Abstract
The tumor suppressor ARF inhibits cell growth in response to oncogenic stress in a p53-dependent manner. Also, there is an increasing appreciation of ARF's ability to inhibit cell growth via multiple p53-independent mechanisms, including its ability to regulate the E2F pathway. We have investigated the interaction between the tumor suppressor ARF and DP1, the DNA binding partner of the E2F family of factors (E2Fs). We show that ARF directly binds to DP1. Interestingly, binding of ARF to DP1 results in an inhibition of the interaction between DP1 and E2F1. Moreover, ARF regulates the association of DP1 with its target gene, as evidenced by a chromatin immunoprecipitation assay with the dhfr promoter. By analyzing a series of ARF mutants, we demonstrate a strong correlation between ARF's ability to regulate DP1 and its ability to cause cell cycle arrest. S-phase inhibition by ARF is preceded by an inhibition of the E2F-activated genes. Moreover, we provide evidence that ARF inhibits the E2F-activated genes independently of p53 and Mdm2. Also, the interaction between ARF and DP1 is enhanced during oncogenic stress and “culture shock.” Taken together, our results show that DP1 is a critical direct target of ARF.
The Ink4a/ARF locus in mice and humans encodes two tumor suppressor proteins, p16Ink4a and ARF (p19ARF in mice and p14ARF in humans) (49, 54, 69). The p16Ink4a protein is an inhibitor of Cdk4/6-cyclin D and is linked to the retinoblastoma (Rb) tumor suppressor pathway (54, 69). The ARF tumor suppressor protein, which is mutated or inactivated in a significant number of human tumors, is linked to the p53 tumor suppressor pathway. In response to oncogenic insult, the p53 tumor suppressor protein is stabilized and activated by ARF by virtue of its ability to inhibit the function of Mdm2 (25, 29, 32, 33, 34, 47, 59, 61, 67), which is a negative regulator of p53 (17, 20, 24, 41, 57). The ARF-p53-Mdm2 pathway therefore serves as a checkpoint that protects cells from oncogene-induced transformation.
Interestingly, a number of studies have demonstrated that ARF possesses p53- and Mdm2-independent functions. For example, ARF has been shown to inhibit the proliferation of cells that lack p53 or both Mdm2 and p53 (7, 13, 62). Moreover, mice lacking both ARF and p53 developed multiple primary tumors of a wider spectrum than that with mice lacking either gene alone (62). In the same study, mice lacking p53, Mdm2, and ARF were found to develop tumors at a higher frequency and of a wider spectrum than those with mice lacking p53 and Mdm2 (62). The reintroduction of ARF into p53− Mdm2− ARF− mouse embryonic fibroblasts (MEFs) caused a delayed G1-phase growth arrest (62). More recently, analyses of p19ARF highlighted the ability of ARF to cause cell cycle arrest independent of Mdm2 relocalization to nucleoli and p53 stabilization (27, 32). These studies point to the facts that the cell cycle arrest and tumor suppression functions of ARF are not entirely elicited through the p53-Mdm2 pathway and that there must be additional cellular factors that are targeted by ARF.
Several p53/Mdm2-independent functions of ARF have been identified. For example, it has been shown that ARF can induce the expression of a number of p53-dependent and -independent antiproliferative genes (30). ARF has also been shown to inhibit the production of rRNA and to retard the processing of the 47/45S and 32S precursor rRNAs into mature 28S, 18S, and 5.8S rRNAs (58). The mechanism of inhibition of rRNA processing remains unclear, although recent reports showing an interaction of ARF with B23 (nucleophosmin), a multifunctional nucleolar protein involved in ribosomal biogenesis, point towards a possible mechanism (5, 21). Interestingly, two studies have provided evidence of the regulation of c-Myc by ARF (9, 48). ARF has been shown to associate with c-Myc and to inhibit the expression of the c-Myc-activated genes. The inhibition of c-Myc-activated genes preceded S-phase inhibition by ARF (9). Moreover, the expression of ARF inhibits the S-phase stimulatory activity of c-Myc. The mechanism by which ARF inhibits c-Myc function is not clear. One group suggested that ARF sequesters c-Myc to the nucleolus, thereby limiting its availability in the nucleoplasm (9), whereas the other group suggested a mechanism in which the c-Myc/ARF complex interacts with the c-Myc-target gene promoters to regulate expression (48). ARF also targets Foxm1b, a proliferation-associated transcription factor that is essential for the development of hepatocellular carcinomas (23). ARF relocalizes Foxm1b to the nucleolus and inhibits its transcription and transformation functions (23).
Interestingly, both human and mouse ARFs interact with certain members of the E2F family of transcription factors (13, 37, 40). One study reported that mouse ARF could bind E2F1 independent of p53 and also relocalized E2F1 from the nucleoplasm to the nucleolus (37). Moreover, ARF induced 26S proteasome-mediated degradation of E2F1 and E2F3 but not E2F6 (37). Furthermore, the ARF-mediated suppression of growth of p53-defective cells could be rescued partially by the ectopic expression of E2F1. Studies with human ARF also provided evidence for an interaction with E2F1 leading to an inhibition of E2F1-activated transcription in cells lacking p53 or both p53 and Mdm2 (13, 40). Members of the E2F family of transcription factors play a vital role in the cell cycle by coordinately regulating genes that are required for the G1-to-S transition and mitosis (12). Therefore, the findings on ARF/E2F are significant, as they point to a biochemical basis for ARF's ability to cause cell cycle arrest in the absence of p53 and Mdm2.
DP1 (DRTF1 polypeptide 1) belongs to a family of factors that were first identified as proteins that bind to E2F DNA binding sites (14). DP1 and E2F1 contain hydrophobic heptad repeats, which are involved in heterodimer formation through coil-coil interactions. The association of DP1 and E2F1 enhances and is critical for both the DNA binding and transcriptional activities of E2F1 (2, 18, 64). Expression of the E2F-regulated genes is crucial for the progression of cells from G1 to S phase of the cell cycle. The activity of DP1 is also regulated by phosphorylation in a cell cycle-dependent manner. It was shown that cyclin A-cdk2 physically interacted with the N terminus of E2F1-3 and phosphorylated both E2F and DP1 (28). Moreover, the phosphorylation of E2F1/DP1 by cyclin A attenuated the affinity of the heterodimer for DNA, which was shown to be critical for proper S-phase progression (28). Recent studies using a transgenic mouse model expressing DP1 under the control of the keratin 5 promoter indicated a possible oncogenic function for DP1 (60). Furthermore, it was shown that DP1 was absolutely required for extraembryonic development and embryonic survival, consistent with the notion that E2F/DP1 plays a key role in the cell cycle (26). DP1 is therefore critical for the activity of the E2Fs and thus plays a crucial role in cellular functions such as regulation of the cell cycle.
Recently, we provided evidence that DP1 could interact with ARF (8). Here we show that ARF directly binds to DP1 through sequences involved in heterodimerization with the E2Fs and that ARF inhibits the interaction between E2F1 and DP1. Importantly, oncogenic stress and “culture shock,” conditions that activate the expression of ARF, enhance the interaction between ARF and DP1. Moreover, ARF's growth-inhibitory activity correlates with its ability to bind DP1, relocalize DP1 to nucleoli, and inhibit E2F1 transcriptional activity. These findings correlate with an ARF-mediated inhibition of E2F-regulated gene expression independently of p53. Our results suggest that ARF inhibits E2F activity by binding and sequestering DP1, providing new insight into the ARF-mediated regulation of E2F function and p53-independent ARF signaling.
MATERIALS AND METHODS
Cell cultures.
U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen). p53−Mdm2− MEFs were obtained from Charles Sherr and were maintained in DMEM containing 10% FBS and supplemented with 1% l-glutamine (Invitrogen) and 1% nonessential amino acids (Invitrogen).
Expression plasmids.
The hemagglutinin-tagged p19ARF mutants D1-5, D6-10, D21-25, and D29-34 have been described before (27) and were obtained from D. E. Quelle, University of Iowa. The DP1 expression plasmid has been described before (8).
DNA transfection and CAT assays.
Transient transfections were carried out by the calcium phosphate method as previously described (42). The E2-CAT reporter plasmid has been described before (8). The chloramphenicol acetyltransferase (CAT) assay was performed as described before (8)
Immunoprecipitation and Western blot analysis.
Wild-type (WT) MEFs (passage 2) were infected with recombinant retroviruses carrying either an empty vector (pBabehygro) or a vector expressing activated Ras (pBabehygroRAS) as described by Groth et al. (15). Forty-eight hours after infection, selection was started by growing the cells in medium containing 50 μg/ml of hygromycin (Mediatech). After 72 h of selection, cells were harvested, and the total cell extract was prepared by incubating the cell pellet on ice for 1 h with NETT 250 buffer (20 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 250 mM NaCl, 0.5% Triton X-100). After incubation, the lysate was centrifuged at 13,000 rpm for 10 min. One milligram of total extract was subjected to immunoprecipitation using a monoclonal antibody against DP1 (1DP06; Labvision). The immunoprecipitates were eluted and subjected to Western blot analysis using a polyclonal p19ARF antibody (R562; GeneTex).
Early (P1)- and late (P4)-passage MEFs were harvested, and total cell extracts were prepared using NETT 250 buffer as described above. One milligram of total extract was subjected to immunoprecipitation with a monoclonal DP1 antibody (1DP06; Labvision). The eluted proteins were subjected to Western blot analysis using a polyclonal p19ARF antibody (R562; GeneTex). The U2OS cells were harvested 48 h after transfection, and total cell extracts were prepared as described before. The total extract (1.5 mg) was subjected to immunoprecipitation using the p19ARF antibody (R562; GeneTex). The bound proteins were subjected to Western blot analysis as described before (8). The immunoprecipitates were assayed for DP1 by Western blot analysis. Bacterial extracts expressing glutathione S-transferase-DP1 (GST-DP1) or GST-DP1 d205-277 were incubated with or without a purified ARF N64 polypeptide for 30 min on ice. An extract expressing GST-E2F1 was then added to the tubes, and the mix was further incubated on ice for 30 min. Subsequently, a monoclonal antibody against DP1 (1DP06; Labvision) was added, and the tubes were further incubated on ice for 1 h. Following incubation, protein G-Sepharose beads were added to the tubes, and the tubes were rocked for 1 h at 4°C. The beads were collected by centrifugation and washed three times with 400 μl of NETN buffer containing 0.1% Triton X-100. The bound proteins were eluted with the gel loading dye, separated in a 12% sodium dodecyl sulfate-polyacrylamide gel, and detected by Western blot analysis using an E2F1 antibody (KH129; 1:200 dilution) (Labvision). U2OS-ARF cells were either left untreated or treated with 1 mM tetracycline for 8 h. The cells were harvested, and total cell extracts were prepared using NETT 250 buffer as described above. One milligram of total protein was then subjected to immunoprecipitation using 1 μg of DP1 antiserum (1DP06; Labvision). The immunoprecipitates were washed with NETT 250 buffer, and the eluates were analyzed for the presence of E2F1 by Western blot analysis using an antibody against E2F1 (KH129; 1:200 dilution) (Labvision).
Protein purification and in vitro pull-down assay.
A synthetic minigene encoding the polyhistidine-tagged N-terminal 64 amino acids (N64) of mouse p19ARF was obtained from Charles Sherr, St. Jude Children's Research Hospital. The ARF N64 polypeptide was purified from bacterial extracts according to a protocol described by Weber et al. (63). For the binding experiment, GST-tagged DP1, E2F1, and DP1 d205-277 and the GST protein alone were initially bound to 10 μl of a 50% slurry of glutathione-Sepharose beads by rocking the beads with the required amount of bacterial extract for 1 h at 4°C. The beads were washed three times with 350 μl of NETN buffer (20 mM Tris-Cl [pH 8], 100 mM NaCl, 1 mM EDTA [pH 8]) containing 0.5% Triton X-100. The beads with the bound GST proteins were then resuspended in binding buffer (50 mM Tris-Cl [pH 8], 150 mM NaCl, 5 mM EDTA [pH 8], 0.5% NP-40) supplemented with 100 μg/ml of bovine serum albumin. Five microliters of the purified ARF N64 polypeptide was added to the tubes, and the tubes were rocked at 4°C for another hour. The beads were recovered by centrifugation and washed four times with 400 μl of the binding buffer. The bound proteins were eluted with the gel loading dye, separated in a 12% sodium dodecyl sulfate-polyacrylamide gel, and detected by Western blot analysis using a polyclonal p19ARF antibody (R562; 1:1,000 dilution).
Immunostaining.
For DP1 localization with either wild-type p19ARF or p19ARF deletion mutants, U2OS cells grown on coverslips were transfected with T7-DP1 (0.2 μg) in combination with either wild-type p19ARF (0.5 μg) or various deletion mutants of p19ARF (D 1-5, D 6-10, D 21-25, and D 29-34) (0.5 μg). The coimmunolocalization assay was performed exactly as described before (8).
Stable cell lines and Northern blot analysis.
U2OS-ARF is a single cell clone derived from the T-REx-U2OS cell line (Invitrogen), which was stably transfected with a plasmid expressing a T7 epitope-tagged p19ARF cDNA under the control of the tetracycline operator. A single clone that expressed the optimal level of the protein was used for the experiments. The stable cell line is regularly maintained in DMEM containing 10% tetracycline system-approved FBS (Clontech), 50 μg/ml of hygromycin B (Mediatech), and 50 μg/ml of Zeocin (Invitrogen). U2OS-ARF-p53GSE cells were constructed by transfecting U2OS-ARF cells with a retroviral construct (p56SN) (45) and selecting the transfected cells with 500 μg/ml of G418 for 10 days. The drug-resistant cells were subsequently pooled, and the pooled cells were maintained in DMEM containing 10% tetracycline system-approved FBS (Clontech), 50 μg/ml of hygromycin B (Mediatech), 50 μg/ml of Zeocin (Invitrogen), and 500 μg/ml of G418 (Mediatech). U2OS-ARF and U2OS-ARF-p53GSE cells were treated with tetracycline at a final concentration of 1 mM. Cells were harvested after various time periods of induction with tetracycline, and total RNAs were prepared using TRIZOL (Invitrogen) as specified by the manufacturer. Twenty micrograms of total RNA was separated in 1% agarose-6% formaldehyde gels and transferred to Hybond-NX membranes (Amersham Pharmacia Biotech) by capillary blotting. After UV cross-linking, membranes were hybridized sequentially to cDNA probes for cyclin A and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that had been labeled with [32P]dCTP by random priming. Northern blot analysis for cyclin A and mRNAs was done with an EcoRI fragment from a cytomegalovirus cyclin A-encoding plasmid. Northern blotting for GAPDH was done using a 1.4-kb PstI fragment from plasmid pBS-GAPDH (N. Hay, University of Illinois at Chicago).
Retroviral infection and small interfering RNA (siRNA)-mediated p19ARF knockdown.
The pSIRIPP and pSIRIPPp19ARFsi retroviruses have been described before (52) and were obtained from T. Jacks, Massachusetts Institute of Technology. p53− Mdm2− MEFs were infected as described earlier (15). Thirty-six hours after infection, selection was started by growing the cells in medium containing 2 μg/ml of puromycin. At 60 h postinfection, cells were split 1:3 into selection medium, and selection was continued for another 48 h. At the end of selection, one set of plates was harvested, and total protein extracts were prepared. The other set of plates was harvested, and total RNAs were prepared using TRIZOL reagent.
ChIP assay.
U2OS-ARF (106) cells were either induced for ARF expression by treatment with 1 mM tetracycline for 18 h or left unstimulated and then processed for chromatin immunoprecipitation (ChIP) assays. Cells from the treated and untreated plates were cross-linked by the addition of formaldehyde to a 1% final concentration, the chromatin was sonicated, and immunoprecipitation was performed using 1 μg of DP1 antibody (1DPO6; Labvision). ChIP was done using a ChIP assay kit (Upstate) according to the manufacturer's protocol. DNAs released from the precipitated complexes were amplified by PCR alongside 0.1% of the input chromatin used to perform the immunoprecipitation. Human dihydrofolate reductase (DHFR) promoter (+446 to −17)-specific primers (DHFR-forward [5′ CTACAAGTTAGAGAAACAGCGTTACTCGAA 3′] and DHFR-reverse [5′TTCTGCTGTAACGCGCGGGCTCGGA3′]) were used to perform PCR. The PCR products were separated in agarose gels and visualized by ethidium bromide staining.
p53− Mdm2− MEFs were infected with the pSIRIPP and pSIRIPPp19ARFsi viruses as described above. Cells (106) from the control or p19ARF siRNA-treated cells were cross-linked and sonicated, and chromatin immunoprecipitation was performed exactly as described above. DNAs released from the immunoprecipitated complexes were amplified by PCR alongside 0.1% of the input chromatin used for the immunoprecipitation. Mouse DHFR promoter-specific primers (DHFR + 962 [5′ CGGCAATCCTAGCGTGAAGGC 3′] and DHFR + 1360 [5′ GGCTCCATTCAGCGACGAAAG 3′]) were used to perform PCR, and the PCR products were visualized as described above.
Quantitative real-time reverse transcription-PCR (RT-PCR).
U2OS-ARF cells were either left untreated or treated with tetracycline (1 mM) for 18 h. Total RNAs were extracted from the treated and untreated cells using TRIZOL reagent. Ten micrograms of the total RNA was then subjected to DNase I treatment using RQ1 RNase-free DNase I (Invitrogen). One microgram of the DNase I-treated RNA was then reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. PCR amplification was performed in triplicate using the following primers: for human dhfr, 5′-ATGCCTTAAAACTTACTGAACAACCA-3′ and 5′-TGGGTGATTCATGGCTTCCT-3′ (annealing temperature, 55°C); and for the human cyclophilin gene, 5′-GCAGACAAGGTCCCAAAGACAG-3′ and 5′-CACCCTGACACATAATCCCTGG-3′ (annealing temperature, 55.7°C). Each PCR mix contained the following: 0.05 μg of cDNA, a 200 nM concentration of each primer, and 1× iQ SYBR green supermix (Bio-Rad) in a 25-μl reaction mix. Real-time PCR was performed using the MyiQ single-color real-time PCR detection system (Bio-Rad). Melting curve analysis was performed for every reaction, and a single sharp peak was observed. To create a standard curve for relative quantification, the sample that was not treated with tetracycline was chosen as a standard control, diluted in water (1×, 0.5×, 0.2×, 0.1×, and 0.05×), and subjected to real-time quantitative PCR in triplicate. The dilution value (starting quantity) of the standard was plotted against the threshold cycle number (CT) at which fluorescence first increased above the background by the use of MyiQ software (Bio-Rad). The expression of the indicated gene in each sample was evaluated with this standard curve. The levels of dhfr mRNA were normalized against the levels of cyclophilin mRNA, which was used as an internal control. The amount of change in the levels of dhfr mRNA was obtained by dividing the normalized values of dhfr mRNA in the tetracycline-treated sample by the normalized values of dhfr mRNA in the untreated sample and is represented in a bar graph. p53− Mdm2− MEFs were infected with retroviruses encoding a control or p19ARF siRNA as described above. Total RNAs were isolated from the cells using TRIZOL reagent. The RNAs were treated with DNase I and reverse transcribed exactly as described above. PCR amplification was performed in triplicate using the following primers: for mouse dhfr, 5′-CTGGTTCTCCATTCCTGAGAAG-3′ and 5′-GCCACCAACTATCCAGACCATG-3′ (annealing temperature, 55°C); and for the mouse cyclophilin gene, 5′-GGCAAATGCTGGACCAAACAC-3′ and 5′-TTCCTGGACCCAAAACGCTC-3′ (annealing temperature, 57.5°C). Real-time PCR and quantitation were done as described above.
RESULTS
ARF directly binds DP1 and inhibits DP1-E2F1 interaction.
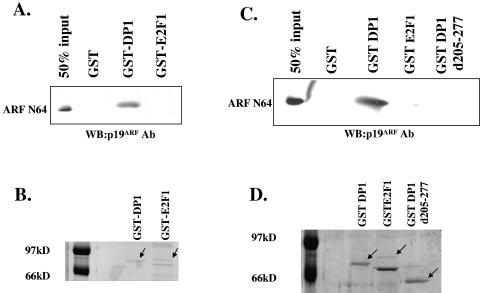
Our previous studies demonstrated that ARF coimmunoprecipitated with DP1 (8). Also, we observed evidence of interactions of endogenous ARF and DP1 in both mouse (p53− Mdm2− MEFs) and human (HeLa) cells (data not shown). However, the nature of the interaction between ARF and DP1 was not clear. DP1 is a heterodimeric partner of E2F1, and it has been demonstrated that both human and mouse ARF proteins can interact with E2F1 (13, 37, 40). Therefore, it is possible that the observed interaction between ARF and DP1 is indirect and is mediated through E2F1. The relevance of this issue is also highlighted by our previous observation that a DP1 mutant lacking the heterodimerization domain, which is required for binding to E2F1, also fails to interact with ARF in coimmunoprecipitation assays (8). In order to determine whether the interaction between ARF and DP1 is direct, we set up an in vitro binding assay using purified proteins. We purified a synthetic minigene-encoded polyhistidine-tagged N64 polypeptide of ARF (syn-ARF N64) from bacterial extracts (63). This polypeptide, representing the N-terminal 62 amino acids of p19ARF, was found to localize to the nucleolus and interact with Hdm2 (63). The purified ARF N64 polypeptide (500 ng) was incubated with GST, GST-E2F1 (175 to 200 ng), or GST-DP1 (200 ng) bound to glutathione-Sepharose beads. The beads were washed, and bound proteins were eluted and subjected to Western blot analysis. As shown in Fig. 1A, GST-DP1 was able to bind ARF. We could detect 40 to 50% of the input ARF N64 in the bound fraction when GST-DP1-containing beads were used (Fig. 1A). GST alone or GST-E2F1 failed to demonstrate any significant binding to the ARF N64 polypeptide. We were unable to detect any band (significantly above the background) for ARF N64 with GST-E2F1 even after an extended exposure of the film. A Coomassie blue-stained gel (Fig. 1B) confirmed that similar levels of the GST-DP1 and GST-E2F1 proteins were used in the binding assay. ARF was shown to bind E2F1 by coimmunoprecipitation experiments (13, 37, 40), and therefore a lack of binding of the recombinant proteins suggests that the interaction might involve another protein. Alternatively, the interaction might depend on a modification of E2F1 that is missing in the GST-E2F1 fusion protein. Also, it is possible that residues of ARF that are missing in the ARF N64 polypeptide are critical for an interaction with E2F1. Nevertheless, the binding of purified ARF to purified DP1 suggests that ARF binds DP1 directly without a requirement for E2F1.
FIG. 1.
DP1 binds to ARF directly through its heterodimerization domain. (A) Glutathione-Sepharose beads were first incubated with bacterial extracts expressing either GST, GST-DP1, or GST-E2F1. After rocking of the beads with the extracts for 1 h at 4°C, the beads were washed with NETN buffer containing 0.5% Triton X-100. The beads were resuspended in binding buffer containing 100 mg/ml of bovine serum albumin and 500 ng of the ARF N64 polypeptide. The beads were rocked for 1 h at 4°C and washed with the binding buffer, and the eluted proteins were subjected to Western blot analysis using a p19ARF (R562; GeneTex) antibody to detect the ARF N64 polypeptide. (B) Glutathione-Sepharose beads were incubated with identical amounts of bacterial extracts expressing either GST-DP1 or GST-E2F1 as those used for the binding assay described above. The beads were washed, and the bound proteins were eluted and stained with Coomassie brilliant blue R250. (C) Glutathione-Sepharose beads were incubated with bacterial extracts expressing either GST, GST-DP1, GST-E2F1, or GST-DP1 d205-277. The binding reaction and Western blot analysis were performed as described above. (D) Glutathione-Sepharose beads were incubated with identical amounts of bacterial extracts expressing either GST-DP1, GST-E2F1, or GST-DP1 d205-277 as those used for the binding assay described above, and the bound proteins were eluted and stained with Coomassie brilliant blue R250.
If the heterodimerization domain in DP1 were required for a physical interaction between ARF and DP1, we predicted that this mutant would be defective in binding to ARF in our in vitro binding assay. In order to investigate that possibility, we constructed a GST-DP1 mutant protein lacking the heterodimerization domain and compared wild-type DP1 and mutant DP1 for the ability to interact with the purified ARF N64 polypeptide. In agreement with our coimmunoprecipitation studies, we found that the mutant DP1 protein was significantly impaired in interacting with ARF (Fig. 1C) under conditions where we could detect robust binding between ARF and GST-DP1 (Fig. 1C). The failure of GST-DP1 d205-277 to interact with the ARF N64 polypeptide could not be attributed to a difference in the levels of GST-DP1 and GST-DP1 d205-277 because comparable levels of the two proteins were used (Fig. 1D).
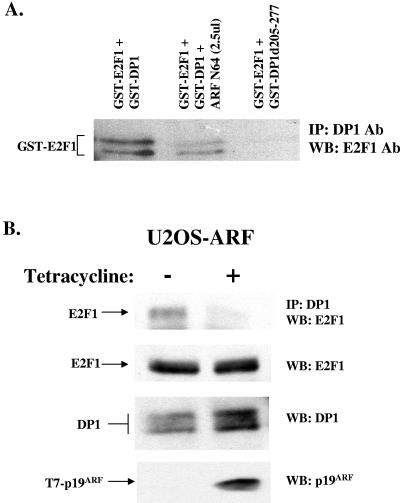
The observation that E2F1 and ARF interact with DP1 through sequences between residues 205 and 277 of DP1 suggested the possibility of a competition between ARF and E2F1 for binding to DP1 when the level of ARF increases. The prediction was that ARF would reduce the interaction between DP1 and E2F1. We failed to design an experiment to assay the effect of ARF on E2F1-DP1 DNA binding activity using a gel retardation assay because of the high nonspecific DNA binding activity of ARF. Therefore, the GST fusion proteins of E2F1 and DP1 were used in coimmunoprecipitation assays to analyze the effect of ARF on the E2F1-DP1 complex. The fusions GST-DP1 (200 ng) and GST-E2F1 (175 to 200 ng) in the presence or absence of the purified ARF N64 (250 ng) polypeptide were immunoprecipitated with a monoclonal antibody against DP1. The immunoprecipitates were then probed for the presence of GST-E2F1 by Western blotting using an antibody against E2F1. Clearly, the presence of ARF reduced the extent of the interaction between E2F1 and DP1 (Fig. 2A). To further investigate the effect of ARF on the interactions between E2F1 and DP1 inside the cell, we employed a U2OS-derived cell line that inducibly expresses ARF (9). The cells were induced or not induced by the addition of tetracycline in the culture medium for 8 h. We do not see any E2F1 or DP1 proteolysis at that early (8 h) time point of induction. As shown in Fig. 2B, the expression of ARF caused a significant reduction in the interaction between E2F1 and DP1. These results are consistent with the notion that ARF competes with E2F1 for binding to DP1. We believe that the result is physiologically relevant because the levels of ARF that resulted in the reduction in E2F1-DP1 association were very similar to the levels of endogenous ARF induced in MEFs following the expression of oncogenic Ras (data not shown).
FIG. 2.
ARF prevents formation of E2F1-DP1 complexes. (A) Extracts expressing GST-DP1 or GST-DP1 d205-277 were incubated with or without the indicated amount of ARF N64 polypeptide, followed by incubation with GST-E2F1-expressing extracts. The mix was then subjected to immunoprecipitation using an antibody against DP1 (1DP06; Labvision). The immunoprecipitates were analyzed for bound GST-E2F1 by Western blot analysis (left panel) using E2F1 antiserum (KH129; Labvision). (B) U2OS-ARF cells were either left untreated or treated with 1 mM tetracycline for 8 h to induce the expression of p19ARF. Total cell extracts were prepared as described in Materials and Methods, and 1 milligram of extract was subjected to immunoprecipitation using a DP1 antibody (1DP06). The immunoprecipitates were subjected to Western blot analysis using an antibody against E2F1 (KH129) to detect bound E2F1 (top panel). The total extracts were also tested for the expression of E2F1, DP1, and T7-p19ARF by Western blot analysis. The apparent increase in the DP1 signal in the “+ tetracycline” lane was a result of a loading difference.
ARF mutants defective in cell cycle inhibition are deficient in DP1 regulation.
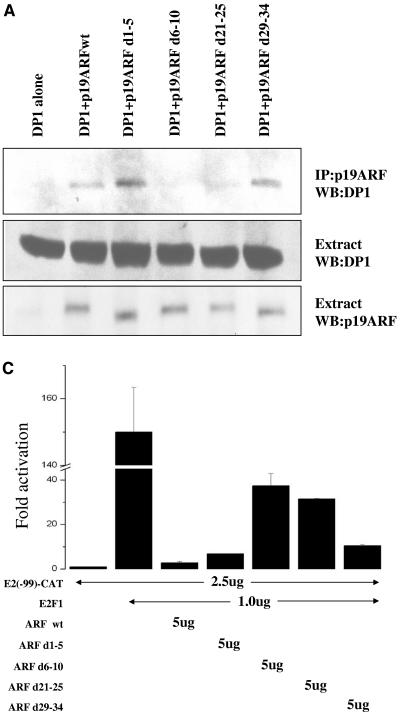
The expression of wild-type ARF in wild-type and p53− Mdm2− ARF− triple-knockout MEFs causes an inhibition of S-phase progression (61). Interestingly, two deletion mutants of mouse p19ARF, which lack residues 6 to 10 (d6-10) or 21 to 25 (d21-25), fail to inhibit S-phase entry in both wild-type MEFs and triple-knockout MEFs (27). Therefore, we considered the possibility that those residues are required for the association with DP1. To test that hypothesis, we compared the abilities of wild-type p19ARF and the deletion mutants to bind to DP1. U2OS cells were transfected with plasmids expressing DP1 alone or cotransfected with plasmids expressing wild-type p19ARF or a panel of deletion mutants. Extracts from the transfected cells were subjected to immunoprecipitation with p19ARF antisera, and the immunoprecipitates were analyzed for DP1 association by immunoblotting with a DP1 antibody (Fig. 3A). Whereas wild-type ARF and two active mutants of ARF that retained growth-inhibitory activity (d1-5 and d29-34) efficiently interacted with DP1, the ARF mutants lacking residues 6 to 10 and 21 to 25 were greatly impaired. Since both of those mutants properly localize to nucleoli and bind Mdm2 (27), their inability to bind DP1 supports the notion that the DP1 association is critical for ARF function.
FIG. 3.
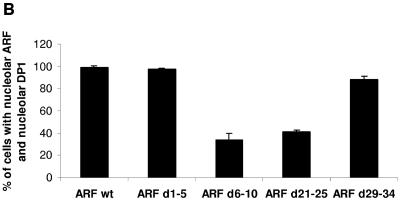
p19ARF d6-10 and d21-25 mutants are impaired in regulating DP1. (A) U2OS cells were transfected with plasmids that expressed either DP1 (5 μg) alone or DP1 (5 μg) along with wild-type p19ARF or various deletion mutants of p19ARF (d1-5, d6-10, d21-25, and d29-34; 5 μg). (Top) Forty-eight hours after transfection, cells were harvested, and total cell extracts were prepared. The total cell extract (1.5 mg) was subjected to immunoprecipitation (IP) with R562 antibody (for p19ARF), and the immunoprecipitates were subjected to Western blot (WB) analysis. The blots were probed with a monoclonal DP1 antibody (WTH-16; Labvision) to detect coimmunoprecipitating DP1. (Middle and bottom) Extracts were also tested for the expression of DP1, wild-type p19ARF, and deletion mutants of p19ARF by probing the blot with DP1- and p19ARF-specific antibodies. (B) U2OS cells grown on coverslips were transfected with plasmids expressing T7-DP1 (0.2 μg) either alone or in combination with either wild-type p19ARF (0.5 μg) or various deletion mutants of p19ARF (d1-5, d6-10, d21-25, and d29-34; 0.5 μg). Twenty-four hours after transfection, the cells were fixed and probed with a T7 epitope tag-specific monoclonal antibody (1:500 dilution) and R562 antibody (p19ARF; 1:200 dilution). Tetramethyl rhodamine isocyanate-labeled anti-mouse and fluorescein isothiocyanate-labeled anti-rabbit antibodies (both at a 1:200 dilution) were used as the secondary antibodies. Immunofluorescence was detected by confocal microscopy as described in Materials and Methods. One hundred cells on two independent coverslips demonstrating nucleolar p19ARF (wild-type or deletion mutant) staining were simultaneously scored for the colocalization of T7-DP1. The numbers of cells which showed colocalization of T7-DP1 and p19ARF (wild-type or mutant) are represented in a bar graph. Averages from two independent coverslips are shown in the bar graph. (C) U2OS cells were transfected with the indicated CAT reporter gene. Where indicated, the transfection mix also contained an expression plasmid for E2F1 or a combination of E2F1 and either wild-type p19ARF or the various deletion mutants of p19ARF (d1-5, d6-10, d21-25, and d29-34). The transfections were carried out by the calcium phosphate method as described in Materials and Methods. A plasmid expressing β-galactosidase was included to control for transfection efficiencies. The activation of CAT gene activity was calculated by dividing the fold activation of CAT gene activity obtained by E2F1 expression alone or by the coexpression of E2F1 and the various ARF constructs by the CAT gene activity obtained when the reporter gene was expressed alone. The average inhibition of the CAT gene activity in three independent experiments is shown.
Given the ability of ARF to govern DP1 nucleolar localization (8), we directly compared wild-type ARF and its mutants for the ability to relocalize DP1 to the nucleolus. U2OS cells were cotransfected with plasmids expressing T7 epitope-tagged DP1 and empty vector, wild-type ARF, or ARF mutants. Immunostaining with ARF antibodies showed that wild-type ARF and its mutants localized to nucleoli with the expected frequency (27). Specifically, wild-type, d1-5, d6-10, and d21-25 cells exhibited complete nucleolar localization in 100% of cells, whereas only 70% of cells expressed the d29-34 mutant in nucleoli due to a partial loss of the nuclear localization signals (27). Quantification of DP1 nucleolar localization, as assessed by staining with a T7 antibody, showed that wild-type ARF efficiently relocalized DP1 from the cytosol to the nucleolus (Fig. 3B; data not shown). The same was true for the growth-inhibitory ARF mutants d1-5 and d29-34, although d29-34 was modestly less effective at relocalizing DP1 to nucleoli due to its own incomplete nucleolar localization pattern (Fig. 3B). Conversely, the inactive d6-10 and d21-25 mutants, which exhibited impaired associations with DP1, showed a dramatic reduction in the ability to mobilize DP1 into nucleoli. These studies establish a tight correlation between ARF-DP1 association and ARF's ability to inhibit growth and relocalize DP1 into nucleoli.
Since DP1 is an essential functional partner of the E2F1 transcription factor, we hypothesized that a deficiency of the ARF mutants to interact with and relocalize DP1 would result in an impairment of the mutants to inhibit E2F1-activated transcription. To test our hypothesis, we measured E2F1 transcriptional activity in the presence of ARF or its deletion mutants in U2OS cells. A CAT reporter plasmid that contains E2F-responsive elements (E2-CAT) was used for these studies. The expression of E2F1 resulted in a significant stimulation of transcription of the reporter gene. The coexpression of ARF mutants capable of binding DP1 and inhibiting growth (i.e., the wild type, d1-5, and d29-34) resulted in a marked inhibition of E2F1-activated transcription (Fig. 3C). Consistent with our prediction, the deletion mutants d6-10 and d21-25 were significantly weaker at inhibiting E2F1-activated transcription than wild-type ARF and d1-5 (Fig. 3C). The impairment was still apparent, albeit not statistically significant, compared to the inhibition evoked by the ARF mutant d29-34. These results directly reflect the ability of each mutant to relocalize DP1 into nucleoli (Fig. 3B), suggesting that ARF's ability to inhibit cell cycle progression may depend, at least in part, on its ability to bind and sequester DP1 in nucleoli.
Oncogenic stress and “culture shock” increase the interaction between ARF and DP1.
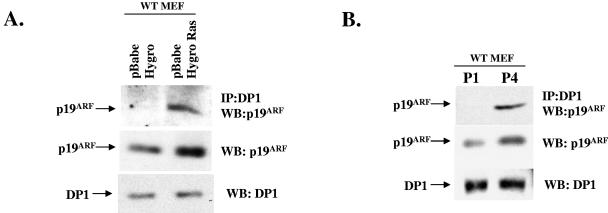
The tumor suppression function of ARF is believed to be important during oncogenic stress (55, 71). Several oncogenes have been shown to activate the expression of ARF (10, 11, 46, 50, 53, 70). The stimulation of ARF expression by oncogenes leads to cellular senescence or apoptosis, depending on the oncogene. The induction of senescence or apoptosis prevents the development of cancer cells. The expression of ARF is also induced by “culture shock” (24). For example, MEFs in culture undergo senescence after six of seven passages, and this is dependent on ARF, as ARF−/− MEFs do not senesce and are immortal (24). The level of ARF increases after passages 3 and 4, which is believed to initiate the senescence program in cultured MEFs. Therefore, we investigated whether oncogenic stress and “culture shock” in MEFs have any effect on the ARF-DP1 interaction. We employed a retrovirus expressing the Ras oncogene to infect MEFs (passage 2). Activated Ras induces a senescence-like phenotype in WT MEFs but not in ARF−/− MEFs (46). Consistent with previous observations (46), there was a threefold increase in the level of ARF protein in Ras virus-infected cells compared to empty virus-infected cells (Fig. 4A). Moreover, there was a significant increase in the interaction between ARF and DP1, as judged by the coimmunoprecipitation of ARF with DP1 (Fig. 4A). The increase in the level of ARF was about threefold, whereas the increase in DP1-ARF interaction was much greater (about sixfold). We think that newly synthesized ARF has a greater opportunity to interact with DP1 that is mainly in the cytoplasm and nucleoplasm before localizing to the nucleolus. However, it is possible that an unknown Ras-induced mechanism modulates the interaction between ARF and DP1. The observation was similar when we compared extracts of early-passage MEFs with those from late-passage MEFs (Fig. 4B). There was an increase in the level of ARF from passage 1 to passage 4, and that was accompanied by a significant increase in binding to DP1. These increased interactions between ARF and DP1 during oncogenic stress and “culture shock” provide strong evidence for a role of the ARF-DP1 interaction in the biological function of ARF.
FIG. 4.
Increase in ARF-DP1 interaction following oncogenic stress and culture shock. (A) WT MEFs were infected with the indicated recombinant retroviruses, and total cell extracts were prepared as described in Materials and Methods. Extracts were subjected to immunoprecipitation with a DP1 antibody (1DP06; Labvision), and the immunoprecipitates were checked for the presence of ARF by Western blot analysis using an antibody against p19ARF (R562). Total cell extracts were also tested for the levels of DP1 and p19ARF by Western blotting using antibodies against DP1 (1DP06; Labvision) (middle panel) and p19ARF (R562) (bottom panel). (B) Early (P1)- and late (P4)-passage WT MEFs were harvested, and total cell extracts were prepared as described in Materials and Methods. The extracts were subjected to immunoprecipitation using an antibody against DP1 (1DP06; Labvision). The immunoprecipitates were washed, and the eluted proteins were subjected to Western blot analysis using a p19ARF antibody to detect the presence of bound ARF (top panel). The cell extracts were also tested for the expression of DP1 (middle panel) and p19ARF (bottom panel) by Western blot analysis.
ARF inhibits expression of cyclin A in a p53-independent fashion.
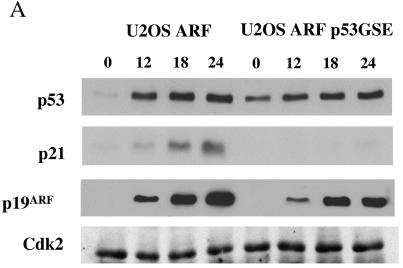
Next, we sought to investigate whether ARF expression had any effect on the expression of the E2F-activated gene cyclin A. We employed an inducible cell line, U2OS-ARF, which expresses a T7-tagged p19ARF protein upon the addition of tetracycline to the growth medium. Since U2OS cells express the wild-type p53 protein, we realized that the expression of ARF in these cells could lead to an inhibition of E2F-activated genes via the p53-p21 pathway. To distinguish between a direct effect of ARF on E2F function and an indirect inhibition of E2F-regulated genes via activation of p53, we stably expressed a dominant-negative form of p53, p53 GSE (p53 genetic suppressor element) (45), in the U2OS-ARF cell line to obtain the U2OS-ARF p53GSE cell line. The two cell lines were compared for the induction of p53 and its downstream gene p21 following the expression of ARF. U2OS-ARF and U2OS-ARF p53GSE cells were induced with tetracycline for various times (0, 12, 18, and 24 h) and analyzed by immunoblotting for the expression of p53, p21, and p19ARF. As shown in Fig. 5A, p53 and its transcriptional target, p21, were induced in response to ARF expression in U2OS ARF cells. In contrast, U2OS-ARF-p53GSE cells expressing the dominant-negative form of p53 failed to show a detectable induction of p21 following ARF expression, confirming the lack of p53 activity in those cells.
FIG. 5.
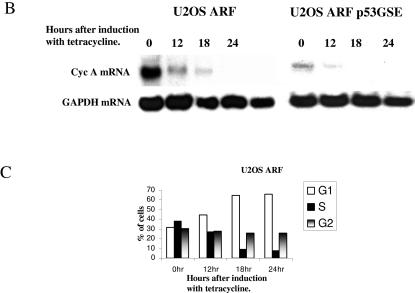
ARF inhibits E2F-activated genes prior to significant inhibition of S phase and in a p53-independent manner. (A) U2OS-ARF (left) and U2OS-ARF-p53GSE (right) cells were induced with tetracycline (1 mM). At each indicated time point following induction, cells were harvested, and total cell extracts were prepared. The extracts were tested for the expression of p53, p21, and p19ARF by Western blot analysis. The blot was probed with antibodies against p53 (Ab-6; Oncogene), p21 (Ab-11; Labvision), and p19ARF (R562; GeneTex). The extracts were also probed for Cdk2 (M2; Santa Cruz Biotechnology) as a loading control. (B) U2OS-ARF and U2OS-ARF-p53GSE cells were induced with tetracycline (1 mM) for the indicated times. Cells were harvested at each of the indicated time points, and total RNAs were prepared as described in Materials and Methods. RNAs were subjected to Northern blot analysis by probing the blot with a 32P-labeled probe against CycA and GAPDH. (C) Plots showing the percentages of cells in various phases of the cell cycle after induction of U2OS-ARF cells with tetracycline.
Total RNAs were prepared from the two cell lines at various time after tetracycline treatment, and Northern blot analysis was performed to assess cyclin A mRNA expression. Although the basal levels of cyclin A mRNA expression differed in the two cell lines, the induction of ARF in both U2OS-ARF and U2OS-ARF-p53GSE cells caused a significant decrease in cyclin A mRNA expression by 12 h after the addition of tetracycline (Fig. 5B). No significant change was observed for expression of the GAPDH transcript. Since the ARF-mediated inhibition of cyclin A mRNA occurred prior to a significant inhibition of S phase in U2OS-ARF cells (Fig. 5C) and in the absence of p21 induction in U2OS-ARF-p53GSE cells, we concluded that ARF inhibits E2F function in a p53-p21-independent fashion and prior to ARF-induced cell cycle arrest.
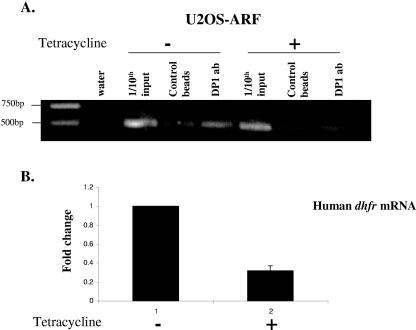
Since we observed a significant decrease in E2F/DP transcriptional activity upon ARF expression, we investigated whether that was due to a direct inhibition of binding of the E2F/DP complex to the promoter of an E2F-regulated gene. We performed ChIP experiments with U2OS-ARF cells to test whether the expression of ARF prevented the binding of DP1 to the endogenous dhfr (dihydrofolate reductase) promoter, a known E2F-regulated gene. U2OS-ARF cells were either induced to express ARF by the addition of 1 mM tetracycline to the cell culture medium for 18 h or left untreated, after which the cells were cross-linked and the chromatin was sonicated and then processed for chromatin immunoprecipitation using a DP1 antibody. As shown in Fig. 6A, the expression of ARF resulted in a significant inhibition of DP1 binding to the dhfr promoter. Consistent with this observation, we found that the expression of ARF resulted in about a threefold decrease in the levels of dhfr mRNA, as measured by quantitative real-time RT-PCR (Fig. 6B). These results clearly suggest that the expression of ARF inhibits E2F transcriptional activity by preventing the association of the E2F/DP complex with the E2F-regulated genes.
FIG. 6.
ARF inhibits association of DP1 with dhfr promoter. (A) U2OS-ARF cells were either left untreated or induced with tetracycline (1 mM) for 18 h. Cells were cross-linked, and the cross-linked chromatin from treated and untreated cells was incubated with an antibody against DP1 or with control beads. Immunoprecipitates from each sample were analyzed by PCR using primers specific for the human dhfr promoter. (B) Bar graph depicting change in the level of human dhfr mRNA in U2OS-ARF cells treated with tetracycline for 18 h, as measured by quantitative real-time RT-PCR. The level of dhfr mRNA was normalized to the level of human cyclophilin mRNA.
Endogenous ARF inhibits expression of E2F-activated genes.
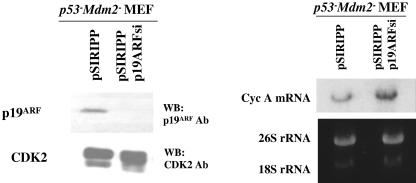
The ARF-induced inhibition of cyclin A and DHFR mRNA described above suggests that ARF is capable of inhibiting endogenous E2F function. However, since these experiments relied on the expression of exogenous ARF, we assessed endogenous DP1 function in the presence or absence of endogenous ARF. The siRNA-mediated knockdown of ARF was performed in p53− Mdm2− MEFs, and the effect of ARF loss on cyclin A expression was determined by Northern blotting (Fig. 7). We designed this experiment based on the observation that p53/Mdm2-null MEFs express higher levels of ARF than their wild-type counterparts, and we hypothesized that this high level of endogenous ARF may repress endogenous E2F function. Hence, knocking down ARF in these cells would cause a derepression of E2F-regulated genes. p53− Mdm2− MEFs were infected with a control empty vector retrovirus or a retrovirus expressing a siRNA against p19ARF (52), and infected cells were selected for 3 days in antibiotic-containing medium. Following selection, total protein and RNA extracts were prepared. Western blotting showed a nearly complete knockdown of endogenous mouse ARF in MEFs by the silencing construct, whereas the expression of an unrelated protein, Cdk2, was not altered. Notably, Northern blot analysis of the same cell populations showed increased levels of cyclin A mRNA in ARF-deficient cells (Fig. 7), consistent with the idea that ARF negatively regulates E2F function.
FIG. 7.
Endogenous ARF inhibits endogenous E2F-regulated genes. p53− Mdm2− MEFs were infected with retroviruses encoding control or p19ARF siRNA as described in Materials and Methods. Following selection, one set of cells was harvested, and total protein extracts were prepared. The extracts were probed for levels of p19ARF and Cdk2 by Western blot analysis. The membrane was first probed with a polyclonal antibody against p19ARF (R562; GeneTex), and later the blot was stripped and reprobed with an antibody against Cdk2 (M2; Santa Cruz Biotechnology) as a loading control. The other set of cells was harvested for the preparation of total RNA as described in Materials and Methods. Twenty micrograms of total cellular RNA was subjected to Northern blot analysis by probing the blot with a 32P-labeled probe against CycA. The levels of 28S and 18S rRNA are shown as loading controls.
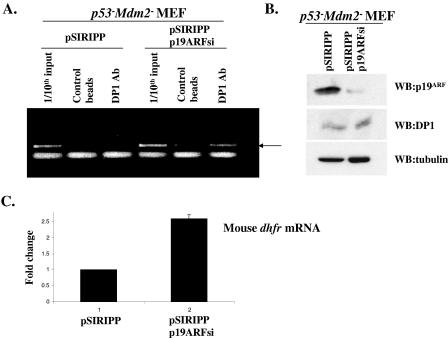
We next investigated whether endogenous ARF was capable of inhibiting the recruitment of the endogenous E2F/DP complex to the promoter of E2F-regulated genes. We performed chromatin immunoprecipitation experiments with p53− Mdm2− MEFs after the siRNA-mediated knockdown of ARF. As shown in Fig. 8B, the infection of p53− Mdm2− MEFs with retroviruses expressing a siRNA against p19ARF resulted in a significant reduction in the levels of endogenous ARF without affecting the levels of endogenous DP1. We then subjected the control and p19ARF siRNA-treated MEFs to chromatin immunoprecipitation using an antibody against DP1. The depletion of ARF resulted in a clear increase in the association of endogenous DP1 with the mouse dhfr promoter, as visualized by the ChIP assay (Fig. 8A). Consistent with this increased association of DP1 with the dhfr promoter, our quantitative real-time RT-PCR analysis showed that knocking down endogenous ARF resulted in a significant increase in the levels of dhfr mRNA (Fig. 8C). These results clearly demonstrate that ARF inhibits E2F function by blocking the recruitment of DP1 to E2F-regulated promoters.
FIG. 8.
Endogenous ARF inhibits recruitment of DP1 to mouse dhfr promoter. (A) p53− Mdm2− MEFs were infected with retroviruses encoding control or p19ARF siRNA as described in the text. Following selection, the cells from each set were cross-linked with formaldehyde, and the cross-linked chromatin was incubated with an antibody against DP1 or with control beads. The immunoprecipitated chromatin was then subjected to PCR amplification using dhfr promoter-specific primers as described in Materials and Methods. A 1/10 dilution of the chromatin used to perform the immunoprecipitation (1/10 input) was also amplified alongside the immunoprecipitated chromatin. The arrow indicates the band corresponding to the amplified dhfr promoter region. (B) (Top panel) Western blot analysis to show the levels of p19ARF (R562 antibody; GeneTex) in extracts prepared from control or p19ARF siRNA-treated MEFs. (Middle panel) Western blot analysis to show the levels of DP1 (1DP06 antibody; Labvision) in extracts prepared from control or p19ARF siRNA-treated MEFs. (Bottom panel) Extracts were subjected to Western blot analysis using an antibody against tubulin as a loading control. (C) Quantitative real-time RT-PCR was performed to detect changes in the level of mouse dhfr mRNA after p19ARF knockdown. The bar graph depicts the change in the level of dhfr mRNA in p53− Mdm2− MEFs following infection with control or p19ARF siRNA-expressing retroviruses. The levels of dhfr mRNA were normalized against that of mouse cyclophilin mRNA.
DISCUSSION
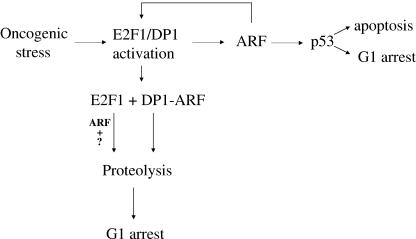
This work is significant in several ways. We show that ARF directly targets DP1 and inhibits the interaction between DP1 and E2F1 and that binding to DP1 correlates with the cell cycle inhibition function of ARF. ARF reduces the binding of DP1 to E2F-activated promoters, as evidenced by ChIP of the dhfr promoter. Moreover, ARF inhibits E2F-activated genes before a significant inhibition of S phase. Also, we show that the interaction between ARF and DP1 increases following oncogenic stress and “culture shock,” conditions that are related to the tumor suppression function of ARF. Our results are consistent with the model that the G1 arrest function of ARF involves an inhibition of the E2F family of transcription factors (E2Fs), at least partly through a direct interaction with DP1 (Fig. 9).
FIG. 9.
ARF attenuates oncogenic stress by modulating functions of the E2F complex. ARF levels are elevated in response to oncogenic stress via activation of E2F1/DP1. ARF in turn stabilizes p53, which by executing its transcriptional programs leads to either cell cycle arrest or apoptosis. ARF can also attenuate the oncogenic stress by directly targeting DP1. The binding of ARF to DP1 results in free E2F1, which could then become a target of ARF-dependent or -independent degradation. An ARF-dependent mechanism could include an association of ARF with E2F1 through other factors (?) or following a posttranslational modification of E2F1. DP1 can itself be a target of ARF-induced proteolysis.
DP1 is an essential functional partner of the E2Fs, and the association of DP1 with the E2Fs is required for efficient DNA binding and transcription activation of the E2F-regulated genes (12, 14, 18). DP1 is therefore an essential component of the cellular E2F activity, which has a critical role in cellular proliferation and apoptosis (reviewed in reference 56). DP1 has also been shown to possess oncogenic potential. For example, transgenic mice expressing DP1 under the control of the keratin K5 promoter undergo mild hyperplasia and proliferation in their basal-layer keratinocytes along with tumor development after treatment with chemical carcinogens (60). These observations are supported by the report that DP1 and DP2 cooperate with activated H-Ras in transforming rat embryo fibroblasts (22).
Previous studies done in our laboratory demonstrated that ARF could bind and induce nucleolar relocalization of DP1 under conditions of overexpression (8). In this study, we provide further evidence of a physical interaction between ARF and DP1. The results of our in vitro binding assay led us to three significant conclusions. Firstly, our observation that ARF could interact with GST-tagged wild-type DP1 in the in vitro binding assay clearly shows that the ARF-DP1 interaction is direct and is not mediated through E2F1. Secondly, the failure of the DP1 mutant lacking amino acids 205 to 277 to interact with ARF suggests that the heterodimerization domain serves as the binding site for ARF. Finally, the lack of an interaction between ARF and GST-E2F1 under conditions where we observe a robust interaction between ARF and GST-DP1 suggests that E2F1 may not be a direct binding target of ARF. Several recent reports demonstrated an interaction between ARF and E2F1 (13, 37, 40). However, those studies relied on cell-based coimmunoprecipitation assays, and therefore it is unclear whether the interaction between ARF and E2F1 is direct or not. In contrast to the results obtained by Mason et al. (40), our in vitro binding assay points to the possibility that the observed interaction between ARF and E2F1 could be mediated through other proteins. Also, it remains possible that a posttranslational modification of E2F1 regulates the ARF-E2F1 interaction. These conclusions led us to propose a new model in which ARF regulates the functions of the E2Fs by targeting DP1 (Fig. 9).
The fact that ARF and E2F1 target the same general region within DP1 to form a physical complex sets up an interesting possibility that ARF and E2F1 could compete for binding to DP1. We tested this possibility, and our results confirmed the notion that in the presence of ARF there is a significant decrease in the level of functional E2F1-DP1 complexes. Given that E2F1 and DP1 are differentially expressed during the cell cycle, the propensity of DP1 to form a complex with ARF or E2F1 might very well depend on the cell cycle phase and the levels of E2F1 and DP1. Interestingly, the recent observation that an E2F3-mediated down-regulation of p19ARF is required for cell cycle progression (1) also points to a scenario where the lowering of ARF levels results in a conducive environment for the formation of a functional E2F1-DP1 complex during the normal cell cycle. ARF is up-regulated in response to oncogenic stimuli (reference 54 and references therein). Our results suggest that an increase in the level of ARF would lead to a decrease in the level of E2F1-DP1 complexes. Thus, the levels of ARF in the cells are expected to determine the abundance of the E2F1-DP1 complex.
ARF has also been shown to induce the proteasome-mediated degradation of E2F1 (37). E2F1 is degraded by the SCF pathway and by multiple ROC-Cullin ligases in both phosphorylation-dependent and -independent manners (38, 43). Moreover, E2F1 is protected from degradation when in complex with Rb (6, 16, 19). DP1, interestingly, is required for the formation of a stable E2F1-Rb complex (18, 64). It is tempting to speculate, therefore, that one possible mechanism that could potentially explain the ARF-mediated degradation of E2F1 could involve the binding and sequestration of DP1 by ARF. This would result in free pools of E2F1, which could then be targeted for degradation via the known ubiquitin-proteasome pathways. Indeed, our previous observations showed that the coexpression of E2F1 and DP1 renders E2F1 more resistant to ARF-induced proteolysis than in a scenario where there is an excess of only free E2F1. However, the possibility that ARF acts as an adaptor to bring E2F1 and the degradation machinery together or that it stimulates a posttranslational modification to induce proteolysis of E2F1 cannot be formally ruled out. ARF is a nucleolar protein, and it has been demonstrated before that it can induce the nucleolar relocalization of some of its binding partners, such as Mdm2 (62, 68) and Foxm1b (23). One possible consequence of a physical interaction between ARF and DP1 could be an ARF-mediated sequestration of DP1 in the nucleolus. We have demonstrated previously that ARF can induce the relocalization of DP1 from the cytosol to the nucleolus when both proteins are overexpressed in cells. Since the formation of an E2F1-DP1 complex is critical for efficient binding of the complex to promoters of E2F-regulated genes and for the induction of their transcription, ARF, by lowering the levels of available DP1, prevents the formation of the functional E2F complex, thereby leading to an inhibition of E2F-regulated genes.
A predicted outcome of this possibility would be an inhibition of E2F-regulated genes in response to ARF expression. We performed experiments to provide evidence in support of this possibility. Using stable cell lines which express the mouse ARF protein upon the addition of tetracycline to the medium, we found that the expression of ARF results in a significant decrease in the levels of cyclin A mRNA in these cells. We also performed a cell cycle analysis of these cells after the induction of ARF to demonstrate that the inhibition in E2F-regulated genes occurs prior to a significant inhibition of S phase following ARF expression. We also observed that the ARF-induced inhibition of E2F-regulated genes is independent of p53. Moreover, we obtained evidence that the ARF-mediated inhibition of E2F-regulated genes is physiologically relevant. siRNA-mediated knockdown of ARF resulted in an increase in the levels of cyclin A and DHFR mRNAs. Another key finding presented in the present study is the evidence of a correlation between the G1 arrest function of ARF and its DP1 regulatory activity.
Several studies (3, 51, 66) using dominant-negative forms of E2F1 and DP1 indicated that the functionality of the E2F/DP1 complex is not essential for cell proliferation. But those studies are in apparent contradiction with the observations made with E2F1, E2F2, and E2F3 conditional triple-knockout mice. MEFs from the triple-knockout embryos are severely defective in proliferation (65). A recent study carried out a careful investigation of the dominant-negative mutant (dnE2F) of E2F1 (35). That study compared the effects of the dnE2F mutant with those of the siRNA-mediated knockdown of DP1 on cell proliferation and the expression of E2F-activated genes. It was demonstrated that the dnE2F mutant had very little effect on the expression of E2F-activated genes because the inhibition of the endogenous E2F function by dnE2F expression was only partial. The DP1 siRNA, on the other hand, eliminated DP1 and clearly inhibited the expression of several E2F-activated genes, causing the cells to exhibit a senescence-like phenotype (35). Those results are clearly in agreement with our observation that ARF-mediated inhibition of DP1 leads to an inhibition of E2F-activated genes, which would contribute to cellular senescence.
The interaction between ARF and DP1 is physiologically significant. The tumor suppression function of ARF is believed to be important during oncogenic stress (55, 71). ARF expression is induced by oncogenic stress causing the cell to undergo replicative senescence or apoptosis. ARF-induced apoptosis or senescence eliminates the possibility of oncogenic transformation. Our observation that oncogenic stress increases the interaction between ARF and DP1 is consistent with the notion that the interaction plays a significant role in the biological function of ARF. That notion is further reinforced by the observation that ARF expression in late-passage MEFs resulted in an accumulation of the ARF/DP1 complex. We believe that by targeting DP1, ARF attenuates the severity of oncogenic insults experienced by the cells. It is now known that the activating E2Fs are directly involved in the up-regulation of ARF in response to oncogenic stimuli (4). Our work now demonstrates that there is a feedback loop mechanism whereby the elevated levels of ARF compete with E2F1 for binding to DP1. The formation of ARF/DP1 complexes leads to free pools of E2F1, which could be degraded by an ARF-dependent or other previously known mechanism of E2F1 degradation. DP1 can itself be a target of ARF-mediated proteolysis. Thus, by binding to DP1, ARF causes a decrease in the levels of the functional E2F-containing complexes, leading to a dampening of the proliferation stimuli from oncogene expression (Fig. 9). It is likely that the ARF/DP1 feedback loop plays a significant role in the tumor suppression function of ARF.
Acknowledgments
We thank C. Sherr and M. F. Roussel at the St. Jude Children's Research Hospital and G. Lozano at the M. D. Anderson Cancer Center for generously providing the valuable constructs and cells used for this study.
This work was supported by grants from the National Cancer Institute (CA77637 and CA100035) to P.R. and from the National Institute on Aging (AG 21842) to R.H.C.
REFERENCES
- 1.Aslanian, A., P. J. Iaquinta, R. Verona, and J. A. Lees. 2004. Repression of the ARF tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 18:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandara, L. R., V. M. Buck, M. Zamanian, L. H. Jonhson, and N. B. LaThangue. 1993. Functional synergy between DP-1 and E2F-1 in the cell cycle regulating transcription factor E2F/DRTF. EMBO J. 12:4317-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargou, R. C., C. Wagener, K. Bommert, W. Arnold, P. T. Daniel, M. Y. Mapara, E. Grinstein, H. D. Royer, and B. Dorken. 1996. Blocking of transcription factor E2F/DP by dominant-negative mutants in a normal breast epithelial cell line efficiently inhibits apoptosis and induces tumor growth in SCID mice. J. Exp. Med. 183:1205-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumor suppressors Rb and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 5.Berwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campanero, M. R., and E. K. Flemington. 1997. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:2221-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4a and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. [DOI] [PubMed] [Google Scholar]
- 8.Datta, A., A. Nag, and P. Raychaudhuri. 2002. Differential regulation of E2F1, DP1, and the E2F1/DP1 complex by ARF. Mol. Cell. Biol. 22:8398-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta, A., A. Nag, W. Pan, N. Hay, A. L. Gartel, O. Colamonici, Y. Mori, and P. Raychaudhuri. 2004. Myc-ARF (alternate reading frame) interactions inhibit the functions of Myc. J. Biol. Chem. 279:36698-36707. [DOI] [PubMed] [Google Scholar]
- 10.DeStanchina, E., M. E. Mccurrach, F. Zindy, S. Y. Sheih, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimri, G. P., K. Itahana, M. Acosta, and J. Campisi. 2000. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14ARF tumor suppressor. Mol. Cell. Biol. 20:273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson, N. 1998. The regulation of E2F by pRb-family members. Genes Dev. 94:2245-2262. [DOI] [PubMed] [Google Scholar]
- 13.Eymin, B., L. Karayan, P. Seite, C. Brambilla, E. Brambilla, C. Larsen, and S. Gazzeri. 2001. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20:1033-1041. [DOI] [PubMed] [Google Scholar]
- 14.Girling, R., J. F. Patridge, L. R. Bandara, N. Burden, N. F. Totty, J. J. Hsuan, and N. B. LaThangue. 1993. A new component of the transcription factor DRTF1/E2F. Nature 362:83-87. [DOI] [PubMed] [Google Scholar]
- 15.Groth, A., J. Weber, B. M. Willumsen, C. J. Sherr, and M. F. Roussell. 2000. Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J. Biol. Chem. 275:27473-27480. [DOI] [PubMed] [Google Scholar]
- 16.Hateboer, G., R. M. Kerkhoven, A. Shvarts, R. Bernards, and R. L. Beijersbergen. 1996. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 10:2960-2970. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Helin, K., C.-L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factor E2F1 and DP1 leads to cooperative transactivation. Genes Dev. 7:1850-1861. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, F., F. Martelli, D. M. Livingston, and Z. Wang. 1996. The retinoblastoma gene product protects E2F1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 10:2949-2959. [DOI] [PubMed] [Google Scholar]
- 20.Honda, R., and H. Yasuda. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itahana, K., K. P. Bhat, K. P. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12:1151-1164. [DOI] [PubMed] [Google Scholar]
- 22.Jooss, K., E. W. F. Lam, A. Bybee, R. Girling, R. Muller, and N. B. La Thangue. 1995. Proto-oncogenic properties of the DP family of proteins. Oncogene 10:1529-1536. [PubMed] [Google Scholar]
- 23.Kalinichenko, V., M. L. Major, X. Wang, J. Kuechle, H. M. Yoder, M. B. Dennewitz, B. Shin, A. Datta, P. Raychaudhuri, and R. H. Costa. 2004. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18:830-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, R. A. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse Ink4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 25.Kamijo, T., J. D. Weber, G. Zambetti, F. Zindy, M. F. Roussel, and C. J. Sherr. 1998. Functional and physical interaction of the ARF tumor suppressor with p53 and MDM2. Proc. Natl. Acad. Sci. USA 95:8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohn, M. J., R. T. Bronson, E. Harlow, N. J. Dyson, and L. Yamasaki. 2003. DP1 is required for extra-embryonic development. Development 130:1295-1305. [DOI] [PubMed] [Google Scholar]
- 27.Korgaonkar, C., L. Zhao, M. Modestou, and D. E. Quelle. 2002. ARF function does not require p53 stabilization or Mdm2 relocalization. Mol. Cell. Biol. 22:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krek, W., G. Xu, and D. M. Livingston. 1995. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell 83:1149-1158. [DOI] [PubMed] [Google Scholar]
- 29.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M., E. J. Duncavage, R. Mathew, W. Besten, D. Pei, D. Naeve, T. Yamamoto, C. Cheng, C. J. Sherr, and M. F. Roussel. 2003. Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res. 63:1046-1053. [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Llanos, S., P. A. Clark, J. Rowe, and G. Peters. 2001. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 3:445-452. [DOI] [PubMed] [Google Scholar]
- 33.Lohrum, M. A., M. Ashcroft, M. H. G. Kubbutat, and K. H. Vousden. 2000. Contribution of two independent MDM2-binding domains in p14ARAF to p53 stabilization. Curr. Biol. 10:539-542. [DOI] [PubMed] [Google Scholar]
- 34.Loughran, O., and N. B. La Thangue. 2000. Apoptotic and growth-promoting activity of E2F modulated by MDM2. Mol. Cell. Biol. 20:2186-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maehara, K., K. Yamakoshi, N. Ohtani, Y. Kubo, A. Takahashi, S. Arase, N. Jones, and E. Hara. 2005. Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional Rb and p53. J. Cell Biol. 168:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reference deleted.
- 37.Martelli, F., T. Hamilton, D. P. Silver, N. E. Sharpless, N. Bardeesy, M. Rokas, R. A. DePinho, D. M. Livingston, and S. R. Grossman. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1:14-19. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Mason, S. L., O. Loughran, and N. B. La Thangue. 2002. p14(ARF) regulates E2F activity. Oncogene 21:4220-4230. [DOI] [PubMed] [Google Scholar]
- 41.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 42.Morosov, A., W. C. Phelps, and P. Raychaudhuri. 1994. Activation of the c-fos gene by the HPV-16 oncoproteins depends upon the cAMP-response element at −60. J. Biol. Chem. 269:18434-18440. [PubMed] [Google Scholar]
- 43.Ohta, T., and Y. Xiong. 2001. Phosphorylation- and SKP1-independent in vitro ubiquitination of E2F1 by multiple ROC-Cullin ligases. Cancer Res. 61:1347-1353. [PubMed] [Google Scholar]
- 44.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumor-suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 45.Ossovskaya, V. S., I. A. Mazo, M. V. Chernov, O. B. Chernova, Z. Strezoska, R. Kondratov, G. R. Stark, P. M. Chumakov, and A. V. Gudkov. 1996. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl. Acad. Sci. USA 93:10309-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumor suppressor p53 to Ras. Nature 395:125-126. [DOI] [PubMed] [Google Scholar]
- 47.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, and R. A. DePinho. 1998. The Ink4a tumor-suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-734. [DOI] [PubMed] [Google Scholar]
- 48.Qi, Y., M. A. Gregory, Z. Li, J. P. Brousal, K. West, and S. R. Hann. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431:712-717. [DOI] [PubMed] [Google Scholar]
- 49.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the Ink4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 50.Radfar, A., I. Unnikrishnan, H.-W. Lee, R. A. DePinho, and N. Rosenberg. 1998. p19ARF induces p53 dependent apoptosis during Abelson virus-mediated B-cell transformation. Proc. Natl. Acad. Sci. USA 95:13194-13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Pepper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19ARF/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 52.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 53.Serrano, M., A. W. Lin, M. E. Mccurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic Ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 54.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 55.Sherr, C. J. 2001. The INK4a/ARF network in tumor suppression. Nat. Rev. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 56.Stevens, C., and N. B. La Thangue. 2003. E2F and cell cycle control: a double-edged sword. Arch. Biochem. Biophys. 412:157-169. [DOI] [PubMed] [Google Scholar]
- 57.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDK2NA locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugimoto, M., M. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 59.Tao, W., and A. J. Levine. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, D., J. Russell, H. Xu, and D. J. Johnson. 2001. Deregulated expression of DP1 induces epidermal proliferation and enhances skin carcinogenesis. Mol. Carcinog. 31:90-100. [DOI] [PubMed] [Google Scholar]
- 61.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 2000. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 62.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. 2000. p53-independent functions of the p19ARF tumor suppressor. Genes Dev. 14:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber, J. D., M.-L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar relocalization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, C. L., L. R. Zukerberg, C. Ngwu, E. Harlow, and J. A. Lees. 1995. In vivo association of E2F and DP family proteins. Mol. Cell. Biol. 15:2536-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Song, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cell proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16Ink4a, TGFβ, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, Y., and Y. Xiong. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175-186. [PubMed] [Google Scholar]
- 70.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zindy, F., R. T. Williams, T. A. Baudino, J. E. Rehg, S. X. Skapek, J. L. Cleveland, M. F. Roussel, and C. J. Sherr. 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc. Natl. Acad. Sci. USA 100:15930-15935. [DOI] [PMC free article] [PubMed] [Google Scholar]