig. 3a to d) demonstrate the cross-like arrangement of the elongated cluster of gene-poor regions overlying another elongated cluster of gene-rich regions (or vice versa) from different perspectives. Morphological analysis of clusters showed that both the gene-rich and gene-poor clusters are similar in size and shape between the nuclei, indicating a nonrandom pattern of chromosomal folding. Gene-rich clusters were 2 μm long (SD = 0.43 μm) and 0.7 μm wide (SD = 0.16 μm). Gene-poor clusters had almost identical sizes (2 μm long [SD = 0.40 μm] and 0.66 μm wide [SD = 0.12 μm]). The angle between the long axes of the gene-rich and the gene-poor clusters was 67o (SD = 19o) (Fig. 3l). Three-dimensional reconstruction showed that clusters of gene-rich and gene-poor regions are spatially separated from each other (Fig. 4a to c). Similar observations of the formation of reproducible structures by gene-rich and gene-poor regions were obtained with the probes that covered a different region of chromosome 2 (Fig. 1, probe set 3, and 4d to f), signifying that nonrandom chromosome folding is not limited to a particular segment of the D. melanogaster genome. Our results indicate that the pattern of chromosome folding in the interphase nucleus is encoded in the genome sequence and that this code is manifested by the profile of gene density along the chromosome. Further, we observed clustering of gene-rich and gene-poor chromosomal segments in distinct nonoverlapping nuclear compartments.
Gene-rich regions are organized in fragile clusters that easily disintegrate.
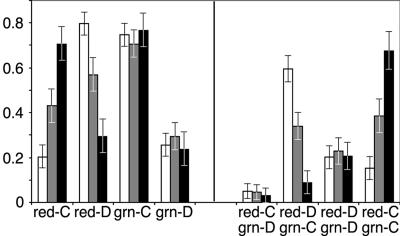
In 68% of nuclei analyzed, the six gene-rich segments of the studied region of chromosome 2 were observed as a single cluster, as well as the six gene-poor segments, as described above. However, there were smaller fractions of nuclei showing different arrangements of signals. In particular, individual FISH signals from both gene-rich and gene-poor regions were dispersed and seen as distinct specks (Fig. 3g) in 21% of nuclei (Fig. 6). In 9% of nuclei, gene-rich regions were dispersed while gene-poor regions still clustered (Fig. 3e); the opposite situation (Fig. 3f) was observed in only 3% of nuclei. Altogether, the gene-rich regions were clustered in 70% and the gene-poor regions in 75% of nuclei, thus showing no significant difference (Fig. 6).
FIG. 6.
Clustering of the gene-rich chromosomal regions but not of the gene-poor regions is affected by treatment of nuclei prior to hybridization. This figure represents the fraction of nuclei in which different distributions of red and green signals were observed for the cells fixed with formaldehyde and psoralen (black) or formaldehyde alone (gray) or preextracted with 2 M NaCl (white). The left panel shows the compact (red-C) versus the dispersed (red-D) distribution of probes for the gene-rich regions (red) and a similar distribution for the green-labeled probes for the gene-poor regions (grn). The right panel demonstrates distribution of nuclei with different arrangements of red and green signals, showing that the two classes significantly affected by pretreatment are the “red dispersed, green clustered” (red-D, grn-C) and “red clustered, green clustered” (red-C, grn-C). Standard error bars are shown.
We suggested that chromatin denaturation known to occur during the in situ hybridization procedure (27) leads to disruption of the compact clusters. Indeed, the observed proportion of nuclei showing the scattered versus clustered signals depended on the procedure for preparation of cells for hybridization. In our studies, we usually treated cells with the cross-linking agents formaldehyde and psoralen before hybridization to preserve the chromatin morphology; this method mostly yields clustered signals, as discussed above. In contrast, fixation of the cells with the formaldehyde alone—the method typically used for studies of chromatin structure—results in an increase in the number of nuclei showing dispersed signals. Specifically, omission of cross-linking with psoralen affects clustering of the gene-rich regions but does not apparently affect gene-poor regions (Fig. 6). The result is an increase in the number of nuclei showing clustered gene-poor regions and dispersed gene-rich regions (34% versus 9% in psoralen-fixed samples) at the expense of the fraction of nuclei showing clustered gene-poor and gene-rich regions (39% versus 68% in psoralen-fixed nuclei). However, in formaldehyde-fixed nuclei with clustered gene-rich and gene-poor segments (Fig. 3i and j), the observed structures were similar to the ones in formaldehyde/psoralen-fixed cells (Fig. 3a to d). Likewise, morphologies of the spots in the nuclei with dispersed signals were similar in the specimens fixed with formaldehyde and psoralen (Fig. 3e to g) and formaldehyde alone (Fig. 3k). Therefore, additional cross-linking with psoralen does not significantly change preservation of chromatin packaging but rather stabilizes three-dimensional chromatin architecture. While packaging of interphase chromosome in large-scale fibers observed in vivo seems to be conserved in fixed specimens (3), the spatial arrangement of such fibers is apparently quite sensitive to chromatin denaturation during the FISH procedure.
Intranuclear locations of dispersed signals from the gene-rich regions relative to the cluster of gene-poor regions and the numbers of individual specks were not conserved between nuclei (Fig. 3e to g and k), in contrast to the highly reproducible arrangement of the clustered signals (Fig. 1h and i and 2a to d, i, j, and l). Moreover, the dispersed FISH signals were observed out of bounds of Hoechst-stained bulk chromatin in 52% of nuclei (SE = 10%) (Fig. 2i). In contrast, clustered signals were within the bounds of the nucleus in 91% of nuclei (SE = 6%). These observations lend further support to the suggestion that dispersed arrangement of gene-rich regions, frequently observed in formaldehyde-fixed nuclei, is mainly a result of disintegration of compact clusters caused by partial chromatin denaturation during the FISH procedure. Breakdown of the gene-rich clusters, with gene-poor regions still being bound together, likely reflects looping of the gene-rich regions towards the surface of the CT or even beyond. This is similar to the observation in mammals where such loops containing active genes have been detected in formaldehyde-fixed specimens, and the likelihood of looping of the genomic segment correlated to increased gene density rather than to activity of genes within the segment (21). Therefore, the relation between the gene density pattern and chromosome folding seems to be conserved between flies and mammals.
Since a gene-rich (and gene-poor) region typically spans about half a megabase of genome and contains multiple genes with different expression patterns, we believe that these structures are simply too large to be adjusted according to contradicting changes in transcription programs of multiple genes contained within. Such adjustments are probably limited to smaller chromatin domains that often contain genes with similar transcription patterns. Therefore, observations of the loops extending into interchromosomal space only upon transcriptional activation of genes contained therein (9, 23, 30) likely reflect decondensation of unusually large chromatin domains, such as in the case of the beta-globin or the hox gene clusters.
While clustering of gene-poor regions is mediated by extensive binding to nuclear matrix (see below), the nature of interactions that bring the gene-rich regions together is not clear yet. It is possible that these regions are ensnared into nuclear compartments enriched with transcription and splicing factors (such as “local euchromatic neighborhoods” [26]) because of their active involvement in gene expression. Alternatively, clustering of gene-rich regions may be a primary process mediated by specific mutual affinity of such regions. This creates nuclear compartments with elevated gene density that, in turn, may recruit transcription and splicing factors. Dissection of the mechanism of clustering of gene-rich regions and of its relation to the distribution of core transcription/splicing machinery within the nucleus appears to represent an exciting avenue of future research.
Gene-poor segments are securely anchored in nuclear matrix.
We observed that the compact structures formed by gene-rich segments are quite fragile and easily disrupted by partial denaturation of chromatin. In contrast, clustering of gene-poor chromosome segments is not apparently affected by formaldehyde fixation of cells compared to the formaldehyde/psoralen procedure (Fig. 6). In both cases, gene-poor segments were clustered in a majority of nuclei. Hence, the interactions that underlie such clustering are more stable than the ones that organize the clusters of gene-rich regions.
We hypothesized that the secure anchoring within the nucleus is probably achieved through extensive binding of nuclear matrix-associated proteins to noncoding DNA in gene-poor regions. These interactions, as well as corresponding noncoding DNA sequences, may be less frequent in the gene-rich regions. In this case, variation of gene density along the chromosome may simply reflect the periodic enrichment of genomic sequence with noncoding DNA regions attached to each other and/or to the nuclear matrix via binding proteins, such as the Su(Hw)/Mod(mdg4) insulator binding complex (6). This model implies regular chromosome looping between the gene-poor regions, with the loops being relatively gene rich.
To test the model, we analyzed the distribution of predicted S/MARs along the 5-Mb segment of chromosome 2 (chromosome positions 3 through 8 Mb in Fig. 1). This region contains 682 genes, and a total of 574 S/MARs were predicted. Correlation between the gene density and the density of predicted S/MARs was substantially negative (−0.52), thus supporting the model. However, MAR prediction algorithms are still far from perfection, and the function of any putative MAR in vivo is questionable until proven experimentally.
To further analyze the involvement of nuclear matrix in localization of the gene-rich and gene-poor regions, we extracted nuclei with 2 M NaCl and then performed in situ hybridization. High-salt treatment removes the majority of nuclear proteins, except for the components of the nuclear matrix, while preserving the overall integrity of CTs (19). The morphology of the structures detected by FISH (Fig. 5) in the salt-extracted nuclei was in striking contrast to the observations made of the “intact” nuclei fixed with formaldehyde/psoralen or with formaldehyde alone (Fig. 2 and 3). Dispersed signals were detected as diffuse staining, sometimes extending beyond the boundaries of DAPI-stained bulk chromatin, rather than as a set of few compact specks. In the cases when the signals were clustered, the cluster was surrounded by a halo giving it a fuzzy appearance, indicating unfolding of chromatin caused by extraction of nuclear proteins. We observed that the removal of nonmatrix proteins leads to dispersion and disintegration of the structures formed by gene-rich regions but leaves the clusters of gene-poor regions largely intact (Fig. 5 and 6). In 59% of salt-extracted nuclei, the gene-rich regions were dispersed while the gene-poor regions were still clustered. This is a significant increase in the proportion of nuclei showing such a pattern compared to the samples fixed with formaldehyde and psoralen (9%) or with the formaldehyde alone (34%) but not extracted with salt. These data are consistent with the model that invokes anchoring of gene-poor regions in nuclear matrix and compaction of gene-rich regions by a different mechanism mediated by nonmatrix proteins.
Gene density profile indicates pattern of chromosome folding in interphase.
We have shown that chromosome folding in the interphase nucleus is highly nonrandom and that the pattern of folding correlates with the pattern of gene density along the chromosome. A model consistent with our data assigns different major roles to the gene-rich and gene-poor chromosome segments. Gene-poor segments are securely bound together by nuclear matrix components. The resulting structure represents the “backbone” of CTs and is responsible for the structural integrity of CTs throughout the cell cycle in vivo (31) and for the preservation of global CT organization after removal of nonmatrix proteins in vitro (19). Gene-rich chromosome segments are probably looping out but also are associated with each other by delicate interactions, thus forming subnuclear compartments with elevated gene density. These compartments probably represent the “local euchromatic neighborhoods” (26) beneficial for active gene expression. This model is consistent with observations in plants, where CTs are organized as compact chromocenters with looping-out euchromatin (13).
An important implication of this work is that certain aspects of chromosome folding are encoded in the chromosome sequence itself. While the code per se is yet to be identified (although our data indicate that it is probably the frequency of matrix attachment regions), we show that gene density can be used as the manifestation of such code. Therefore, chromosome folding can be inferred through the in silico analysis of the chromosome sequence—somewhat similar to the predictions of secondary protein structure from the primary polypeptide sequence. According to the presented results, gene-poor regions may be assigned a status of “CT backbone,” and gene-rich segments are expected to form elements of the “gene-rich compartment.” We have shown that neighboring backbone elements associate with each other, as do the gene-rich segments, and that the resulting composite structure is reproducible between the nuclei. These observations indicate the existence of a preprogrammed “tertiary” chromosome structure built from the backbone elements and gene-rich segments. However, any predictions of three-dimensional parameters of this structure are far beyond the scope of the presented model. In other words, we may identify chromosome segments as elements of the “backbone” or “gene-rich compartment,” but we still cannot predict the shape and location of the “backbone” and “gene-rich compartment” within the chromosome territory.
It should be noted here that the gene-poor regions do contain a considerable number of active genes; hence, they are not located in a nuclear environment hostile for transcription, such as heterochromatin. On the other hand, gene-rich regions do contain genes that are silent in particular tissues. This is not surprising, because an average gene-rich region on the D. melanogaster chromosome arm 2R is 390 kilobases long and contains about 59 genes, and gene-poor regions are even larger, averaging 500 kb. Despite the observed clustering of coexpressed genes on chromosomes of higher eukaryotes (2, 4, 18, 24), genomic segments of this size are inevitably composed of genes with different expression patterns; thus, the position of whole segments in the nucleus cannot be adjusted to fit the transcriptional activity of every gene in every tissue. Therefore, chromosome folding described here reflects the stable rather than the dynamic aspect of nuclear architecture, suggesting that chromatin remodeling relevant to transcriptional activation or repression of particular genes occurs at the smaller scale of individual chromatin domains.
Acknowledgments
This work was supported by grant GM61549 from the U.S. Public Health Service.
We are indebted to W. Brunken, J. Fitch, E. Gibney, and M. Nurminskaya for valuable comments and careful reading of the manuscript, and we sincerely thank W. Brunken for kindly providing access to the imaging equipment.
REFERENCES
- 1.Alvarez, M., S. J. Rhodes, and J. P. Bidwell. 2003. Context-dependent transcription: all politics is local. Gene 313:43-57. [DOI] [PubMed] [Google Scholar]
- 2.Arbeitman, M. N., E. E. Furlong, F. Imam, E. Johnson, B. H. Null, B. S. Baker, M. A. Krasnow, M. P. Scott, R. W. Davis, and K. P. White. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297:2270-2275. [DOI] [PubMed] [Google Scholar]
- 3.Belmont, A. S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250-257. [DOI] [PubMed] [Google Scholar]
- 4.Boutanaev, A. M., A. I. Kalmykova, Y. Y. Shevelyov, and D. I. Nurminsky. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420:666-669. [DOI] [PubMed] [Google Scholar]
- 5.Brown, K. E., S. S. Guest, S. T. Smale, K. Hahm, M. Merkenschlager, and A. G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845-854. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, K., and V. G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bystritskiy, A. A., and S. V. Razin. 2004. Breakpoint clusters: reason or consequence? Crit. Rev. Eukaryot. Gene Expr. 14:65-77. [PubMed] [Google Scholar]
- 8.Chambeyron, S., and W. A. Bickmore. 2004. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16:256-262. [DOI] [PubMed] [Google Scholar]
- 9.Chambeyron, S., and W. A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 11.Csink, A. K., and S. Henikoff. 1998. Large-scale chromosomal movements during interphase progression in Drosophila. J. Cell Biol. 143:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietzel, S., A. Jauch, D. Kienle, G. Qu, H. Holtgreve-Grez, R. Eils, C. Munkel, M. Bittner, P. S. Meltzer, J. M. Trent, and T. Cremer. 1998. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 6:25-33. [DOI] [PubMed] [Google Scholar]
- 13.Fransz, P., J. H. De Jong, M. Lysak, M. R. Castiglione, and I. Schubert. 2002. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99:14584-14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisch, M., K. Frech, A. Klingenhoff, K. Cartharius, I. Liebich, and T. Werner. 2002. In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res. 12:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grogan, J. L., M. Mohrs, B. Harmon, D. A. Lacy, J. W. Sedat, and R. M. Locksley. 2001. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14:205-215. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, D. A. 2003. The anatomy of transcription sites. Curr. Opin. Cell Biol. 15:311-317. [DOI] [PubMed] [Google Scholar]
- 17.Kurz, A., S. Lampel, J. E. Nickolenko, J. Bradl, A. Benner, R. M. Zirbel, T. Cremer, and P. Lichter. 1996. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 135:1195-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lercher, M. J., A. O. Urrutia, and L. D. Hurst. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31:180-183. [DOI] [PubMed] [Google Scholar]
- 19.Ma, H., A. J. Siegel, and R. Berezney. 1999. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J. Cell Biol. 146:531-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahy, N. L., P. E. Perry, S. Gilchrist, R. A. Baldock, and W. A. Bickmore. 2002. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157:579-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahy, N. L., P. E. Perry, and W. A. Bickmore. 2003. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moen, P. T., Jr., C. V. Johnson, M. Byron, L. S. Shopland, I. de la Serna, A. Imbalzano, and J. B. Lawrence. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell 15:197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragoczy, T., A. Telling, T. Sawado, M. Groudine, and S. T. Kosak. 2003. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11:513-525. [DOI] [PubMed] [Google Scholar]
- 24.Roy, P. J., J. M. Stuart, J. Lund, and S. K. Kim. 2002. Chromosomal clustering of muscle-specific genes in Caenorhabditis elegans. Nature 418:975-979. [DOI] [PubMed] [Google Scholar]
- 25.Schardin, M., T. Cremer, H. D. Hager, and M. Lang. 1985. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum. Genet. 71:281-287. [DOI] [PubMed] [Google Scholar]
- 26.Shopland, L. S., C. V. Johnson, M. Byron, J. McNeil, and J. B. Lawrence. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162:981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solovei, I., A. Cavallo, L. Schermelleh, F. Jaunin, C. Scasselati, D. Cmarko, C. Cremer, S. Fakan, and T. Cremer. 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 276:10-23. [DOI] [PubMed] [Google Scholar]
- 28.Stein, G. S., J. B. Lian, A. J. van Wijnen, J. L. Stein, A. Javed, M. Montecino, S. K. Zaidi, D. Young, J. Y. Choi, S. Gutierrez, and S. Pockwinse. 2003. Nuclear microenvironments support physiological control of gene expression. Chromosome Res. 11:527-536. [DOI] [PubMed] [Google Scholar]
- 29.Versteeg, R., B. D. van Schaik, M. F. van Batenburg, M. Roos, R. Monajemi, H. Caron, H. J. Bussemaker, and A. H. van Kampen. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpi, E. V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R. D. Campbell, M. Goldsworthy, S. H. Powis, J. Ragoussis, J. Trowsdale, and D. Sheer. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565-1576. [DOI] [PubMed] [Google Scholar]
- 31.Zink, D., T. Cremer, R. Saffrich, R. Fischer, M. F. Trendelenburg, W. Ansorge, and E. H. Stelzer. 1998. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 102:241-251. [DOI] [PubMed] [Google Scholar]