Abstract
SMARCB1 is a core unit of the BAF chromatin remodelling complex and its functional impairment interferes with the self-renewal and pluripotency of stem cells, lineage commitment, cellular identity and differentiation. SMARCB1 is also an important tumour suppressor gene and somatic SMARCB1 pathogenic variants (PVs) have been detected in ~ 5% of all human cancers. Additionally, germline SMARCB1 PVs have been identified in patients with conditions as clinically diverse as Rhabdoid Tumour Predisposition Syndrome type 1 (RTPS1), schwannomatosis and neurodevelopmental disorders such as Coffin-Siris syndrome (CSS). RTPS1 is characterized by the occurrence of highly malignant atypical teratoid rhabdoid tumours (AT/RT) affecting mostly infants, whereas SMARCB1-related schwannomatosis is generally diagnosed after the age of 30 and is characterized by benign schwannomas. Patients with germline SMARCB1 PVs and neurodevelopmental disorders do not usually develop SMARCB1-deficient tumours but instead exhibit severe intellectual disability and congenital malformations. It is intriguing how germline SMARCB1 PVs can be responsible for these very different pathologies. However, a network of different factors has emerged that play important roles in this context. Thus, the tumour phenotype associated with germline SMARCB1 PVs is determined by the nature and location of the SMARCB1 mutation and the timing of SMARCB1 inactivation in specific progenitor cells. Biallelic complete loss of SMARCB1 function during a narrow time window of early embryonic development in neural crest cells is essential for AT/RT development. By contrast, hypomorphic SMARCB1 PVs during later developmental stages affecting more differentiated Schwann cell precursors give rise to schwannomas. However, the loss of the wild-type SMARCB1 allele is insufficient for schwannoma growth which appears to be dependent upon concomitant somatic NF2 PVs in patients with SMARCB1-related schwannomatosis according to the four-hit/three-step model of tumorigenesis. In patients with neurodevelopmental disorders such as CSS, germline PVs would appear to cluster within the C-terminal SMARCB1 domain, interfering with the nucleosomal interactions of SMARCB1 but not with its tumour suppressor activity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10689-025-00486-4.
Introduction
The protein product of the SMARCB1 gene (also known as hSNF5, INI1 and BAF47) is a core member of the mammalian SWItch/Sucrose Non-Fermentable (SWI/SNF) complex, also termed the BAF (Brg/Brahma-associated factor) chromatin remodelling complex. The human SMARCB1 gene (MIM #601607) was originally cloned as the homolog of yeast SWI/SNF complex member SNF5 [1]. Subsequently, SMARCB1 was found to act as a bona fide tumour suppressor in malignant rhabdoid tumours (MRTs) and many other tumour types. Indeed, both inherited (germline) and acquired (somatic) SMARCB1 mutations have been implicated in causing the highly aggressive intracranial atypical teratoid/rhabdoid tumour (AT/RT) [2–8, reviewed in 9]. AT/RTs are characterized by the biallelic loss of SMARCB1 function [2, 10] and are highly malignant, developing in the main in infants and very young children, frequently leading to death within the first few years of life [reviewed by 11]. Approximately 25–35% of patients with AT/RT carry a germline SMARCB1 alteration that defines the Rhabdoid Tumour Predisposition Syndrome type 1 (RTPS1; MIM #609322) [12, 13, reviewed by 14]. AT/RTs are intracranial MRTs but MRTs may also arise in extracranial tissues such as kidney or soft tissues. In many instances, MRT develop before the age of three [15]. Less frequently, MRTs may also arise in adults [reviewed by 16].
SMARCB1 functions as a classical tumour suppressor in cases of MRT and complete loss of nuclear SMARCB1 protein expression is characteristic of this type of malignancy [reviewed by 17]. In addition to MRTs, many other tumour types exhibit somatic SMARCB1 gene inactivation or loss of expression; this group of malignancies has been collectively defined as SMARCB1-deficient tumours [reviewed by 18–20]. Astonishingly, 5% of all human cancers have pathogenic variants (PVs), albeit mostly somatic, in the SMARCB1 gene [21] highlighting its general importance in tumorigenesis.
The SMARCB1 protein is an important component of the BAF complexes, which are chromatin remodelers comprising multiple subunits mobilizing nucleosomes and regulating gene expression [22, reviewed by 20]. It turns out that approximately 20% of all human cancers have mutations in one of the BAF complex subunits [18]. The analysis of MRTs and other SMARCB1-deficient malignant tumours has indicated the consequences of complete SMARCB1 protein loss including profound changes in epigenetic architecture, aberrant activation of transcriptional and metabolic programs that promote cell growth, deregulation of stem cell maintenance and suppression of terminal differentiation [23–26]. In SMARCB1-deficient malignancies, the dysregulation of the BAF complex-dependent chromatin remodelling machinery leads to reprogramming and a blockage of differentiation that drives these cells to malignancy [27].
In 2007, germline pathogenic SMARCB1 variants were identified for the first time as predisposing to familial schwannomatosis [28]. This came as some surprise since schwannomatosis is characterized by the occurrence of mainly benign tumours and a median age at diagnosis of 40 years (range, 16–70 years) [29]. This is in stark contrast to the involvement of SMARCB1 in the tumorigenesis of highly malignant aggressive tumours such as pediatric AT/RT associated with a very poor prognosis. Non-NF2-related schwannomatosis is characterized by the development of multiple benign schwannomas of the spinal, peripheral and cranial nerves in the absence of intra-dermal schwannomas, ependymomas and ophthalmic features [29–32]. Since the first discovery of SMARCB1 PVs causing late-onset Schwann cell-derived tumours, it became clear that SMARCB1-related schwannomatosis (SWN) is one of the major forms of schwannomatosis [33–43]. The fact that germline SMARCB1 PVs not only predispose to SWN but also to highly malignant pediatric tumours in the context of RTPS1, indicates that cells of different origin must be vulnerable to the complex cellular, molecular and developmental disturbances resulting from SMARCB1 loss. In addition to its role as a tumour suppressor, SMARCB1 also plays an important role during neurodevelopment [reviewed by 44]. Thus, germline PVs in SMARCB1 may also cause neurodevelopmental disorders associated with severe intellectual disability such as Coffin-Siris syndrome (CSS, MIM #135900), which is not associated with the development of pediatric malignancies such as MRTs [45–49]. In turn, severe intellectual disability is not observed in patients with SMARCB1-related SWN or in carriers of germline SMARCB1 PVs in RTPS1 families. To make matters even more complicated, in families with RTPS1, carriers of pathogenic SMARCB1 variants have been identified without clinical symptoms. In most of these families, mosaicism cannot account for the lack of penetrance [8, 33, 50–56].
The different pathologies associated with germline SMARCB1 PVs are likely to be caused by a number of different determinants including the type of pathogenic SMARCB1 variant and its position within the different regions/domains of the gene/protein, the timing of the loss of the second SMARCB1 allele, the type of mutation associated with the loss of the second SMARCB1 allele (intragenic PV, large deletion, loss of chromosome 22q), the cellular origin of the tumour progenitor cells and the possible concomitant loss of other tumour suppressor genes. Furthermore, complex epigenetic and transcriptome changes caused by SMARCB1 mutation may play an important role in defining the clinical phenotype associated with SMARCB1 loss.
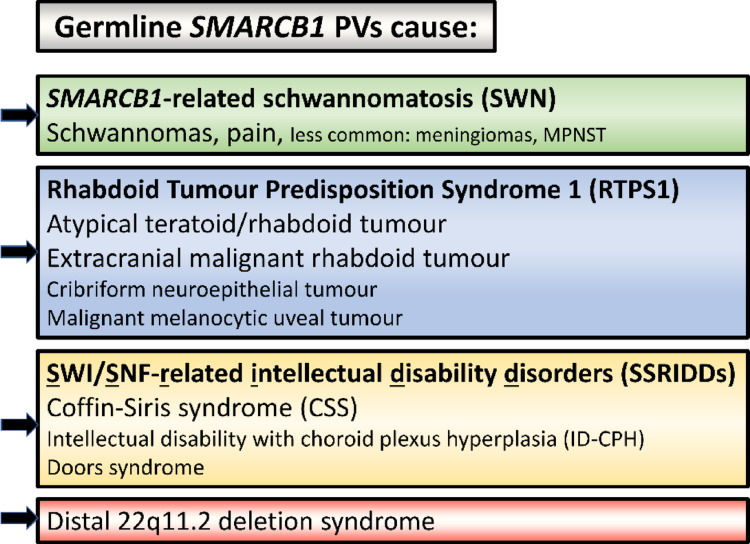
This review focuses on the germline pathogenic SMARCB1 variants responsible for a number of completely different diseases including schwannomatosis, RTPS1 and syndromic neurodevelopmental disorders (Fig. 1) as well as the functional impact of SMARCB1 loss in the context of these very different pathologies. Furthermore, particular attention is paid to the pathogenic consequences of SMARCB1 loss including disturbances in cellular differentiation and lineage specification of neural crest cells underlying the tumorigenesis of either poorly differentiated pediatric rhabdoid tumours or more differentiated adult tumours such as schwannomas.
Fig. 1.
Pathologies associated with germline SMARCB1 pathogenic variants (PVs)
Clinical spectrum of SMARCB1-related schwannomatosis and other SMARCB1-associated phenotypes
Schwannomatosis (SWN)
The autosomal dominant inherited tumour predisposition syndromes, schwannomatosis (collectively termed non-NF2-related SWN) and neurofibromatosis type-2 (NF2) (now designated as NF2-related schwannomatosis), predispose affected individuals to the development of schwannomas. These benign, well-circumscribed nerve sheath tumours only very rarely undergo malignant transformation [54, 57–60]. They contain clonal populations of Schwann cells and most schwannomas are sporadic [reviewed by 61]. However, some cases are associated with a genetic predisposition and occur in the context of a form of schwannomatosis (SWN). Despite the clinical overlap between non-NF2-related SWN and NF2-related SWN, it became clear quite early on that they are distinct clinical and genetic entities since patients with non-NF2-related SWN do not exhibit bilateral vestibular schwannomas, which is a hallmark feature of NF2-related SWN [30, 32, 62]. Furthermore, patients with non-NF2-related SWN do not have germline pathogenic variants (PVs) in the NF2 gene. Instead, tumour-specific PVs in the NF2 gene characterize the schwannomas of patients with non-NF2-related SWN [35, 62, 63]. Until the identification of the schwannomatosis-causing genes LZTR1 and SMARCB1, schwannomatosis was mainly identified by clinical criteria and by the exclusion of germline NF2 PVs [31, 64] (Supplementary Table 1). Germline pathogenic SMARCB1 variants were identified in 2007 as a cause of non-NF2-related SWN [28]. Seven years later, in 2014, LZTR1 became the second gene to be identified as causing non-NF2-related SWN [65]. In view of the clinical overlap between the different types of schwannomatosis, an update of the diagnostic criteria and the nomenclature for these schwannoma predisposition syndromes became necessary, including the need for genetic testing to arrive at the correct differential diagnosis [66].
SMARCB1-related SWN in relation to other types of SWN
The term schwannomatosis comprises a group of neurogenetic disorders that differ in terms of their genetic predisposition (Table 1). The most common form of schwannomatosis is NF2-related SWN, which is caused by heterozygous pathogenic NF2 gene variants. The non-NF2 related forms of SWN are less common and include LZTR1-related SWN and SMARCB1-related SWN. Affected individuals harbour germline or first hit PVs in the LZTR1 gene or SMARCB1 gene, respectively (Table 1). In addition to LZTR1 and SMARCB1, other SWN predisposing genes may exist since in 14–30% of patients with familial SWN, and in 60% of sporadic SWN cases, no germline PVs have been detected in LZTR1, SMARCB1 or NF2 by the application of conventional mutation screening assays [33–36, 38, 40, 67–69]. So far, only a few other genes have been identified as putative SWN-causing genes since pathogenic variants were identified in these genes in SWN patients without germline PVs in NF2, SMARCB1 or LZTR1. However, none of them would appear to account for a significant number of SWN cases since pathogenic variants have only been identified in single patients or families [43, 70–75] (Supplementary Table 2).
Table 1.
Classification of the different types of schwannomatosis (SWN) following Plotkin et al. [66]
| SWN type (MIM #) |
Gene (MIM #) [Accession number] |
Specification |
|---|---|---|
| NF2-related SWN (101000) | NF2 (101000) [NM_000268.4] | Autosomal dominant inherited syndrome caused by heterozygous germline PVs in the NF2 gene, which is located on chromosome 22q12.2, and which encodes the Merlin protein. Mosaic NF2-related SWN is caused by somatic PVs in NF2 that are not present in all cells of the patient in question. |
| LZTR1-related SWN (615670) |
LZTR1 (600574) |
Autosomal dominant condition caused by heterozygous germline PVs in the LZTR1 gene, located on chromosome 22q11.21. |
| SMARCB1-related SWN (162091) | SMARCB1 (601607) [NM_003073.5] | Autosomal dominant condition caused by heterozygous germline PVs in the SMARCB1 gene, located on chromosome 22q11.23. |
| 22q-related SWN | Unknown | Classification intended for patients with multiple schwannomas with common molecular findings on chromosome 22q such as tumour-specific loss of heterozygosity/ large (multi-gene) deletions. |
| SWN not otherwise specified (NOS) | Unknown | Classification intended for patients who exhibit clinical features of NF2-related SWN or non-NF2-related SWN but have not been subjected to molecular analysis. |
| SWN not elsewhere classified (NEC) | Unknown | Classification intended for patients in whom molecular analysis of blood and tumours has failed to detect a PV in one of the known SWN genes. |
PV: pathogenic variant; SWN: schwannomatosis
Somatic mosaicism for a SOX10 (MIM #602229) indel PV was identified in two patients with segmental SWN but they lacked germline PVs in SMARCB1 or LZTR1 [76]. Further, heterozygous somatic SOX10 indel PVs were detected in 29% of sporadic non-vestibular schwannomas [77] and in 93% of sporadic gastrointestinal schwannomas [78]. These findings indicate that a subgroup of sporadic schwannomas can arise in the context of disturbed cellular differentiation of Schwann cells resulting from mutated SOX10 [77]. So far, there are no hints as to a putative role for pathogenic SOX10 variants during tumorigenesis in patients with SMARCB1-related SWN or any other type of SWN.
Prevalence of SMARCB1-related SWN and other SWN types
Epidemiological studies from the UK have indicated that NF2-related SWN has a prevalence of 1 in 61,332 individuals [79]. The proportion of de novo cases among those with NF2-related SWN was found to be very high (72%) [79]. The second most common type of SWN was found to be LZTR1-related SWN. According to the UK data, the prevalence of LZTR1-related SWN is 1 in 527,000 [79]. Much less common is SMARCB1-related SWN with a prevalence of just 1 in 1.1 million individuals [79]. Consequently, LZTR1- and SMARCB1-related SWN are 8.4–18.4 times less frequent than NF2-related SWN.
Diagnostic criteria for SMARCB1-related SWN
The clinical overlap between the different forms of SWN represents a challenge in differentiating these disorders [32]. This made an update of the diagnostic criteria and nomenclature for the different types of SWN both necessary and urgent [66]. Crucial to differential diagnosis in SWN is genetic testing including blood as well as tumour tissue [66, 80, 81]. Genetic testing also ensures the timely diagnosis of SWN [82]. The updated diagnostic criteria for SMARCB1-related SWN are listed in Table 2.
Table 2.
Diagnostic criteria for SMARCB1-related SWN after Plotkin et al. [66]
| A diagnosis of SMARCB1-related SWN may be made when an individual meets one of the following criteria: | ||||
|---|---|---|---|---|
| At least one pathologically confirmed schwannoma or hybrid nerve sheath tumour and a SMARCB1 pathogenic variant in an unaffected tissue such as blooda | ||||
| A shared SMARCB1 PV in at least two schwannomas or hybrid nerve sheath tumours | ||||
| Pattern of genetic changes in unaffected and tumour tissues in SMARCB1-related SWN | ||||
| Gene | Unaffected tissueb | Tumour 1 | Tumour 2 | Comment |
| SMARCB1 | ||||
| Allele 1 | PV1c | PV1 | PV1 | Shared SMARCB1 pathogenic variant |
| Allele 2 | WT | LOH | LOH | LOH typically presents as a deletion of the 22q region encompassing the LZTR1/SMARCB1/NF2 genes |
| NF2 | ||||
| Allele 1 | WT | PV2 | PV3 | Tumour-specific pathogenic NF2 variant in cis to the pathogenic SMARCB1 variant |
| Allele 2 | WT | LOH | LOH |
Tumour-specific partial loss of 22q in the trans position, LOH typically presents as a deletion of the 22q region encompassing the LZTR1/SMARCB1/NF2 genes |
LOH, loss of heterozygosity; PV, pathogenic variant; WT, wild-type
aIf a likely pathogenic variant is identified, tumour analysis may help to classify the PV
bTissues unaffected by tumours e.g. blood or skin
cIf the variant allele fraction is clearly < 50%, then the diagnosis is mosaic SMARCB1-related SWN
Particularly difficult can be the differential diagnosis between mosaic NF2-related SWN and non-NF2-related SWN, specifically LZTR1-related SWN [83, 84]. This is because unilateral vestibular schwannomas may occur in patients with mosaic NF2-related SWN but also in patients with germline LZTR1 PVs (Table 3). Furthermore, some patients with mosaic NF2-related SWN may not develop vestibular schwannomas at all and only present with peripheral schwannomas. Additionally, LZTR1 variant classification can be impeded by the high number of LZTR1 loss-of-function variants in the general population [69, 85]. This might not however be as much of a problem in the context of SMARCB1, since this gene is highly loss-of-function intolerant [86].
Table 3.
Frequency of tumours in the different types of schwannomatosis (SWN) after Evans et al. [32]
| Tumour type/ location | Non-NF2-related SWNa | NF2-related SWN |
|---|---|---|
| Peripheral nerve schwannomas | 81%b | 38.5% |
| Spinal schwannomas | 74%c | 66% |
| Trigeminal nerve schwannoma | 11% | 27% |
| Lower cranial nerve schwannoma | 4.5% | 15.5% |
| Vestibular schwannoma | 7–16%d | 94% |
| Facial schwannoma | 9% | 19.5% |
| Meningioma | 4.5%e | 53%f |
| Ependymoma | 0% | 19% |
| Malignant peripheral nerve sheath tumour (MPNST) | Reportedg | 0% without radiation |
aNon-NF2-related SWN affects patients who fulfil the diagnostic criteria for SWN according to [64] (Supplementary Table 1)
bIntra-dermal schwannomas are common in patients with NF2-related SWN but very rare or even absent in patients with non-NF2-related SWN
cSpinal schwannomas are significantly more common in patients with LZTR1-related SWN (5/5, 100%) than in patients with SMARCB1-related SWN (6/15, 40%) according to [104]
dUnilateral vestibular schwannomas have so far only been observed in patients with LZTR1-related SWN [32, 68, 81, 83, 93, 96]. Histologically confirmed vestibular schwannomas have not been reported in patients with SMARCB1-related SWN
eOnly observed in patients with SMARCB1-related SWN, not in patients with LZTR1-related SWN
fMeningioma is often the first tumour detected in a child with NF2-related SWN with an early presentation of symptoms
gMPNSTs have been reported in patients with SMARCB1-related SWN but not in other types of SWN [34, 38, 40, 54, 102, 140]. Hence the MPNST risk is increased in SMARCB1-related SWN but not in other types of SWN without prior irradiation
Mosaic NF2-related SWN mimicking non-NF2-related SWN
Mosaic NF2-related SWN often exhibits substantial clinical overlap with non-NF2-related SWN resulting in the misdiagnosis of at least 9% of non-NF2-related SWN cases [32]. Remarkably, 57% of patients clinically diagnosed with non-NF2-related SWN but without germline LZTR1 or SMARCB1 lesions exhibit mosaic NF2-related SWN as determined by identical pathogenic NF2 variants detected in two independent schwannomas [32, 84]. It should be noted that somatic mosaicism is quite frequent in NF2-related SWN cases, being observed in at least 33% of de novo patients with bilateral vestibular schwannomas and in up to 60% of de novo patients with unilateral vestibular schwannomas [87–90].
Mosaic NF2-related SWN can be difficult to identify by genetic testing without at least two tumours being available for analysis in addition to blood [32, 43, 64, 81, 89, 91, 92]. No fewer than 43% of the patients with at least one non-vestibular schwannoma, and who did not meet the clinical criteria for NF2-related SWN, exhibited somatic mosaicism for an NF2 PV and hence had mosaic NF2-related SWN [81]. Conspicuously, 1.8% of patients with apparently sporadic vestibular schwannomas actually had mosaic NF2-related SWN, whilst 3% had a germline LZTR1 PV [93]. Taken together, these findings emphasize the importance of comprehensive genetic testing of tumour and blood samples for the differential diagnosis of patients with schwannomas.
Clinical presentation of patients with non-NF2-related SWN
In many studies assessing the clinical symptoms of patients with schwannomatosis (SWN), no strict distinction was made between LZTR1-related or SMARCB1-related or any other type of non-NF2-related SWN. Instead, the patients were analysed as a miscellaneous group having ‘schwannomatosis’. In the following, we shall refer to this genetically heterogeneous group as patients with non-NF2-related SWN. Whenever possible, the clinical differences between SMARCB1-related SWN compared to other types of SWN will be emphasized.
Age at initial referral, diagnosis and life expectancy
Patients with non-NF2-related SWN are most commonly diagnosed in adulthood [94, 95]. The median age at initial symptoms was 30 years (range: 8–59 years) and the median age at diagnosis was 40 years (range: 16–70 years) according to the study of Merker et al. [29].
Isolated schwannomas in patients with SMARCB1-, LZTR1- or NF2-related SWN may already be present in early childhood or in young adults [96]. In a total of 153 patients aged younger than 24 years with an isolated schwannoma, genetic testing indicated that 15 (9.8%) patients had a germline NF2 pathogenic variant, six patients (3.9%) had a germline SMARCB1 PV, and 10 patients (6.5%) had a germline LZTR1 PV [96]. A total of 13 patients (8.5%) with an isolated schwannoma had mosaic NF2-related SWN, while somatic mosaicism for either SMARCB1- or LZTR1-related SWN was not observed [96]. These findings indicate that both SMARCB1- and LZTR1-related schwannomatosis can present already in childhood or in young adulthood with an isolated schwannoma and should be suspected in addition to NF2-related SWN [97].
Remarkably, the life expectancy is significantly higher in patients with non-NF2-related SWN (mean age at death, 76.9) as compared to patients with NF2-related SWN (mean age at death, 66.2) [32]. Early age at diagnosis, truncating NF2 PVs and the presence of intracranial meningiomas were associated with increased mortality in NF2-related SWN [98–100]. Further, the presence of lower cranial nerve schwannomas is a poor prognostic factor in NF2-related SWN [101]. In patients with SMARCB1-related schwannomatosis, the increased malignancy risk must be considered which may contribute to reduced life expectancy [97, 102, 103] (Sect. Malignancy risk in patients with SMARCB1-related SWN).
Pain
The most common symptom reported by patients with non-NF2-related SWN is chronic pain, affecting 67–94% of patients [29, 31, 95, 104, 105]. Pain without a visible or palpable tumour affects 35–46% of SWN patients [29, 32]. Pain associated with a schwannoma has been reported by 11% of SWN patients [29]. However, not all schwannomas are painful. In non-NF2 SWN, a significant association was observed between high tumour volume and high levels of pain [104, 106]. SWN-associated pain can include local, multifocal or diffuse pain, which might be regarded as systemic neuropathic pain irrespective of the location of the schwannomas [29, 107]. SWN patients often have persistent or recurrent pain despite the surgical removal of schwannomas, and exhibit generalized whole-body pain [29]. There is no medication that is broadly effective in treating SWN-associated pain [108]. Surgical removal of painful schwannomas may result in complete resolution of pain symptoms but some painful schwannomas have a significant neuropathic component and drugs such as tricyclic antidepressants and gabapentinoids may help to improve the quality of life of affected patients [103]. According to the guidelines published by the European Reference Network (ERN) for Genetic Tumour Risk Syndromes (GENTURIS), the permanent use of opioids to reduce the pain in patients with schwannomatosis is not recommended owing to their poor effect on neuropathic pain and associated dependency and hyperalgesia [103].
Importantly, pain appears to correlate with the germline PV in patients with SWN. Pain-associated quality of life is significantly worse in patients with LZTR1-related SWN as compared to patients with SMARCB1-related SWN [104]. In this study, high pain levels correlated with increased whole-body tumour volume but not with the number of tumours. Further, the tumour location appears unlikely to be the primary driver of pain. Pain and pain-related quality of life were not significantly different between patients with and without spinal schwannomas [104].
Reduced intra-epidermal nerve fibre density in patients with non-NF2-related SWN
The molecular mechanisms underlying tumour-associated or neuropathic pain in patients with schwannomatosis have not so far been fully elucidated. Patients with non-NF2-related SWN exhibit a markedly lower intra-epidermal nerve fibre density (IEND) in skin biopsies as compared to controls [109]. The reduced IEND may reflect a reduction in C-fibres causing small fibre neuropathy associated with neuropathic pain [109]. Hence, patients with SWN may suffer from a form of small fibre neuropathy associated with chronic neuropathic pain. The study of Farschtschi et al. [109] included 15 patients with LZTR1-related SWN, one patient with SMARCB1-related SWN and 16 schwannomatosis patients without a pathogenic variant in either gene as determined by the analysis of blood samples. Misra et al. [110] analysed a cohort of 88 patients with small fibre neuropathy (SNF) and identified two patients with likely pathogenic variants in the LZTR1 gene. Whether this is a hint that LZTR1 plays an important role in the etiology of SNF or whether this is random co-occurrence is unclear. In this context, it should be remembered that the frequency of loss of function LZTR1 variants in the general population is quite high (0.36%) [69].
Schwannomas with and without pain in non-NF2-related SWN
Remarkably, some schwannomas in patients with SWN are not painful, whereas others are associated with severe pain, and both types may occur in the same patient. Mansouri et al. [111] observed that a significant proportion of painful schwannomas in patients with SWN affected the lower extremities, occurring predominantly in females and particularly in those with germline PVs in LZTR1. Furthermore, 16% of the very painful schwannomas were positive for the somatic SH3PXD2A-HTRA1 gene fusion [111] (Sect. Recurrent SH3PXD2A-HTRA1 fusion in SWN-schwannomas). Notably, painful schwannomas in SWN patients exhibit a significantly upregulated RAS/MAPK pathway. This is likely to be caused by LZTR1 deficiency in these tumours. Normally, the LZTR1 protein facilitates the polyubiquitination-mediated degradation of RAS proteins via the proteasomal degradation systems, leading to the inhibition of RAS/MAPK signalling activity [112, 113]. Further, the ERBB2/HER2 and VEGF pathways are significantly upregulated in painful schwannomas from patients with germline LZTR1 PVs. Moreover, painful schwannomas from patients with LZTR1- and SMARCB1-related SWN have been found to contain a significantly higher proportion of mast cells than pain-free schwannomas [111]. Mast cells are known modulators of nociceptive pain [114, 115].
Remarkably, some painful schwannomas from SWN patients would appear to secrete different factors that act on nearby nerves to augment nociception by neuronal sensitization or spontaneous neuronal firing. Thus, it may be concluded that some painful schwannomas exhibit a specific ’secretome’ [116]. This has been examined using immortalized cell lines established from primary painful and non-painful schwannomas of patients with SWN [117]. Importantly, the cell lines demonstrated the same gene expression pattern as the schwannomas they were derived from, as confirmed by microarray expression analysis [117]. Conditioned medium (CM) collected from cell lines of painful schwannomas, but not that from cell lines derived from non-painful schwannomas, contained increased amounts of multiple cytokines [116]. Furthermore, culturing mouse dorsal root ganglion neurons with CM derived from painful schwannomas led to the upregulated expression of known inflammatory pain-related genes and an increased responsiveness to noxious agonists (capsaicin and/or cinnamaldehyde) of TRPV1 and TRPA1 calcium channels [116]. In this model system, substances secreted by painful schwannomas would seem to sensitize neurons and alter neuronal gene expression [116]. However, the schwannomas analysed by Ostrow et al. [116] were not classified by the type of germline pathogenic variant causing SWN. In a follow-up study, Rubright et al. [118] included the classification by germline PV and analysed cell lines established from schwannomas of patients with either SMARCB1-related SWN or LZTR1-related SWN. These authors also included schwannomas derived from patients with NEC-related SWN, in whom molecular analysis of blood and tumours had failed to detect a PV in either LZTR1 or SMARCB1 (Table 1). Their analysis confirmed previous findings since CM from painful schwannoma cell lines contained elevated levels of specific inflammatory cytokines (IL-6, IL-8, VEGF), compared with CM collected from cell lines derived from non-painful schwannomas. Remarkably, the CM from painful schwannoma-derived cell lines of patients with NEC-related SWN (termed NEC-CM) contained higher levels of IL-8, CCL2 and CCL20 than CM from painful schwannoma cell lines of patients with SMARCB1-related SWN (SMARCB1-CM) and CM from painful schwannoma cell lines of patients with LZTR1-related SWN (LZTR1-CM). Painful LZTR1-CM contained higher levels of GDF-15, CXCL1 and GM-CSF than painful NEC-CM and painful SMARCB1-CM. These findings indicate an association between distinct profiles of secreted cytokines and chemokines in schwannomas of patients with germline PVs in different SWN genes [118]. These authors also investigated the pain response behaviour of mice after CM injection. All CM from painful schwannomas caused an increase in licking and flinching compared to control media. However, only painful LZTR1-CM caused a significantly increased acute pain response compared to non-painful LZTR1-CM. Furthermore, the increase in pain response after injection of painful LZTR1-CM was higher compared to the response after injection of painful SMARCB1-CM and NEC-CM.
Pre-treatment of cultured mouse neurons with CM from painful schwannoma cell lines enhanced their responsiveness to noxious TRPV1 and TRPA1 agonists. However, this responsiveness was different when comparing LZTR1-CM, SMARCB1-CM and NEC-CM. Painful SMARCB1-CM and LZTR1-CM enhanced the response to low-dose capsaicin more than NEC-CM. Conversely, painful NEC-CM evoked a significantly higher response to low-dose cinnamaldehyde than painful LZTR1-CM and SMARCB1-CM.
Taken together, CM from painful schwannomas sensitized mice to painful stimuli. The injection of CM from painful schwannomas in mice evoked more acute pain than did CM from non-painful schwannomas of patients with non-NF2 SWN. Further, the behavioural effects of CM injection were different when comparing CM derived from schwannomas of patients with PVs in different SWN-related genes. Additionally, the cytokine and chemokine content of CMs were different comparing schwannomas derived from patients with different forms of SWN [118].
Schwannomas
Schwannomas are the most common tumours in all types of SWN [29, 32]. In patients with non-NF2-related SWN, schwannomas of the peripheral nerves have been observed in 81–89% of patients whereas spinal schwannomas have been noted in 74% of these patients [29, 32]. Both peripheral nerve and spinal schwannomas are less common in NF2-related SWN than in non-NF2-SWN [32] (Table 3). Intra-dermal schwannomas are common in NF2-related SWN but very rare in patients with non-NF2-related SWN [119] and even considered to be completely absent in non-NF2-related SWN [29, 31, 64].
Bilateral vestibular schwannomas are the hallmark feature of NF2-related SWN, and are present in 88% of patients with germline NF2 PVs older than 30 [79, 83, 120]. Bilateral vestibular schwannomas are an important diagnostic criterion for NF2-related SWN [32, 121] and appear to be absent or are at least extremely rare in non-NF2-related SWN [32, 65, 122]. So far, only one patient with a germline pathogenic LZTR1 variant has been reported with bilateral vestibular schwannoma. The clinical presentation of this patient was however atypical and distinct from patients with NF2-related SWN as hearing was never lost and the second tumour formed quite late in life at the age of 47 [65]. In contrast to bilateral vestibular schwannomas, unilateral vestibular schwannomas (UV) may occur more often in LZTR1-related SWN. UV have been observed in 7–16% of LZTR1-related SWN patients [32, 68, 81, 83, 93, 96]. Although there is a single case report of an apparent UV in a family with a germline SMARCB1 PV, this potential association has not yet been validated [123]. The study of larger cohorts of patients with SMARCB1-related SWN imply that vestibular schwannomas do not occur in this group of patients [32, 40, 83, 120, 122]. Remarkably, patients with pathogenic LZTR1 germline PVs appear to have a significantly higher prevalence of spinal schwannomas as compared to patients with SMARCB1-related SWN [104].
Segmental schwannomas
About one third of patients with non-NF2-related SWN develop segmental schwannomatosis, with schwannomas apparently confined to a body segment such as a limb or several spinal nerve roots [29, 124]. As yet, there is no evidence for a causal relationship between the segmental presentation of SWN and genetic mosaicism of either SMARCB1 or LZTR1 PVs. Most of the patients with segmental SWN and a pathogenic variant identified in blood harboured LZTR1 PVs [124, 125]. However, segmental schwannomas have also been reported in a patient with a SMARCB1 PV (c.92 A > G, p.Glu31Gly). The patient had intradural extramedullary schwannomas only in a region of the thoracic spine (T9–T12) associated with severe pain. Interestingly, her mother possessed the same germline SMARCB1 PV but exhibited generalized SWN with multifocal (non-segmental) painless extradural neurogenic tumours in various parts of her body [126]. The observed intrafamilial phenotypic heterogeneity suggests that in addition to the nature of the germline PV, other factors such as the timing of the somatic inactivation of the second allele also determine whether schwannomatosis presents as generalized or segmental disease.
Meningiomas
Germline mutations in SMARCB1 also predispose to the development of cranial meningiomas [32, 37, 55, 127–130]. Meningiomas are observed as single tumours in 4–5% of patients with SMARCB1-related SWN [32, 38] occurring predominantly in the anterior falx cerebri [55, 131]. Importantly, germline SMARCB1 PVs do not appear to be a frequent cause of multiple meningiomas even though some families with multiple meningiomas and SMARCB1-related SWN have been reported [127, 128, 132]. It should be noted that meningiomas have not so far been observed in patients with LZTR1-related SWN. By contrast, 53% of patients with NF2-related SWN develop meningiomas [32, 133], which are among the earliest clinical features to become evident in these patients [100] (Table 3). Taken together, SMARCB1 germline PVs probably represent an occasional cause of meningioma predisposition [130]. Somatic SMARCB1 mutations not present in the germline may sometimes occur in sporadic meningiomas but these are essentially rare events [134–137].
Leiomyomas
Whether germline SMARCB1 PVs may also predispose to leiomyomas remains unclear. So far, only one patient with SMARCB1-related SWN has been reported with a leiomyoma of the cervix uteri [138]. Remarkably, chromosome 22q deletions are frequent in patients with sporadic uterine leiomyomas [139].
Malignancy risk in patients with SMARCB1-related SWN
Importantly, patients with SMARCB1-related SWN but not those with other types of SWN have an increased risk of malignancy. Although malignant rhabdoid tumours are uncommon in patients with SMARCB1-related SWN [29, 32], malignant peripheral nerve sheath tumours (MPNSTs) have been reported to occur in these patients [34, 38, 40, 54, 102, 140] (Table 3). MPNSTs are rare in the general population but they occur at an increased frequency in patients with Neurofibromatosis type 1 (NF1). An estimated 20–50% of patients with MPNSTs have NF1 [141, 142]. The lifetime risk of an MPNST in NF1 is 8–13% [102]. In addition to patients with NF1 or SMARCB1-related SWN, MPNSTs also occur with higher frequency in carriers of germline TP53 mutations. However, MPNSTs are not observed at an increased rate in other tumour predisposition syndromes [102]. In contrast to SMARCB1-related SWN, MPNSTs are extremely rare in NF2-related SWN and almost never occur in the absence of radiation treatment [102, 143]. Furthermore, in patients with LZTR1-related SWN, MPNSTs have not so far been reported.
An association between specific pathogenic SMARCB1 variants and the occurrence of MPNSTs in schwannomatosis patients has not been identified. It is likely that additional genetic alterations drive malignant transformation of schwannomas in patients with SMARCB1-related SWN. In addition to an increased risk of MPNST, a more extended malignancy phenotype may be associated with SMARCB1-related SWN. Eelloo et al. [140] reported a 51-year old female patient with a germline pathogenic SMARCB1 variant and multiple benign and malignant tumours including schwannomas, follicular lymphoma (WHO grade II), neurofibroma, uterine leiomyoma, MPNST and a neuroendocrine carcinoma of the kidney. The patient had a single base-pair deletion in SMARCB1 exon 1 causing a frameshift (c.38del; Lys13Serfs*3). Pathogenic variants in SMARCB1 exon 1 are quite frequent in patients with SMARCB1-related SWN [40] (Sect. Hypomorphic or semi-functional SMARCB1 PVs in patients with SWN).
Other malignant tumours observed in patients with SMARCB1-related SWN include papillary renal cell carcinoma [144] and leiomyosarcoma [145]. Owing to the increased risk of malignancy in patients with SMARCB1-related SWN, it has been recommended that a growing tumour, especially one causing increasingly severe functional impairment, should be immediately investigated for possible malignant transformation [103].
Rhabdoid tumour predisposition syndrome type 1 (RTPS1)
Approximately 25–35% of patients with malignant rhabdoid tumours (MRTs) carry a germline SMARCB1 alteration, which defines the Rhabdoid Tumour Predisposition Syndrome type 1 (RTPS1) [12–15, 146, 147]. In rare cases, patients with MRT harbour germline PVs in SMARCA4; this causes RTPS2 and is much less common than RTPS1 [15, 148]. Biallelic loss of function of SMARCB1 drives malignancy in MRTs, which comprise a group of highly aggressive embryonal tumours. MRTs occur predominantly in young children, frequently leading to death within the first few years of life [2–6, 8, 149]. MRTs commonly arise in the central nervous system and are termed atypical teratoid/rhabdoid tumours (AT/RT). Other anatomical sites of MRTs are extracranial including head and neck, paravertebral muscles, liver, bladder, mediastinum, retroperitoneum, extremities, pelvis, heart and kidney [reviewed by 150]. MRTs contain rhabdoid cells, characterized by eccentric vesicular nuclei with prominent nucleoli and glassy eosinophilic, inclusion-like cytoplasmic structures, which are aggregates of intermediate filaments [151]. Loss of nuclear SMARCB1 protein expression is the diagnostic hallmark of AT/RT which make up 1–2% of all CNS tumours in children, primarily affecting infants (> 70%) [reviewed by 9]. They belong to the WHO grade 4 embryonal CNS tumours group [152] and represent 40–50% of CNS tumours occurring during the first year of life [11]. Although AT/RTs develop in a variety of brain regions, the posterior fossa seems to be a frequent location [149, 153]. AT/RTs are highly malignant cancers with substantial clinical heterogeneity, poor prognosis and low overall survival rates [149, 154–160, reviewed by 11]. Even though AT/RTs occur mainly in infants and very young children, they also develop rarely as primary tumours in adults and then predominantly in the sellar region [161–170].
In patients with RTPS1, tumours develop at an earlier age than in patients without germline SMARCB1 PVs [12, 155, 171]. The very young age of patients with RTPS1 may account for their poorer prognosis compared to patients with somatic SMARCB1 PVs and AT/RT [12]. However, the association between germline predisposition to MRT and prognosis is controversial. Upadhyaya et al. [172] did not observe an association between germline predisposition by a SMARCB1 PV and a poor prognosis of AT/RT. However, in contrast to this, other studies confirmed just such an association [53, 56, 173]. Likewise, Frühwald et al. [15] reported that in addition to young age (younger than one year), a germline SMARCB1 PV is a negative prognostic factor for the survival of patients with AT/RT. Familial penetrance of RTPS1 is approximately 90% by the age of 5 [150]. The median age of diagnosis of a rhabdoid tumour in patients with RTPS1 is between 4 and 7 months. By contrast, the age at diagnosis in sporadic patients with AT/RT is 18–30 months [150]. Synchronous tumours occur in one third of patients with RTPS1, with the kidney as the most common synchronous site [146, 173–176].
Co-occurrence of RTPS1 and schwannomatosis in families
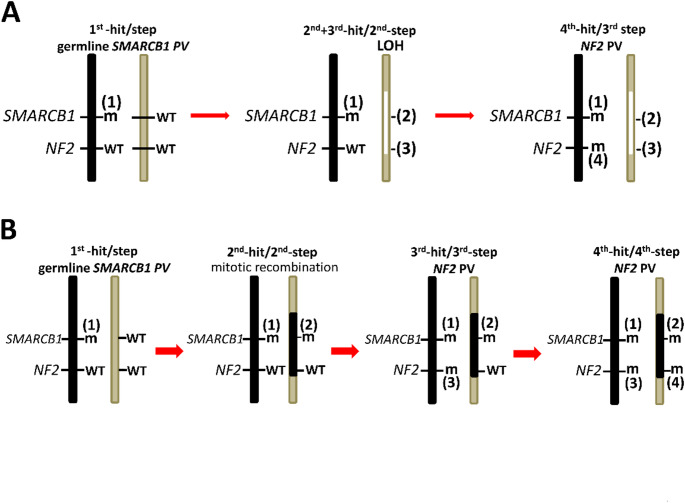
An estimated one-third of patients with MRT and germline SMARCB1 PV are familial cases [13]. In these families, the pathogenic heterozygous SMARCB1 variant detected in the child with MRT is also present in the blood of one of their parents [13]. Four families have so far been reported, with carriers of germline pathogenic SMARCB1 variants having either schwannomatosis or MRT [13, 52, 54, 177]. A common pattern observed in these families was that the parent who carried the SMARCB1 PV had schwannomatosis whereas their offspring had MRT. Importantly, in familial cases of MRT, at least 17 unaffected SMARCB1 PV carriers have been reported [8, 33, 50–56]. These SMARCB1 PV carriers were unaffected in the sense that they did not develop MRT or symptomatic schwannomas. However, it is unclear whether these apparently unaffected SMARCB1 PV carriers had clinical signs of SMARCB1-related SWN since they were not investigated by MRI, especially later in life, in order to exclude the occurrence of asymptomatic schwannomas. The incomplete penetrance observed in these families in terms of MRT development is caused by the fact that a narrow developmental window exists during which neural crest cells are sensitive towards the complete loss of the SMARCB1 protein thereby initiating rhabdoid tumour growth [178, 179, reviewed by 180]. If this sensitive period is completed prior to biallelic SMARCB1 inactivation, MRT may not develop at all (Sect. The time window of SMARCB1 inactivation in rhabdoid tumour development). However, schwannomatosis is highly likely to occur later in life in these SMARCB1 PV carriers without MRT since the penetrance of SMARCB1-related SWN is high, most likely 100% [103]. This estimate cannot be given more precisely since SMARCB1-related SWN is often diagnosed after the age of 30 and can only be confirmed or excluded by comprehensive MRI investigation. It should be noted that the co-occurrence of MRT and SMARCB1-related schwannomatosis in the same patient has been observed in long-term survivors of AT/RT thereby substantiating this hypothesis.
RTPS1 long-term survivors may develop schwannomas
Patients with RTPS1 have a very poor prognosis due to malignancy in infancy or early childhood. Long-term survival in children with AT/RT is very rare [51, 181, 182]. However, with improved treatment strategies, patients have been reported who survived the childhood MRT. Several of them developed additional primary SMARCB1-deficient tumours after being cured of the initial MRT. The tumours observed in AT/RT survivors that developed beyond the age of 5 included extracranial MRT [12, 147], AT/RT [183], MRT of the kidney [184], epitheloid sarcoma [185], schwannoma [52, 147, 186], chrondrosarcoma [187], myoepithelioma and meningioma [51], epitheloid malignant peripheral nerve sheath tumour (epitheloid MPNST) [54, 147], MPNST [188], primitive neuroectodermal tumour (PNET) [5] and adult sellar AT/RT [167]. This indicates that patients with RTPS1 remain at elevated risk for developing SMARCB1-deficient tumours after the peak age of MRT in early childhood. Consequently, clinical surveillance of RTPS1 patients beyond the age of 5 is very important [147]. It may be argued that in these MRT long-term survivors, tumour stem cells persist that are vulnerable to a critical second hit, which would drive malignancy later in life.
Cribriform neuroepithelial tumour (CRINET)
In addition to rhabdoid tumours such as AT/RT or extracranial MRT, patients with germline SMARCB1 PVs and RTPS1 appear to be predisposed to rare non-rhabdoid tumours such as cribriform neuroepithelial tumours (CRINETs). CRINETs are rare embryonal CNS tumours mostly diagnosed in children younger than 2.5 years [152, 189, 190]. Only rarely do children older than 2.5 years develop CRINETs [190, 191]. CRINETs also exhibit biallelic SMARCB1 loss as observed in MRTs [190–193]. In CRINETs, SMARCB1 loss leads to high tyrosinase expression, strikingly resembling the AT/RT-TYR subgroup based on DNA methylation and gene expression profiles [190] (Sect. AT/RT subgroups). Moreover, CRINETs and AT/RT-TYR both harbour large heterozygous losses of chromosome 22, with accompanying intragenic pathogenic variants of the other SMARCB1 allele, which is uncommon in other AT/RT subgroups. Nevertheless, CRINETs exhibit distinct histopathological features and a more favourable long-term outcome than tumours of the AT/RT-TYR subgroup [190]. In the majority of patients with CRINET, SMARCB1 PVs are somatic and accompanied by loss of heterozygosity of the other allele. However, two patients with CRINETs and germline SMARCB1 PVs have been reported [190] suggesting that CRINETs belong to the spectrum of tumours that may occur in patients with RTPS1.
Malignant melanocytic uveal tumour
Another tumour type that may expand the spectrum of RTPS1-associated tumours is melanocytic uveal tumour [194]. These authors reported two cases of aggressive intraocular tumours in two children with germline SMARCB1 PVs and biallelic SMARCB1 loss in tumour tissue. The genomic profiles as well as the transcriptome and DNA-methylation profiles of these SMARCB1-deficient malignant melanocytic uveal tumours were clearly different from MRT and uveal melanomas [194]. One of the two patients identified by Cyrta et al. [194] was treated at the age of 15 months for a localised AT/RT. After intensive treatment, she achieved complete remission. Surveillance revealed no sign of recurrence until 11 years of age, when she presented with an asymptomatic lesion of the left eye on a systematic follow-up MRI. The lesion initially showed slow growth, but underwent progression at the age of 14 to a malignant melanocytic uveal tumour with complete loss of SMARCB1 protein expression.
The second patient reported by Cyrta et al. [194] had not developed an MRT during her early years and had no family history of RTPS1. At the age of 23, however, she was diagnosed with a malignant uveal tumour. Blood analysis indicated a de novo germline 3.1 Mb deletion on 22q11.2 encompassing 38 genes including SMARCB1. The patient did not exhibit dysmorphic features or intellectual disability as observed in some patients with large deletions in the distal 22q11.2 region including SMARCB1 [156, 195]. Such large deletions in distal 22q11.2 encompassing 2-3-Mb are not frequent in patients with RTPS1 and only 10 patients with deletions of this type and AT/RT have been reported to date [196–201]. In the tumour tissue of the patient with the 3.1-Mb distal deletion in 22q11.2, complete loss of SMARCB1 expression was observed [194]. Thus, SMARCB1-deficient malignant melanocytic intraocular tumours would appear to be part of the spectrum of RTPS1-associated tumours.
Neurodevelopmental disorders caused by germline SMARCB1 PVs
Coffin-Siris syndrome (CSS)
Germline PVs in SMARCB1 cause the clinically and genetically heterogeneous Coffin-Siris syndrome (CSS, MIM #135900) [45–49, 202–209]. It is estimated that 7% of all patients with CSS carry a germline pathogenic SMARCB1 variant. PVs in other genes encoding members of the BAF complex also cause CSS: ARID1B (MIM #614556), ARID1A (MIM #603024), SMARCA4 (MIM #603254) and SMARCE1 (MIM #603111) [45, 210, 211]. Approximately 60% of all patients with CSS harbour PVs in genes encoding members of the BAF chromatin remodelling complex [46, 212–214].
Pathogenic variants in SMARCB1 generally cause a very severe CSS phenotype with global developmental delay and in most instances, severe intellectual disability [47, 48, 213, 214]. CNS abnormalities (mainly agenesis of the corpus callosum), seizures and absence of speech are common features in SMARCB1-related CSS [213–215]. Cardiovascular defects (septal defects, pulmonal artery stenosis, and/or dextrocardia), gastrointestinal problems (mainly gastro-esophageal reflux or pyloric stenosis) and genitourinary complications are also frequent in patients with CSS caused by germline SMARCB1 PVs. Feeding difficulties, postnatal growth retardation, sparse scalp hair, severe scoliosis and hypoplastic 5th fingers/toes and hypoplastic 5th fingernails/toenails are also common in these patients [47, 48, 213, 214]. Patients with SMARCB1-related CSS also have a marked progressive coarseness of the face with dysmorphic facial features including hypertelorism, thick eyebrows, a depressed and broad nasal bridge, anteverted nares and a large mouth with macroglossia [213, 214, reviewed by 216].
Intellectual disability with choroid plexus hyperplasia (ID-CPH)
A recurrent missense pathogenic variant in the N-terminal part of SMARCB1 causes severe intellectual disability and choroid plexus hyperplasia with resultant hydrocephalus, termed ID-CPH [217]. The pathogenic de novo missense SMARCB1 variant (c.110G > A; p.Arg37His) responsible was first identified in an individual with a clinical presentation overlapping with Kleefstra syndrome (KS) (MIM #610253) [218] and subsequently in an additional three unrelated individuals. The four patients showed a similar clinical phenotype including severe intellectual disability, hydrocephalus due to choroid plexus hyperplasia, various congenital anomalies, severe feeding difficulties, anemia, sleep apnea and ophthalmological problems. Some similarities were noted between individuals with CSS and the four patients with ID-CPH such as severe intellectual deficits, congenital heart defects, kidney anomalies, and feeding difficulties. However, other features frequently observed in patients with CSS caused by SMARCB1 PVs including impaired growth, microcephaly, fifth digit anomalies, dystrophic scoliosis and epilepsy were absent in the patients with ID-CPH.
The most distinctive phenotypic feature associated with the p.Arg37His variant in patients with ID-CPH was the enlargement of the central cerebrospinal fluid spaces, often leading to high-pressure hydrocephalus associated with choroid plexus hyperplasia and overproduction of cerebrospinal fluid [217]. This clinical feature has not been observed in patients with CSS implying that ID-CPH and CSS are different clinical entities within the spectrum of syndromes associated with PVs in SMARCB1 [217].
DOORS syndrome
DOORS syndrome (Deafness, Onychodystrophy, Osteodystrophy, mental Retardation, Seizures) is characterized mainly by sensorineural deafness, shortened terminal phalanges with small nails on hands and feet, increased urinary 2-oxoglutaric acid excretion, intellectual deficiency and seizures [219]. Pathogenic variants within the TBC1D24 gene (MIM #613577) are observed in approximately 50% of the patients exhibiting all the aforementioned clinical features. The genetic analysis of 32 families (36 patients) with DOORS syndrome indicated TBC1D24 PVs in 13 individuals from 10 families [219]. Subsequent whole exome sequencing in patients from the cohort without TBC1D24 PVs indicated the de novo SMARCB1 PV (c.1130G > A; p.Arg377His) in two unrelated patients. Remarkably, this PV is also known to cause Coffin-Siris syndrome (Table 4, Sect. Molecular pathogenesis of Coffin-Siris syndrome (CSS)). In contrast to the other patients in this cohort with DOORS syndrome, the patients with SMARCB1 PVs did not have seizures and 2-oxoglutaric aciduria. They also exhibited coarse facial features, 5th finger hypoplasia and cardiovascular malformations which occur more frequently in patients with CSS than in those with TBC1D24-deficient DOORS syndrome. The differential diagnosis was however impaired by the very young age of both patients with the pathogenic SMARCB1 variant - one even died during the neonatal period [219]. In view of the overlap of clinical symptoms between these two patients with DOORS syndrome and those with CSS [46–48, 213, 214], it might be possible that a small subgroup of patients with atypical CSS may also exhibit symptoms associated with DOORS syndrome.
Table 4.
SMARCB1 pathogenic variants identified in 35 patients with Coffin-Siris syndrome (CSS)
| Pathogenic variant (PV) | Exon | PV type | Amino acid change | Reference | Patient ID | Age at diagnosis | Gender | Inheritance |
|---|---|---|---|---|---|---|---|---|
| c.31G > A | 1 | missense | p.Gly11Arg | 204 | 88_S3 | 1–5 y | F | De novo |
| c.31G > A | 1 | missense | p.Gly11Arg | 209 | ID-28 | ns | ns | De novo |
| c.806 A > G | 7 | missense | p.His269Arg | 205 | 156 | ns | ns | De novo |
| c.1052dup | 8 | frameshift | p.Leu352Thrfs*9 | 205 | 235 | ns | ns | De novo |
| c.1066_1067del | 8 | frameshift | p.Leu356Aspfs*4 | 208 | Fetus 1 | ns | ns | De novo |
| c.1087 A > G | 8 | missense | p.Lys363Glu | 205 | 180 | ns | ns | De novo |
| c.1089G > T | 8 | missense | p.Lys363Asn | 48 | 43 | ns | ns | De novo |
| c.1091 A > C | 8 | missense | p.Lys364Thr | 207 | 1 | 33 y | M | paternal |
| c.1091 A > C | 8 | missense | p.Lys364Thr | 207 | 2 | 36 y | F | paternal |
| c.1091_1093del | 8 | ifd | p.Lys364del | 45 | 4 | 21 y | F | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 45 | 21 | 7 y | F | ns |
| c.1091_1093del | 8 | ifd | p.Lys364del | 45 | 22 | 2 y | M | ns |
| c.1091_1093del | 8 | ifd | p.Lys364del | 46 | 29 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 46 | 37 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 46 | 48 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 48 | 5 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 48 | 18 | ns | ns | ns |
| c.1091_1093del | 8 | ifd | p.Lys364del | 48 | 37 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 392 | ns | 28 y | F | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 205 | 174 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 205 | 136 | ns | ns | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 393 | 10 | 7.5y | F | De novo |
| c.1091_1093del | 8 | ifd | p.Lys364del | 209 | ID-29 | ns | ns | De novo |
| c.1096 C > T | 8 | missense | p.Arg366Cys | 49 | K2588 | ns | ns | De novo |
| c.1096 C > T | 8 | missense | p.Arg366Cys | 202 | MR2788 | 11 months | M | De novo |
| c.1096 C > T | 8 | missense | p.Arg366Cys | 206 | 4 | 13 y | F | De novo |
| c.1096 C > G | 8 | missense | p.Arg366Gly | 205 | 88 | ns | ns | De novo |
| c.1107 C > G | 8 | missense | pAsp369Glu | 394 | 43 | ns | ns | De novo |
| c.1113 C > G | 8 | missense | p.Asn371Lys | 203 | 8 | 7 months | F | De novo |
| c.1121G > A | 9 | missense | p.Arg374Gln | 282 | ns | 26 y | M | De novo |
| c.1121G > A | 9 | missense | p.Arg374Gln | 49 | K2426 | 4 months | M | De novo |
| c.1121G > A | 9 | missense | p.Arg374Gln | 205 | 230 | ns | ns | De novo |
| c.1130G > A | 9 | missense | p.Arg377His | 45 | 11 | 7 y | F | De novo |
| c.1130G > A | 9 | missense | p.Arg377His | 394 | 44 | ns | ns | De novo |
| partial deletion | 8 + 9 | 9 kb deletion | from intron 8 extending into the flanking DERL3 gene | 205 | 076 | ns | ns | De novo |
ifd: in-frame deletion; ns: not specified; y: year(s); M: male, F: female
Distal 22q11.2 deletion syndrome
Chromosome 22q11.2 contains regions of multiple low copy repeat (LCR) sequences that mediate non-allelic homologous recombination (NAHR) and predispose to pathogenic deletions and duplications [reviewed by 220]. Eight LCRs located at chromosome 22q11.2, designated as LCR22 A–H, have been identified [220] (Fig. 2). Patients with germline distal deletions in 22q11.2, encompassing sub-bands 22q11.22-q11.23 and mediated by NAHR between LCR22 D-H, have an increased risk of MRT (Fig. 2). Importantly, these distal deletions (type III) [195] encompass the SMARCB1 gene which explains the predisposition to rhabdoid tumours in these patients [12, 13, 199]. So far, 17 patients with type III deletions have been reported and many of them exhibit congenital anomalies including dysmorphic features, cardiac defects, developmental delay and microcephaly [12, 195–199, 221, 222]. A proportion of these patients also exhibit intellectual deficits, language delay and psychiatric or behavioural problems suggesting that SMARCB1 deficiencies are not only responsible for the increased MRT risk in these patients but also for disturbances during neurodevelopment. In addition to large deletions in the distal 22q11.2 region, other structural rearrangements such as ring-chromosome 22 also predispose to the development of AT/RT [223].
Fig. 2.
Schematic representation of the proximal chromosome 22q region (22q11.2) indicating the locations of the LZTR1 and SMARCB1 genes and the low copy repeats 22 (LCR22 A-H). The nucleotide numbering is given according to GRCh38.p14/hg38. The genomic positions of the LCRs are: LCR22-A: 18,156,276 − 19,035,473; LCR22-B: 20,141,014–20,377,631; LCR22-C: 20,667,276 − 20,738,272; LCR22-D: 21,009,379 − 21,565,091; LCR22-E: 22,617,530 − 22,707,515; LCR22-F: 23,307,813 − 23,477,813; LCR22-G: 24,234,032 − 24,304,032; LCR22-H: 24,599,033 − 24,684,063. The NF2 gene is located telomeric to LZTR1 and SMARCB1 at 22q12.1
Molecular pathogenesis of SMARCB1-related schwannomatosis and other SMARCB1-associated phenotypes
SMARCB1
SMARCB1 (SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily b, member 1) encodes a core subunit of the BAF chromatin-remodelling complex responsible for regulating gene expression and development by positioning/remodelling nucleosomes at genomic regulatory regions in an ATP-dependent manner [224, 225]. BAF complex activity is essential for pluripotency in embryonic stem cells, regulating accessibility and transcription at promoters, enhancers and pluripotency factor binding sites [226–229, reviewed by 230]. SMARCB1 is involved in cell growth regulation and cytoskeleton reorganization [231–235]. SMARCB1 loss in MRT cells alters the translation efficiency of specific mRNAs [236]. SMARCB1 is ubiquitously expressed in the nuclei of all normal cells [237] and acts as a tumour suppressor in pediatric AT/RT and extracranial MRTs. In adult tumours, SMARCB1 may have a more multifaceted, even oncogenic role [238, 239].
Structure of the SMARCB1 gene
The SMARCB1 gene is located on chromosome 22q11.23, approximately 5.9-Mb centromeric to the NF2 gene. SMARCB1 encompasses nine exons but a complex pattern of splice variants of SMARCB1 exists with at least eight mRNA isoforms [240]. The most common ones are isoform 1 with a full-length exon 2 (exon 2 L, 139 bp), and isoform 2, which contains a shorter exon 2 lacking the last 27 nucleotides at its 3′ end (exon 2 S, 112 bp) [240, 241]. The other six isoforms represent a small proportion of all SMARCB1 transcripts and their functional significance remains unclear. Intriguingly, there is some functional redundancy between the two major SMARCB1 isoforms 1 and 2 [37]. Indeed, compensatory expression is observed, such that knockdown of either isoform alone has no effect on cell survival [242]. Pronounced functional differences between the major isoforms have not been reported in comparative studies [243, 244]. This is important because pathogenic variants that affect only one SMARCB1 isoform may be compensated, or partially compensated for, by the other isoform, which may result in residual SMARCB1 function. Such hypomorphic pathogenic SMARCB1 variants may explain why patients with these variants and SMARCB1-related SWN exhibit benign schwannomas during adulthood but usually do not develop the highly malignant pediatric AT/RT characterized by the complete loss-of-function of SMARCB1 (Sect. Hypomorphic or semi-functional SMARCB1 PVs in patients with SWN).
SMARCB1 as a core subunit of BAF and PBAF
The SMARCB1 protein (also termed BAF47) is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters [245–247]. As a core subunit, SMARCB1 is included in two of the BAF complex family members, the canonical BAF complex (cBAF or simply BAF) [248] and the polybromo-associated BAF complex (PBAF) [249]. However, SMARCB1 is not included in the non-canonical BAF complexes [250–252]. The BAF subcomplexes contain 12–15 subunits encoded by 29 genes [reviewed by 253]. As the catalytic subunit, BAF and PBAF subcomplexes each contain a single ATPase (encoded by either SMARCA2 or SMARCA4) as well as three core subunits, including SMARCB1, and additional accessory subunits [reviewed by 254]. The specific composition of the subunits varies between different tissues. BAF complexes use energy derived from ATP hydrolysis to destabilize histone-DNA interactions and alter nucleosome positions, thereby increasing the accessibility of DNA-binding factors to their genomic target sites and activating gene expression. A model of BAF complex-dinucleosome interaction implies that one nucleosome occupies a large pocket on the surface of the BAF complex and stimulates its ATPase-driven DNA translocase activity. The nucleosome in the pocket retains all of its histones, although its structure is altered, while a neighbouring nucleosome in the path of the mobilized nucleosome-BAF complex is evicted from the DNA [255, 256].
Within the BAF complex, SMARCB1 acts as an anchor that binds to the nucleosome acidic patch via its highly basic C-terminal alpha helical domain, whereas the ATPase subunit SMARCA2 or SMARCA4 binds to the opposing side of the nucleosome [257–260]. With the nucleosome held on both sides, the ATPase subunit is able to slide DNA along the nucleosome [20].
As determined by the analysis of SMARCB1 re-expression in SMARCB1-deficient MRT cell lines, the BAF complex facilitates the acetylation of histone 3 lysine 27 (H3K27) through its interaction with histone acetyltransferases. This leads to increased gene and enhancer activity and antagonizes the effect of the polycomb repressive complex (PRC2) which is responsible for the trimethylation of H3K27 [22, 245, 246, 261, 262] (see Sect. BAF and Polycomb complex antagonism).
The three BAF subcomplexes (cBAF, PBAF and ncBAF) have distinct functions determined by the incorporation of complex-specific subunits [250, 263]. For example, the BAF subcomplexes play different roles in macrophage responses to bacterial endotoxins, regulating chromatin accessibility and enhancer activation, thereby influencing the expression of inflammatory response genes [264]. Further, PBAF (but not cBAF) is essential to the maintenance of genomic integrity during mitosis. Additionally, PBAF but not cBAF plays an important role during DNA-damage-induced transcriptional repression involving the polycomb repressive complexes 1 and 2 [265] and the ubiquitination of proliferating cell nuclear antigen (PCNA) induced upon DNA damage [266].
Emerging evidence also points to distinct localization patterns for each BAF subcomplex. The BAF complex preferentially binds to active distal enhancers, which are located some distance away from the gene they regulate [246]. By contrast, PBAF is enriched at proximal promoter regions [246, 267, reviewed by 20].
Some of the BAF/PBAF subunits are tissue- or cell-specific and the combinatorial assembly of tissue-specific complexes plays an important role during cell lineage acquisition [reviewed by 254]. During differentiation, the BAF-complex composition changes in a tightly regulated manner. Many of the resulting complexes are responsible for tissue-specific regulation of neural development and function, heart development, muscle development or embryonic stem cell pluripotency [reviewed by 268]. The timely expression of specific BAF subunits directs stem cell fate in neurogenesis as well as in skeletal myogenesis [reviewed by 216]. The localization of BAF complexes coincides with the major fate-determining factors of the cell lineage [reviewed by 253]. Indeed, embryonic stem cells have a BAF complex comprised of a specialized subunit composition not found in other cell types (designated as esBAF) which binds strongly to pluripotency factors [226]. During neurogenesis, the esBAF complex undergoes sequential developmental changes in subunit composition and is reconstituted in neural stem cells to the neuronal progenitor BAF (npBAF) complex in order to confer multipotency, while maintaining proliferative ability. As neural development moves on and neural progenitors differentiate into neurons, the npBAF complex switches subunits to form the neuron-specific nBAF complex found only in postmitotic neurons and required for dendritic morphogenesis [263, 269, 270]. SMARCB1 is an essential subunit within these BAF complexes, which play important roles during neural development, neural tissue specification, neuronal migration, maturation and dendritic morphogenesis [reviewed by 254]. Pathogenic variants in genes encoding the BAF complex subunits, including SMARCB1, are responsible for different neurodevelopmental disorders. This serves to emphasize the essential role of BAF complex activity during neural development (Sect. Molecular pathogenesis of neurodevelopmental disorders caused by germline SMARCB1 PVs).
Domains of the SMARCB1 protein
SMARCB1 is a 47 kDa nuclear protein encompassing 385 amino acids. Structurally, it has four distinct domains and a key region designated as an intrinsically disordered region (IDR) (Fig. 3).
Fig. 3.
Structure of the SMARCB1 protein: WHD: winged helix domain; IDR: intrinsically disordered region; RPT1 and RPT2: tandem repeat domains; NES: nuclear export signal; CTD: C-terminal coiled-coil domain. The numbers indicate the amino-acid positions
Winged-helix DNA-binding domain (WHD)
The N-terminal WHD of the SMARCB1 protein exhibits structural similarity to other winged-helix domains found in many different DNA-binding proteins. Nuclear magnetic resonance (NMR) studies using recombinant SMARCB1 WHD expression indicated its ability to bind to double stranded DNA [271]. Within the canonical BAF and the PBAF complex, the WHD appears to adopt different conformations because in BAF, the WHD is located distal to the nucleosome, whereas in PBAF, the WHD is located proximal to the nucleosome [260, 272, 273]. The cryo-EM structure of the nucleosome-bound BAF-complex modelled by He et al. [260] indicated that the SMARCB1-WHD binds the ARM (armadillo repeat) domain of ARID1A (T-rich interactive domain-containing protein 1 A) and is located more than 40Å away from nucleosomal DNA implying a role independent of its DNA-binding ability. Furthermore, the WHD is in close proximity to the actin-related protein (ARP) module, suggesting a role in regulating intermodular interactions [260].
Intrinsically disordered region (IDR)
SMARCB1 consists of two globular functional domains that are connected by an intrinsically disordered region (IDR). IDRs enable conformation flexibility and adaptability within proteins in order to facilitate regulation via post-translational modifications, scaffolding and recruitment of transient binding partners, and complex assembly [274]. Within the IDR is a cluster of loss-of-function intolerant residues (aa122, aa123 and aa125) [273]. Missense PVs affecting these residues may impair the flexibility of the SMARCB1 protein within the BAF complex [273].
Repeat domains (RPT1 and RPT2)
The tandem repeat domains RPT1 and RPT2 contain two highly conserved imperfect repeat regions encompassing approximately 60 amino acids [275]. RPT1 is necessary for MYC and HIV-1 integrase binding to SMARCB1 [1, 276, 277]. RPT2 interacts with exportin 1 (XPO1) via a nuclear export signal [278]. Normally, this nuclear export signal (NES-residues 259–276) is masked by the C-terminal domain (residues 319–385) of SMARCB1 [278]. Introduction of a truncated SMARCB1 lacking the C-terminal domain into an MRT cell line led to the cytoplasmic localization of SMARCB1. This cytoplasmic localization is dependent upon exportin 1 (XPO1), which directly interacts with the NES sequence of SMARCB1 potentiating its cytoplasmic localization. Importantly, the BAF complex exerts its biological function only in the nucleus. Thus, the cytoplasmic localization of SMARCB1 eliminates its tumour suppressor function. In AT/RTs with pathogenic variants in the C-terminal region of SMARCB1, cytoplasmic SMARCB1 staining is highly enriched in the absence of any nuclear staining of SMARCB1 [279]. Some 19% of all AT/RTs exhibited a cytoplasmic localization for C-terminally mutated SMARCB1 [279]. By contrast, the majority of AT/RT and extracranial MRT exhibit a complete loss of SMARCB1 protein expression [280]. Aberrant cytoplasmic deposition of mutant SMARCB1 protein is frequent only in some AT/RT, specifically in the subgroup with the AT/RT-TYR molecular signature [281] (see Sect. AT/RT subgroups).
C-terminal coiled-coil domain (CTD)
The C-terminal coiled-coil domain (CTD) of SMARCB1 contains an alpha-helical region of densely packed basic amino acids (aa 357–377), which physically interact with the nucleosome acidic patch opposite the ATPase catalytic subunit within the BAF complex [258]. Importantly, pathogenic variants of single amino acid residues within the highly basic SMARCB1 alpha helix of the CTD disrupt nucleosome binding and reduce the remodelling efficiency of the BAF complex [258]. Interestingly, these C-terminal mutations have little effect on global BAF localization, suggesting that the specific interaction with nucleosomes is not critical for the binding of BAF complexes to their target genes. PVs in the highly basic SMARCB1 C-terminal alpha-helical region cause Coffin-Siris syndrome, a neurodevelopmental disorder associated with severe intellectual disability [48, 213, 214] (Sect. Molecular pathogenesis of Coffin-Siris syndrome (CSS)). However, pathogenic variants in the CTD have also been identified in different types of tumour including meningiomas, adenocarcinomas and schwannomas [134, 282, 283].
SMARCB1-containing BAF complexes regulate enhancers
SMARCB1 is critical for genome-wide BAF complex binding to enhancers as well as for enhancer activation [245–247, 284]. The reintroduction of SMARCB1 into SMARCB1-deficient MRT cell lines increases BAF localization, particularly at distal enhancers, and promotes active enhancer histone modification marks facilitating gene expression [245–247]. SMARCB1 protein deficiency in MRT cells destabilizes the association of BAF complexes on chromatin, without drastically impairing complex stability or assembly [22, 246, 250]. However, conflicting results have been reported concerning BAF complex stability in the absence of SMARCB1 since some studies have suggested dissociation of the complex due to SMARCB1 loss [247, 285]. Nevertheless, accumulating evidence is suggestive of BAF complex stability in the absence of SMARCB1 [246, reviewed by 20].
In SMARCB1-deficient MRT cell lines, re-expression of SMARCB1 resulted in widespread recruitment of the BAF complex to previously unoccupied enhancers, the activation of these enhancers and the resolution of bivalency at promoters towards an active state [245–247]. This certainly holds true for typical enhancers, but the activity of SMARCB1 at super-enhancers is as yet unclear. Super-enhancers comprise clusters of highly active enhancers and are master regulators of cell identity [reviewed by 286, 287]. Although Nakayama et al. [246] reported significant enhancer activation upon SMARCB1 re-expression in MRT cells at both typical enhancers and super-enhancers, Wang et al. [247] observed that SMARCB1 was dispensable for super-enhancer activation. The results of Alver et al. [245] suggested that the BAF complex within intact SMARCB1 is a major regulator of typical distal enhancer and lineage-specific enhancer activity. However, super-enhancers appeared to be refractory to SMARCB1 loss and less dependent upon BAF complex activation [245]. Further analyses are necessary in order to clarify this organization.
In contrast to MRT cells, which exhibit genome-wide loss of enhancer activity upon SMARCB1 loss [246, 247, 284], SMARCB1 knockdown in human embryonic stem cells (hESCs) resulted in widespread transcriptional upregulation and increased expression of bivalent genes [228]. Thus, in differentiating hESCs, the SMARCB1 protein acts as a transcriptional repressor particularly of bivalent genes. Langer et al. [228] also showed that SMARCB1 is essential for hESC super-enhancer silencing during neural differentiation thereby enabling the pluripotent cells to differentiate along this lineage. Loss of SMARCB1 activity in hESCs inhibited neural induction during differentiation assays, a finding which is consistent with its role as a tumour suppressor in the central nervous system [228, 229]. Taken together, SMARCB1 would appear to have differential effects on enhancer and super-enhancer accessibility in a stage- and lineage-specific manner.
During cellular differentiation, regulatory regions such as enhancers become activated by chromatin opening and binding of pluripotency and pioneer transcription factors that confer locus specificity [288, reviewed by 289]. Current models that serve to explain the relationship between chromatin accessibility and transcription include that transcription factors recruit nucleosome remodelers such as the BAF complexes to evict nucleosomes and to facilitate RNA polymerase II (RNAPII) binding. As concluded from murine embryonic stem cell models, RNAPII promoter-proximal pausing promotes BAF occupancy and ATP-dependent nucleosome remodelling, which leads to nucleosome removal and increased DNA accessibility [290]. However, effective chromatin remodelling occurs only at active regulatory regions where simultaneous binding of DNA-sequence-specific transcription factors drives nucleosome eviction [reviewed by 291]. Among these are transcription factors from the Activating Protein 1 (AP-1) family, which, together with lineage-specific transcription factors, bind to nucleosome-occluded enhancers and recruit the BAF complex to induce nucleosome remodelling and establish an accessible chromatin conformation [292, 293]. SMARCB1 inactivation in MRTs results in genome-wide loss of enhancer activity important for normal development [246, 247, 284]. Furthermore, SMARCB1 protein deficiency also impairs the association of lineage-specific transcription factors with enhancers [294]. Re-expression of SMARCB1 in AT/RT cell lines indicated that BAF complexes with active SMARCB1 subunits are necessary to determine the epigenetic regulatory roles of lineage-specific transcription factors [294]. The AP-1 family of transcription factors plays a central role in this process. Loss of SMARCB1 in a subgroup of AT/RT (AT/RT-MYC, see Sect. AT/RT subgroups) has been shown to lead to the specific loss of expression of the AP-1 subunit c-JUN, which normally organises the expression of lineage-specific transcription factors [294]. Importantly, in melanoma cellular models, loss of c-JUN or other members of the AP-1 transcription factor network is associated with a poorly differentiated state [295]. Thus, the cooperativity between the BAF complex and lineage-specific transcription factors indicates that both are important regulators of cellular identity [294].
Differential regulation by BAF and PBAF complexes has been observed at enhancers and promoters, respectively, suggesting distinct functions of each complex that are perturbed upon SMARCB1 loss in MRT cells [246, 296]. By contrast, both BAF and PBAF complexes are important in activating bivalent promoters during development. Upon SMARCB1 loss-of-function, this process is significantly impaired, resulting in the repression of key tumour suppressor and lineage-specific differentiation genes [246] (see Section BAF and Polycomb complex antagonism).
Epigenetic and transcriptome changes in rhabdoid tumours
Much of our knowledge about the molecular and pathogenetic consequences of biallelic SMARCB1 protein loss-of-function derives from the analysis of MRT and AT/RT tissue and cell lines. Indeed, biallelic SMARCB1 inactivation is prevalent in these tumours; other frequently recurring genomic changes including deletions, duplications and pathogenic variants are not observed [10, 24, 26, 297]. Remarkably, rhabdoid tumours are among the tumours with the lowest mutational burden [24, 298, 299]. Despite this lack of genetic heterogeneity, AT/RT exhibit massive changes of their epigenome as is evident from the depletion of H3K27 trimethylation and H3K27 acetylation marks associated with a quiescent genomic state [284, 300, 301].
AT/RT subgroups
AT/RTs exhibit heterogeneity in terms of their DNA methylation signatures associated with specific gene expression profiles that may be used to classify AT/RTs into three different subgroups distinguishable by their epigenetic and transcriptome signatures [24, 26, 302–304]. At least three distinct AT/RT molecular subgroups exist: AT/RT-SHH, AT/RT-TYR and AT/RT-MYC [24, 26, 281, reviewed by 305]. The AT/RT-TYR subgroup was named after the enzyme tyrosinase, which is overexpressed in AT/RT-TYR cases, but not in the other AT/RT subgroups, suggesting that tyrosinase immunohistochemistry is a well-suited diagnostic marker for AT/RT-TYR cases [306]. The AT/RT-SHH subgroup displays overexpression of both sonic hedgehog (SHH) and Notch pathway members. Both pathways are conserved key regulators of development [reviewed by 307, 308]. Protein expression of achaete-scute homolog 1 (ASCL1), a neuronal differentiation transcription factor, has been suggested as an immunohistochemical marker for this subgroup [25, 309]. The AT/RT-MYC subgroup exhibits elevated expression of the MYC oncogene (MIM #190080) as opposed to the MYCN oncogene (MIM #164840), which is overexpressed in the AT/RT-SHH subgroup.
Even if a similar and low mutational burden is common to all AT/RT subgroups, they exhibit substantial differences in their epigenetic profiles. The AT/RT-TYR subgroup, and to a lesser extent the AT/RT-SHH subgroup, exhibit global and promoter hypermethylation comparable to other pediatric brain tumours and normal pediatric brain tissue [24, 304]. By contrast, tumours of the AT/RT-MYC subgroup are characterized by a hypomethylated signature as compared with normal pediatric brain samples [24, 304]. Further, the AT/RT subgroups also exhibit differences in SMARCB1 mutation patterns, clinical features including patient age, tumour location and neuroradiological imaging results [24, 281, 310]. Several studies have also indicated higher immune cell infiltration in AT/RT-MYC and AT/RT-TYR than in AT/RT-SHH [304, 311, 312].
An association between the AT/RT subgroup, age of the patient and survival has been observed. AT/RT-SHH and AT/RT-MYC DNA-methylation signatures as well as age younger than one year are all negative prognostic factors (5-year overall survival rate: 0%) [15]. By contrast, patients with tumours of the AT/RT-TYR subgroup who were older than one year had a much better prognosis and a 5-year overall survival rate of ~ 70% [15]. Likewise, Upadhyaya et al. [172] observed that infants with tumours in the AT/RT-TYR subgroup had the highest survival rate.
Cell-of-origin of AT/RT subgroups
The molecular diversity among the AT/RT subgroups is likely to be associated with a different cell-of-origin. Increasing evidence suggests that AT/RT-SHH derive from neural progenitor cells [281, 313, 314]. Similarities between the DNA methylation and gene expression profiles between extracranial MRT and tumours of the AT/RT-MYC subgroup are suggestive of common dysregulated developmental programs and that they arise from a neural crest-derived lineage shared with Schwann cells and blocked on their way to mesenchymal differentiation [175, 178, 179, 304, 315]. By contrast, the genetically engineered mouse models and single cell transcriptome analyses performed by Graf et al. [316] suggested that AT/RT-MYC as well as extracranial MRTs with the AT/RT-MYC expression profile originate from fetal primordial germ cells (PGCs). Smarcb1 loss in murine PGCs may cause reversal of germ cell specification, misguided migration to various body locations and finally tumorigenesis [316]. However, neural crest cells and PGCs represent different developmental lineages [317]. Thus, the origin of AT/RT-MYC remains unclear, although the neural crest-derived hypothesis has received the most support [reviewed by 305].
The cellular origin of tumours with the AT/RT-TYR signature also remains unresolved. Although the role of overexpression of tyrosinase in AT/RT tumorigenesis remains to be established, it is remarkable that several other components of the melanosomal pathway are also upregulated in the AT/RT-TYR subgroup, which may indicate a neural crest or neuroectodermal origin for this AT/RT subgroup [281, 318].
It remains to be determined if the AT/RT subgroup-specific characteristics are caused by different cells-of-origin [reviewed by 305]. Organoid and mouse models have indicated a specific early developmental time frame (E6–10) during which SMARCB1 loss-of-function leads to the formation of malignant rhabdoid tumours (see Sect. The time window of SMARCB1 inactivation in rhabdoid tumour development). Consequently, the pool of potential cells of origin has been narrowed down to early embryonic development [178, 179, 314, 316, 319]. Nevertheless, the cell-of-origin of AT/RTs is still a matter of intensive investigation. Terada et al. [320] established a xenograft model of AT/RT using human SMARCB1-deficient pluripotent stem cell-derived neural progenitor-like cells (NPLCs). They observed that that the AT/RT cells-of-origin are undifferentiated cells at a very early developmental stage, before their differentiation into neural progenitor cells. Their analysis also showed that SMARCB1−/− cells are still able to differentiate into neural progenitor cells (NPCs). Importantly, the subsequent neuronal differentiation of NPCs is blocked due to SMARCB1 protein loss-of-function. These findings accord with those of Carmel-Gross et al. [229] who showed that the complete loss-of-function of SMARCB1 in human embryonic stem cells does not impair their capacity to differentiate in vitro and in vivo into NPCs. However, in similar vein to the findings of Terada et al. [320], SMARCB1 deficiency impairs the neuronal differentiation of NPCs. Various other studies have suggested that AT/RT derive from NPCs, in particular those AT/RTs with the AT/RT-SHH molecular signature [281, 313, 314, 319].
It is most likely that cell-of-origin is only one of several important factors during AT/RT development. It has been suggested that AT/RT development may follow a “three-hit-model” which requires that the differentiation stage, cell-of-origin and the type of SMARCB1 inactivation (intragenic pathogenic variant, broad deletion and chromosome 22 loss in different combinations) should combine in such a way as to induce tumour development [305].
Blocked neural differentiation in AT/RTs
DNA hypermethylation in AT/RT disrupts the binding of transcription factors to DNA which impairs the expression of genes involved in neural differentiation [284, 321]. Importantly, AT/RTs exhibit DNA hypermethylation in the regulatory regions of pioneer transcription factors such as Neurogenin 1 (NEUROG1) and Neuronal differentiation 1 (NEUROD1) [314, 321]. These transcription factors are among the master regulators of neurogenesis and neural differentiation [322-324, reviewed by 289] and the blockage of neural differentiation is causally associated with AT/RT tumorigenesis [313, 319]. Thus, DNA hypermethylation in AT/RTs perturbs neural differentiation which drives malignancy [reviewed by 305]. Indeed, disturbances in cellular differentiation that result in the unlocking of phenotypic plasticity are among the hallmarks of cancer [325]. Halted neural differentiation in AT/RT by loss of BAF complex function is in line with the important role of functional BAF chromatin remodelling and the targeted opening of chromatin during neural development [254]. Murine embryonic stem cell models have indicated that Smarcb1 plays a crucial role in the development of the nervous system [327, 326, reviewed by 44]. An inducible SMARCB1 loss-of-function system in human induced pluripotent stem cells (iPSCs) has shown that SMARCB1 loss during neuronal differentiation leads to a failure in maturation causing resistance to terminal differentiation [319]. In another tumour model using human SMARCB1-deficient pluripotent stem cell-derived neural progenitor-like cells (NPLCs), brain tumours could be induced after the NPLCs were transplanted into the mouse brain [320]. Activation of an embryonic stem cell (ESC)-like signature was associated with rhabdoid histology in these SMARCB1-deficient NPLC-derived tumours. In accord with this, primary human AT/RT samples also exhibit an ESC-like gene expression DNA-methylation signature [320]. Thus, the AT/RT genome exhibits hypermethylated patterns resembling that of pluripotent stem cells. These stem-like DNA hypermethylation patterns affect the regulatory regions of multiple genes involved in neural differentiation. Hence, SMARCB1 loss impairs the removal of DNA methylation and blocks the regular progression of lineage commitment [319–321]. These findings were substantiated by further studies indicating that partial or complete SMARCB1 loss-of-function in human embryonic stem cell lines impairs neuronal differentiation [228, 229]. The blockage of neural differentiation driven by SMARCB1 loss-associated epigenetic dysregulation is essential for AT/RT tumorigenesis [328]. However, it is unclear if the mechanisms regulating differentiation blocks differ between AT/RT subgroups, as suggested by the differential subgroup-specific vulnerabilities to inhibitors and therapeutic drugs [26, 329–332].
Molecular subgroups of extracranial MRTs and cell-of-origin
RNA-sequencing indicated that two distinct subgroups of extracranial MRTs exist that are distinguishable by virtue of nearly 1000 differentially expressed genes [23]. In subgroup 1, the most significantly over-expressed genes were those encoding immunoglobulins and genes associated with BMP-signalling as well as differentiation. In subgroup 2, significantly overexpressed genes were linked to cell adhesion and migration, WNT signalling and cellular differentiation. In both subgroups, a significant proportion of overexpressed genes was linked to neural crest development and neural differentiation suggesting that the cells-of-origin of MRTs derive from the neural crest lineage [23]. MicroRNA profiles of the extracranial MRTs indicated pronounced similarities to those of pheochromocytomas and paragangliomas, which are also neural crest-derived tumours [23]. Furthermore, DNA methylation analysis of extracranial MRTs indicated two subgroups with distinct methylation profiles that correlated with age at diagnosis. Subgroup A exhibited higher overall promoter methylation at CpG-islands compared to the other subgroup B [23]. Correlating methylation subgroups to clinical patient data revealed an overrepresentation of patients older than one year in subgroup A [23]. Interestingly, the promoters of homeobox genes and tumour suppressor genes were disproportionately represented among those that acquired methylation in subgroup A [23].
Comparing the different subgroups of AT/RTs with extracranial MRTs revealed distinct similarities between MRTs and ATRT-MYCs including global DNA hypomethylation and overexpression of HOX genes and genes involved in mesenchymal development, distinguishing them from other AT/RT subgroups [304] (Sect. AT/RT subgroups).
In order to determine the origin of pediatric extracranial MRTs, Custers et al. [315] reconstructed the developmental relationship between MRT cells and normal tissues from the distribution of somatic mutations. These analyses indicated that pediatric extracranial MRT cells are phylogenetically related to the neural crest lineage, in particular to neural crest-derived Schwann cells, and that MRTs originate during fetal life [315]. Re-expression of SMARCB1 in patient-derived MRT organoids consistently resulted in more differentiated cell types and promoted neural to mesenchymal conversion [315]. Thus, differentiation of neural crest cells and exit from pluripotency appear to be strongly dependent upon SMARCB1 activity [reviewed by 180].
Mechanisms underlying SMARCB1 activity as a tumour suppressor
Transcriptomic and epigenomic analyses of AT/RT samples have characterized the epigenetic alterations that take place following SMARCB1 loss [24, 26, 284, 304]. The multitude of these changes implies a wide variety of mechanisms by which SMARCB1 loss initiates cellular transformation and malignancy. A few of them have been identified as summarized in the following.
BAF and Polycomb complex antagonism
One of the mechanisms by which SMARCB1 loss in MRT precursor cells leads to tumorigenesis is the disturbed balance between the BAF complex and the Polycomb repressive complex 2 (PRC2). This balance is however necessary to maintain chromatin topology [261, 284]. Indeed, SMARCB1-deficient MRT cells are unable to remove repressive PRC2-mediated histone modifications such as H3K27Me3 from tumour suppressor genes, for example CDKN2A (MIM #600160) [261]. The CDKN2A gene encodes p16 which is involved in the suppression of proliferation and acts as a cyclin-dependent kinase inhibitor that binds to CDK4/6 thereby impairing the activation of the CDK4/6-cyclin D1 complex [231, 333]. The active CDK4/6-cyclin D1 complex normally phosphorylates the retinoblastoma protein, which releases the transcription factor E2F1 to promote gene expression associated with S phase progression [334]. SMARCB1-deficient MRT cells exhibit considerably reduced p16 expression causing increased cellular proliferation due to unchecked S-phase progression [335, 336 reviewed by 20]. Upon re-expression of SMARCB1 in MRT cells, p16 expression increases due to restored BAF chromatin remodelling activity at regulatory genomic regions of p16 [233, 336]. Thus, SMARCB1 loss in MRT cells causes imbalance of the activity of BAF and PRC2 complexes, leading to an increase in repressive epigenetic marks by PRC2 [253, 261]. It is likely that, in addition to CDKN2A, other tumour suppressor genes are also repressed in SMARCB1-deficient cells in a similar manner. For example, the glioma pathogenesis-related protein 1 gene (GLIPR1; MIM #602692), which also acts as a tumour suppressor, shows strong promoter hypermethylation and is downregulated in AT/RT [24].
The balance between PRC2 and BAF activity is essential for tumour suppression by SMARCB1 [reviewed by 291, 337-339]. The Enhancer of zeste homolog 2 (EZH2; MIM #601573) encodes a histone-lysine N-methyltransferase and is an important component of PRC2. The inactivation of Ezh2 in a conditional mouse model completely blocked tumour formation caused by Smarcb1 inactivation [261]. In MRT cells, SMARCB1 loss leads to the increased expression and recruitment of EZH2 to PRC2 target genes which are H3K27-trimethylated and consequently in a repressed state [261, 284, 340–342]. In addition to EZH2, other genes encoding protein components of the PRC2 complex are overexpressed in AT/RT [24].
Remarkably, the active BAF complex is able to promote gene expression within a few minutes by removing PRC2 and its repressive H3K27me3 mark from promoters and enhancers [22]. Thus, the activity of the BAF complex opposes PRC2 on a minute-by-minute basis without any need for replication, polymerase occupancy or transcription in order to provide rapid epigenetic plasticity [reviewed by 20].
SMARCB1 inhibits the activation of MYC target genes
The SMARCB1 protein binds via its RPT1 domain to the C-terminus of the MYC protein, a master regulator of genome-wide transcription that potentiates oncogenic transformation when overexpressed [276, 343, 344]. The tumour-suppressor functions of SMARCB1 are mediated in part by inhibition of MYC binding to its target genes [320, reviewed by 345]. The analysis of MRT-derived organoids indicated that SMARCB1 loss during neural crest development prevents the inactivation of certain MYC enhancers, which is essential for proper lineage specification [346]. It has been suggested that SMARCB1 loss in MRTs leads to increased looping of these enhancers to the MYC promoter, thereby potentially activating its transcription [346]. Upon SMARCB1 reintroduction into MRT cells, MYC is displaced from chromatin genome-wide [347]. This activity of SMARCB1 is independent of its effects on chromatin remodelling within the BAF complex. Instead, SMARCB1 induces RNA polymerase pausing at genes regulated by MYC [348]. A key transcriptional function of MYC is to modulate release of paused RNA polymerases at MYC target genes and this activity is impaired by SMARCB1. Independent of any changes in MYC protein expression, the loss of SMARCB1 activates MYC at a functional level, leading to the activation of MYC target genes in MRT [348]. Thus, SMARCB1 antagonizes MYC and SMARCB1 loss drives malignancy via MYC overexpression.
SMARCB1 loss leads to WNT/beta-catenin hyperactivation
Smarcb1 deficiency in the developing limb mesenchyme of conditional knock-out mice is responsible for the aberrant activation of the canonical Wingless-related integration site (WNT)-signalling pathway and leads to defects consistent with WNT/beta-catenin overexpression [349]. Re-expression of SMARCB1 in SMARCB1-deficient MRT cell lines results in the down-regulation of beta-catenin target genes [349]. Transcriptome analysis of WNT pathway genes in AT/RT primary tissues and AT/RT cell lines indicated that the WNT family member 5B gene (WNT5B; MIM #606361) is significantly upregulated as compared with non-tumour brain samples [350]. The WNT5B protein binds to the protein product of the Frizzled class receptor 1 gene (FZD1; MIM #603408) and regulates the differential expression of downstream pathway genes [350]. WNT inhibitors decrease the proliferation of AT/RT cells suggesting that they might have future therapeutic potential [350].
In human embryonic stem cells (hPSCs), SMARCB1 loss-of-function leads to disturbed actin cytoskeleton organization, cell-cell interaction and cell-extracellular matrix (ECM) interaction associated with a significant reduction in beta-catenin levels. Thus, SMARCB1 is important for the regulation of cell-cell and cell-ECM interactions in hPSCs, at least in part mediated by the WNT signalling pathway [229].
SMARCB1 loss activates the hedgehog-GLI1 pathway
In a specific subgroup of AT/RTs, the SHH signal pathway is activated and hence SMARCB1 loss is associated with aberrant activation of this pathway [24, 351]. Hedgehog (HH) signalling has critical functions in cell proliferation and differentiation during development [reviewed by 307]. In mammals, there are three hedgehog genes: sonic hedgehog (SHH; MIM #600725), Indian hedgehog (IHH; MIM #600726) and desert hedgehog (DHH; MIM #605423). They are expressed in different tissues and at different stages of development, suggestive of different biological activities [reviewed by 352]. However, SHH is the most potent of these ligands and is widely expressed in adult tissues [reviewed by 307]. SHH signalling is crucial during embryonic development and for the maintenance of tissue polarity. Aberrant SHH signalling has been implicated in tumorigenesis for many different cancer types [reviewed by307, 353]. Hedgehog signal transduction is initiated by the binding of the HH ligands to the Patched-1 receptor encoded by the PTCH1 gene (MIM #601309). The glioma-associated oncogene family zinc finger-1 (GLI1) is an important downstream effector in this signalling cascade. Importantly, SMARCB1 protein deficiency leads to aberrant activation of the HH-GLI1 pathway [351]. By using affinity purification–mass spectrometry and chromatin immunoprecipitation, SMARCB1 was found to localize upstream to the transcriptional start sites of the GLI1 and PTCH1 genes indicating very specific interactions [351]. Furthermore, small-hairpin-RNA-mediated knockdown of Smarcb1 in mouse TM3 cells causes the upregulation of Gli and Ptch1 expression, which leads to the activation of hedgehog signalling [351]. In accordance with this, re-expression of SMARCB1 in MRT cells represses GLI1 expression [351]. Therefore, the SMARCB1 protein acts as an important regulator of GLI1 gene expression. In the subgroup of AT/RT with the molecular signature termed AT/RT-SHH, an enrichment of gene expression associated with SHH signalling pathway activation has been observed [24, 26]. The reason why SMARCB1 loss does not activate the SHH pathway to the same extent in the other AT/RT subgroups remains unclear but is most likely related to different cellular origins for these subgroups [281] (Sect. Cell-of-origin of AT/RT subgroups).
UPR activation and ER stress in Smarcb1-deficient cells
Several studies have suggested that BAF complexes are involved in the rewiring of cancer metabolism [reviewed by 354]. Epigenetic abnormalities deregulate metabolic enzymes or signalling pathways that are supportive of the survival and rapid proliferation of cancer cells. However, significant upregulation of protein anabolism can render cells susceptible to disruption of their proteostatic machinery. Importantly, SMARCB1-deficient MRT cells are highly sensitive to disturbances of protein homeostasis (proteostasis) as shown by treatment with proteasome inhibitors [355, reviewed by 330]. In order to investigate this in greater detail, Carugo et al. [356] generated embryonic murine mosaic models of liver MRT by introducing a tissue-specific Cre recombinase expressed from the murine albumin promoter via trans-uterine adenoviral injection in Smarcb1LoxP/LoxP embryos at embryonic day E12.5. This was necessary in order to avoid early embryonic and perinatal lethality of classical or other conditional Smarcb1 knockout mice which has impaired the analysis of the role of Smarcb1 loss during tissue specification and mouse organogenesis [178, 357, 358]. Genetic mosaicism in their embryonic murine MRT model enabled Carugo et al. [356] to study the malignant properties of Smarcb1-deficient cells by bypassing the early lethality and allowing the tissue-specific time-restricted activation of a reporter gene and the quantification of tumour burden [356]. Specifically, the in utero mosaic Cre-mediated loss of Smarcb1 targeted to E12.5 epithelial liver progenitor cells resulted in liver hyperplasia with severe dysplastic, degenerative changes and disruption of normal liver architecture [356]. The livers also had tumours that exhibited histopathological features of MRT with high proliferation activity. Transcriptome and protein expression analysis of these liver tumour cells indicated massive activation of the unfolded protein response (UPR) [356]. UPR becomes activated in response to the accumulation of unfolded or misfolded proteins in the lumen of the endoplasmic reticulum in order to restore normal cellular functions by preventing protein translation and the degradation of misfolded proteins. Upon UPR activation, signalling pathways are upregulated that lead to an increase of chaperones involved in protein folding. The activation of UPR is likely to be a by-product of the MYC-induced hypermetabolic state in the Smarcb1-deficient tumour cells [356]. In addition to massive UPR activation, Smarcb1 loss also induced a robust ER stress response and autophagy [356]. Transcriptome analysis of Cre-induced Smarcb1-null liver tumours indicated significant upregulation of p53 pathways involved in the regulation of protein metabolism, ER stress adaptation and autophagy compared to controls. Based upon their findings, Carugo et al. [356] concluded that Smarcb1 loss leads to the activation of different oncogenic pathways and induces UPR stress. Under these circumstances, p53 activation regulates cellular homeostasis, cell-cycle progression, cell survival, protein biosynthesis and removal by autophagy. Of note, rhabdoid tumours do not exhibit pathogenic p53 alterations [298]. Thus, p53 has a context-specific, pro-survival role in Smarcb1-deficient tumour cells and is an important regulator of proteostasis [356]. In accordance with this, interference with the cellular proteostatic machinery by drugs is highly lethal in the Smarcb1-deficient liver tumours and may represent a promising therapeutic target for MRTs [329, 356, reviewed by 330]. It should be noted that the survival of SMARCB1-deficient MRT cell lines is dependent upon DDB1-CUL4 Associated Factor 5 (DCAF5), a substrate receptor of the CUL4-DDB1 E3 ubiquitin-protein ligase complex, which targets specific proteins for ubiquitylation and degradation [reviewed by 359]. In the absence of SMARCB1, DCAF5 mediates the degradation of residual BAF complexes which is essential for the proliferation of SMARCB1-deficient MRT cells [359].
Mutational profile in patients with SMARCB1-related SWN and RTPS1
Hypomorphic or semi-functional SMARCB1 PVs in patients with SWN
The strikingly different tumour phenotypes in patients with SMARCB1-related SWN as compared to patients with RTPS1 indicate that the types and consequences of germline pathogenic SMARCB1 variants must be different between these patient groups and will affect different tumour precursor cells. Additionally, the timing of the tumour-specific inactivation of the SMARCB1 wild-type allele and the loss of additional tumour suppressor genes are responsible for the differences in tumorigenesis in SWN compared with RTPS1.
The pathogenic SMARCB1 variants identified in patients with SWN are predominantly non-truncating mutations, including missense variants, in-frame deletions or splice‐site variants, with a tendency to accumulate in the 5′‐region or 3′‐end of the gene [28, 33–43]. By contrast, pathogenic SMARCB1 variants in MRTs are located in all parts of the gene or delete all, or at least large parts of, the coding sequence [7, 10, 13, 40, 41, 297, 360, 361]. In most MRTs, the complete loss of nuclear SMARCB1 protein expression is a diagnostic marker [reviewed by 17]. According to the data available, there is a strong genotype/phenotype correlation in the sense that complete loss-of-function SMARCB1 PVs are characteristic of MRT [24, 40, 41]. In contrast to this, most PVs in patients with SMARCB1-related SWN are likely to be semi-functional or may not affect all isoforms of SMARCB1 leading to reduced SMARCB1 protein expression levels or only partial loss of SMARCB1 protein function [33–40]. The most common pathogenic SMARCB1 variant identified so far in patients with SWN (c.*82C > T) is located within the 3’UTR [34, 38, 40, 42, 43, 362]. This 3’UTR variant leads to reduced SMARCB1 expression levels due to lower mRNA stability [43, 363].
Even though most germline pathogenic SMARCB1 variants causing SWN are non-truncating, germline SMARCB1 PVs that generate a premature termination codon (PTC) have also been identified in patients with SWN. The majority of these are located in SMARCB1 exon 1. PTCs located in SMARCB1 exon 1 (c.30delC and c.34 C > T) of patients with classical SWN lead to transcripts that are not completely degraded but instead result in N-terminally truncated SMARCB1 proteins by translational reinitiation at a downstream AUG codon [364]. Furthermore, immunohistochemical analysis has indicated that N-terminally truncated SMARCB1 proteins are expressed in schwannomas of the respective patients harbouring exon 1 PVs [364]. The retention of partial activity of N-terminally truncated SMARCB1 has also been detected in a comprehensive deep mutational scanning study of SMARCB1, encompassing 8,418 amino acid substitutions, performed in order to assess their functional impact [273]. The residual activity of N-terminal nonsense PVs is due to alternative methionine start sites at residues 1–4, 27 and 38 in the N-terminus of SMARCB1, which enable downstream read-through [273]. This is in accordance with the reduced efficiency of nonsense-mediated decay (NMD) observed within 200 nucleotides of translational start codons [365].
This retention of partial function by N-terminal truncations mediated by the use of alternative methionine start sites may also explain the mosaic SMARCB1 protein expression pattern in schwannomas harbouring these variants. It is likely that these truncated SMARCB1 proteins are not fully stable thereby resulting in a mosaic SMARCB1 staining pattern. This has been observed and studied in detail in schwannomas of patients with the c.30delC and c.34 C > T pathogenic SMARCB1 variants. Remarkably, pathogenic variants causing PTCs in SMARCB1 exon 1 have not been reported in MRTs. These findings are in line with the concept that in contrast to the complete absence of SMARCB1 expression in MRT, altered SMARCB1 proteins with modified activity and reduced expression are responsible for a mosaic SMARCB1 expression pattern in the tumours of patients with schwannomatosis [364]. Indeed, several studies have observed that schwannomas in patients with germline SMARCB1 PVs exhibit a mosaic SMARCB1 protein expression pattern as determined by immunohistochemistry [28, 363, 366, 367]. This mosaic pattern results from mixed immuno-positive and -negative nuclei, consistent with the expression of the SMARCB1 protein in a subset of tumour cells. This mosaic SMARCB1 expression is most likely related to the hypomorphic nature of the pathogenic variants in SWN patients that encode stable mRNA transcripts giving rise to detectable amounts of SMARCB1 protein. Since the wild-type SMARCB1 allele is often lost in schwannomas of patients with schwannomatosis, the SMARCB1 protein detected in schwannoma cells must be encoded by the mutant allele. Our inability to detect mutant proteins in all tumour cells by immunostaining is most likely a consequence of the instability of mutant SMARCB1 proteins [364]. This instability results in immunologically non-reactive SMARCB1 protein degradation products in a proportion of the schwannoma cells. Since this degradation is probably a random process, some cells may still express detectable amounts of SMARCB1 protein resulting in a mosaic expression pattern when analysing schwannoma tissue sections. [144, 364, reviewed by 368].
SMARCB1 missense variants in MRT
In contrast to schwannomas, complete loss of SMARCB1 protein expression as determined by immunohistochemistry is frequently observed in MRTs. In many instances, this is due to truncating PVs or loss of the complete SMARCB1 gene leading to biallelic SMARCB1 inactivation. However, missense PVs in specific SMARCB1 domains have the potential to be similarly destructive [369]. This has been shown by deep mutational scanning of SMARCB1 performed in order to assess the functional impact of 8,418 amino acid substitutions [273]. After prioritization, thirteen SMARCB1 amino acid residues intolerant to missense PVs were identified by expression of constructs containing these variants in SMARCB1-deficient tumour cell lines. Six of these missense PVs were located within the WHD domain (positions: R52, A55, I63, K77, L90 and L91), three within the IDR (positions: E122, Q123 and A125) and four within the RPT2 domain (positions: D277, W281, E300 and I315) [273]. Not unexpectedly, these missense PVs have not been observed in those patients with SWN that have been analysed to date. Of particular interest were the loss-of-function intolerant residues that are located in RPT2, which appear to facilitate important intramolecular interactions of SMARCB1 [273]. Remarkably, expression constructs containing the SMARCB1 W281P and I315R missense variants exhibited functional properties similar to constructs with nonsense variants at the same positions as determined by reduced proliferation of MRT cell lines transfected with the respective expression constructs. In contrast to constructs with SMARCB1 nonsense variants, the expression of the constructs with either the W281P or the I315R missense variants was readily detectable at both the RNA and protein level. Thus, these missense variants would appear to perturb the ability of SMARCB1 to enable BAF complex assembly and chromatin remodelling, without necessarily leading to complete protein degradation. Molecular dynamic modelling revealed that these missense mutants disrupt the flexibility of the N-terminal winged-helix domain of SMARCB1, suggesting a novel mechanism by which the SMARCB1 tumour suppressor function is disrupted. Indeed, these missense variants caused altered chromatin remodelling patterns, due to significant reduction in BAF complex activity, as well as changes in gene expression profiles in line with severely disturbed SMARCB1 protein function [273]. Thus, certain SMARCB1 PVs result in loss-of-function even if they do not lead to complete loss of mutant SMARCB1 protein expression.
SMARCB1 functional loss in MRT due to nuclear export
In a subgroup of cranial MRT, the AT/RT-TYR subgroup, PVs leading to truncation or mutation of the C-terminal part of SMARCB1 are quite common (44%) [279]. Most of these PVs cause C-terminally truncated SMARCB1 proteins that are localized in the cytoplasm [279]. By contrast, wild-type SMARCB1 is a nuclear protein and loss of nuclear SMARCB1 staining is very frequent in AT/RT. Importantly, the SMARCB1 protein harbours a nuclear export signal (NES) within the RPT2 region [278] (Fig. 3). Remarkably, C-terminal truncation of SMARCB1 leads to the unmasking of the nuclear export sequence causing the cytoplasmic localization of SMARCB1 associated with the loss of tumour suppressor function [278, 279]. It has been estimated that 19% of all AT/RT exhibit cytoplasmic localization of SMARCB1 [279]. Whether nuclear export of mutant SMARCB1 also contributes to the development of schwannomas or other tumours in patients with SMARCB1-related SWN is currently unknown.
Non-coding SMARCB1 PVs in patients with SWN
Pathogenic variants (PVs) in LZTR1 or SMARCB1 are detected in approximately 86% of familial and ∼40% of sporadic schwannomatosis cases utilizing standard clinical mutation analysis including exons and intronic segments at exon boundaries (typically ± 20 nucleotides) [33–36, 38, 40, 42, 65, 67, 68, 362, 363]. It has been argued that PVs in deep intronic or regulatory regions of both genes, regions that are not evaluated by exon-based sequencing strategies, might have been missed in the aforementioned studies and that this has inevitably resulted in a reduced pathogenic variant detection rate. In order to investigate whether PVs in deep intronic or regulatory regions of the NF2, SMARCB1 and LZTR1 genes contribute to the pathogenesis in patients with schwannomatosis, Piotrowski et al. [43] investigated 33 SWN patients without germline first-hit PVs in NF2, SMARCB1 and LZTR1 as determined by initial clinical exon sequencing. The analysis of the entire genomic region of these genes indicated deep intronic but clearly pathogenic SMARCB1 variants in two of the 33 SWN patients (Supplementary Table 3). These authors identified five further intronic likely pathogenic variants in the three genes, one of them also in SMARCB1. Smith et al. [370] also identified a deep intronic SMARCB1 PV (Supplementary Table 3) whilst Tauziède-Espariat et al. [371] identified a SMARCB1 deep intronic pathogenic variant in intron 1 present in the germline of two unrelated young children with AT/RT and RTPS1. These findings indicate that deep intronic SMARCB1 PVs can be disease-causing and that these regions should therefore be included in molecular diagnostic panels.
In addition to intronic PVs, those located in the 3’UTR are also an important cause of SMARCB1-related SWN indicating that this region should be included in the comprehensive screening of SWN patients in order to increase the pathogenic variant detection rate [34, 38, 40, 42, 43, 81, 362, 372].
Somatic mosaicism in SMARCB1-related SWN and RTPS1
Remarkably, the SMARCB1 germline PV detection rate is much higher in familial SWN patients than in patients with sporadic SMARCB1-related SWN [32, 40]. Indeed, germline SMARCB1 PVs account for up to 48% of familial SWN cases and 9.5% of sporadic SWN [33–36, 38, 40, 65, 67, 68, 363]. One possible explanation for this could be a high proportion of somatic mosaicism for SMARCB1 PVs in sporadic patients, with variant allele frequencies being too low to be detected in blood. By contrast, the SMARCB1 PV should be readily detectable in independent tumours of the patient harbouring somatic mosaicism for a SMARCB1 PV. In order to address this, Smith et al. [81] analysed two independent schwannomas from 53 SWN patients who did not fulfil the diagnostic criteria for NF2-related SWN. Remarkably, 43% of patients with no identified germline NF2 PVs in blood had low-level mosaicism for a pathogenic NF2 gene variant and hence had mosaic NF2-related SWN. However, somatic or mosaic SMARCB1 or LZTR1 PVs were not identified in the remaining patients without germline PVs in one of their SWN genes. These findings suggest that somatic mosaicism for SMARCB1 (and LZTR1) is uncommon in SWN patients.
By contrast, somatic SMARCB1 PVs causing mosaicism are more frequent than previously assumed in children with rhabdoid tumours [183]. It is estimated that 6–21% of these children exhibit somatic mosaicism for a SMARCB1 PV as determined by sequence analysis with high sensitivity to detect low-level mosaicism [373, 374]. Gonadal mosaicism of the parents of patients with rhabdoid tumours is also not uncommon and has been observed in several studies [5, 6, 12, 13, 53, 375]. A case of maternal germ line mosaicism has also been reported in SMARCB1-related SWN [376].
Mouse models of Smarcb1 loss leading to either schwannomas or rhabdoid tumours
The time window of SMARCB1 inactivation in rhabdoid tumour development
Both the type of pathogenic SMARCB1 variant and whether it leads to the complete loss or only the partial loss-of-function determines which type of tumour develops, either rhabdoid tumour (RT) or schwannoma. The time window of complete SMARCB1 inactivation during development is also important in this context. This has been ascertained through the analysis of conditional Smarcb1 knockout-mouse models [178, 179]. Homozygous germline inactivation of Smarcb1 in mice is embryonic lethal; nullizygous animals (Smarcb1−/−) are not born after heterozygous inter-crosses. Smarcb1−/− embryos develop to the blastocyst stage but die shortly after implantation before E6.5 [357, 377, 378]. Heterozygous Smarcb1+/− mice are born and appear to be normal but, starting at the age of a few weeks, they develop extracranial sarcomas resembling human RTs mainly in soft tissues derived from the first branchial arch with a long latency and weak penetrance [357]. Only tissue- and developmental stage-specific conditional Smarcb1 knockout mouse models using different promoters to induce Cre-mediated Smarcb1 deletion succeeded in generating a murine model for AT/RT [178, 179, reviewed by 180]. By means of temporal control of tamoxifen injection in Smarcb1flox/flox;Rosa26-CreERT2 mice, the phenotypes associated with Smarcb1 inactivation at different developmental stages could be investigated [178]. Injection before E6, at birth or when the mice were two months of age, caused lethality, hepatic toxicity or development of T-cell lymphomas [178]. However, tamoxifen injection and thus biallelic Smarcb1 loss between E6 and E10 resulted in viable mice which developed mainly intracranial tumours with high penetrance and rapid onset. These tumours exhibited anatomical, morphological and gene expression profiles comparable to those of human AT/RTs [24, 178]. By using the protein zero (P0)-promoter to activate the Cre recombinase and thus Smarcb1 loss at E9.5 in neural crest cells, Vitte et al. [179] also succeeded in generating a conditional knockout mouse model of cranial rhabdoid tumours that resembles human AT/RTs. These P0-CreC; Smarcb1flox/flox mice were viable and developed RTs in cranial nerves and meninges. The tumours displayed typical histological and immunohistochemical features of human RTs [179]. Expression profiling indicated that tumours of the P0-CreC;Smarcb1flox/flox mice recapitulate the molecular diversity of human AT/RTs [179]. Importantly, both human AT/RTs and the rhabdoid tumours from the P0-CreC; Smarcb1flox/flox mice exhibit specific gene expression markers of early neural crest formation indicative of an early neural crest cell as the cell-of-origin [179].
Taken together, the development of AT/RT is time-dependent in the sense that a specific developmental time window exists during which the tumour progenitor cell is vulnerable to complete Smarcb1 loss initiating rhabdoid tumour growth. It is highly likely that during human embryonic development, SMARCB1 loss in neural crest cells or neural progenitor cells during a specific window of time associated with the disturbance of neural differentiation is a critical step in the tumorigenesis of AT/RTs [178, 179, reviewed by 180] (see Sect. Blocked neural differentiation in AT/RTs). The existence of an early narrow spatio-temporal window during which complete Smarcb1 loss results in malignant transformation may explain why some SMARCB1 mutation carriers in RTPS1 families do not develop rhabdoid tumours (Sect. Co-occurrence of RTPS1 and schwannomatosis in families). If a sensitive time period during the early development of neural crest cells is completed without biallelic complete SMARCB1 inactivation, AT/RT tumorigenesis is not initiated. In other words, the absence of AT/RT in carriers of loss-of function germline SMARCB1 PV is explicable in terms of the retention of the SMARCB1 wild-type allele and its activity in neural crest cells or neural progenitor cells during a vulnerable early stage of embryonic development.
Mouse model of SMARCB1-related schwannomas
In schwannomas of patients with schwannomatosis, somatic biallelic NF2 gene inactivation is very frequent [34, 35, 65]. Indeed, loss-of-function of NF2 appears to be important for schwannoma growth. This conclusion is in accord with the finding that conditional Smarcb1 knockout mice do not develop schwannomas, indicating that biallelic Smarcb1 loss is not on its own sufficient for the growth of schwannomas [178, 179]. In order to investigate the co-involvement of Smarcb1 and Nf2 in the pathogenesis underlying schwannomas, Vitte et al. [179] generated conditional knockout mouse models of Smarcb1 and concomitant Nf2 gene loss. First, they created P0-CreC; Smarcb1flox/flox;Nf2flox/flox mice which were not viable, indicating that the loss of both Smarcb1 and Nf2 during early development (E9.5) is lethal. However, by using a different CRE-promoter (mGFAP) which is expressed later in embryonic development than the P0-promoter, mice with the combination of Smarcb1 and Nf2 inactivation became viable. In these mGFAP-Cre; Smarcb1flox/flox;Nf2flox/flox mice, Cre is expressed in the dorsal root ganglia starting at E13.5. The mGFAP-Cre; Smarcb1flox/flox;Nf2flox/flox mice developed tumorlets consisting of Schwann cells in the dorsal root ganglia, reminiscent of the schwannoma tumorlets found in NF2 patients [179]. These tumorlets are considered to be small schwannomas exhibiting biallelic NF2 inactivation and occur mainly in the spinal nerve roots [379]. Importantly, schwannoma tumorlets were not found in the dorsal root ganglia of mGFAP-Cre; Smarcb1flox/flox;Nf2flox/+ mice, indicating that biallelic NF2 loss is essential for schwannoma formation [179]. Similar to the situation in schwannomatosis patients, the loss of both Smarcb1 and Nf2 did not increase the malignancy of the tumours in mGFAP-Cre; Smarcb1flox/flox;Nf2flox/flox mice as compared to tumours in mGFAP-Cre; Nf2 flox/flox mice with biallelic Nf2 loss but retention of Smarcb1 activity.
Taken together, the conditional knockout mouse models established by Vitte et al. [179] indicated that Smarcb1 loss at a later developmental stage (starting at E13.5) in the Schwann cell lineage, in addition to biallelic Nf2 gene inactivation, results in schwannomas in mGFAP-Cre; Smarcb1flox/flox;Nf2flox/flox mice. This mouse model of schwannoma development impressively reproduces the genetic profile of schwannomatosis-associated schwannomas with concomitant loss of both Smarcb1 and Nf2 [179]. Smarcb1 loss and biallelic Nf2 inactivation at later stages of Schwann cell development (starting at E13.5) lead to benign schwannomas but not rhabdoid tumours. By contrast, conditional biallelic Smarcb1 knockout during a narrow time window of early neural crest cell development (E6-E10) results in rhabdoid tumour growth [178, 179]. Thus, Schwann cell differentiation suppresses Smarcb1-driven malignant tumorigenesis in this mouse model.
Co-involvement of several tumour suppressor genes in schwannoma development
The molecular mechanism underlying the tumorigenesis of schwannomas in patients with SMARCB1- (and LZTR1-) related SWN is clearly not in agreement with the classic Knudson two‐hit model hypothesis involving the biallelic inactivation of a single tumour suppressor gene. Instead, tumorigenesis of schwannomas in patients with SMARCB1‐ (and LZTR1-) related SWN appears to follow a four‐hit/three‐step model that includes somatic biallelic inactivation of the NF2 gene [35, 368]. In the following, this model will be explained with a focus on SMARCB1-related SWN although it may also be applied in an analogous manner for LZTR1-related SWN. The basis of the four-hit/three-step model is that all three tumour suppressor genes known to be relevant to schwannoma development, namely NF2, LZTR1 and SMARCB1, are located on chromosome 22q (Fig. 2). Furthermore, chromosome 22q loss-of-heterozygosity (LOH) is a frequent somatic event in schwannomas [33–35, 42, 65, 81, 380]. In patients with SMARCB1-related SWN, the germline SMARCB1 PV is considered to be the first hit (first step). The second and third hit (second step) then involves loss-of-heterozygosity (LOH) of 22q associated with the loss of the wild-type SMARCB1 allele and one of the two NF2 alleles (Fig. 4A). The tumour-specific 22q LOH is caused by deletions of different sizes on the long arm of chromosome 22 (22q) [28, 33–35, 42, 65, 380]. Importantly, the 22q LOH affects the chromosome harbouring the wild-type SMARCB1-allele and hence occurs in trans to the germline SMARCB1 PV which is retained in the schwannoma. Finally, the fourth hit (third step) involves a tumour-specific pathogenic variant of the NF2 gene located in cis to the SMARCB1 germline PV that leads to biallelic NF2 inactivation driving schwannoma development (Fig. 4A). Schwannomas in patients with schwannomatosis frequently exhibit tumour-specific intragenic NF2 PVs that are different in anatomically distinct schwannomas of a given patient, indicative of their somatic origin [33–35, 62, 381]. The chronological order of the tumour-specific alterations, namely 22q LOH and somatic NF2 PVs, is probably not fixed and may be interchangeable. Moreover, these different genetic events may influence each other in the sense that biallelic loss of SMARCB1 and a dosage loss (haploinsufficiency) of half of the NF2 gene due to 22q LOH may cause Schwann cell proliferation that then accelerates mutagenesis giving rise to the somatic intragenic NF2 PV [380]. In any case, the key point to be taken from the four-hit/three-step model is that biallelic SMARCB1 loss is insufficient for schwannoma growth in patients with SMARCB1-related SWN. Additional inactivation of the NF2 gene is also necessary for schwannoma development. The co-involvement of SMARCB1 and NF2 in the pathogenesis underlying schwannomas in patients with SMARCB1-related SWN is substantiated by mouse models of schwannoma development [179] (Sect. Mouse model of SMARCB1-related schwannomas). The fact that NF2 loss is important for the development of schwannomas is also reflected in the observation that at least 50–75% of sporadic schwannomas exhibit somatic pathogenic NF2 variants [380, 382, 383].
Fig. 4.
A Four-hit/three-step model of tumorigenesis in patients with a germline SMARCB1 pathogenic variant (PV) (first hit and step). The second step involves loss of heterozygosity (LOH) of 22q which serves to remove the wild-type SMARCB1 allele and one of the two NF2 alleles located in trans to the germline SMARCB1 PV. The third step is the somatic mutation of the other NF2 allele located on the chromosome harbouring the germline SMARCB1 mutation. B If mitotic recombination were to represent the second step, this would lead to a reduplication of the chromosomal region with the germline SMARCB1 PV allele. However, the NF2 gene would not be deleted by this event. Instead, two wild-type NF2 alleles would still be present. The biallelic inactivation of NF2 would require two independent NF2 PVs (third and fourth mutational steps). Thus, mitotic recombination is not compatible with the four-hit/three-step model of tumorigenesis. Instead, a four-hit/four-step model of tumorigenesis would have to be postulated. m: mutant allele; WT: wild-type allele
In patients with SMARCB1-related SWN, the four-hit/three‐step model appears to underlie the vast majority of schwannomas (Fig. 4A). In the study of Piotrowski et al. [65], all 17 schwannomas from 9 patients with SMARCB1-related SWN exhibited somatic chromosome 22q LOH leading to SMARCB1 and NF2 loss. Further, all 17 schwannomas also exhibited somatic intragenic NF2 PVs. It is likely that the four-hit/three-step model of tumorigenesis also accounts for other tumours in patients with SMARCB1-related SWN including meningioma and leiomyoma [55, 138].
Several studies have indicated that the proportion of schwannomas in patients with SMARCB1-related SWN exhibiting chromosome 22q LOH (chr.22q-LOH) is very high [34, 35, 65]. Of note, mitotic recombination has been excluded as the mechanism causing 22q LOH in schwannomas of patients with germline SMARCB1 PVs [380]. Mitotic recombination as a causative mechanism would in any case not be compatible with the four-hit/three-step model of tumorigenesis because a reduplication of the chromosomal region with the germline SMARCB1 PV allele by mitotic recombination would require two independent NF2 PVs in both alleles for complete inactivation of NF2 (Fig. 4B). Consequently, a four-hit/four-step model of tumorigenesis would have to be postulated which is possible but less likely than the four-hit/three-step model (Fig. 4B). More likely, as suggested by Hadfield et al. [380], a three-step model of schwannoma development, comprising the biallelic loss of SMARCB1 and loss of one NF2 allele mediated by 22q deletion, would serve to accelerate Schwann cell proliferation thereby driving the somatic intragenic NF2 PV that represents the third step.
Instead of mitotic recombination, the frequent chromosome 22q LOH in schwannomas is most likely due to differentially sized deletions of parts of 22q. The mechanism underlying these deletions is unknown. It is however unlikely that non-allelic homologous recombination (NAHR), mediated by the multiple duplicated sequences on chromosome 22, the low copy repeats 22 (LCR22), is responsible for the deletions because the deletion breakpoints do not coincide with the positions of the LCR22 repeats which are located within a 6.5-Mb region on 22q11.2 [reviewed by 220] (Fig. 2). The majority of the somatic deletions of chromosome 22q observed in schwannomas of patients with SMARCB1-related SWN extend beyond this 6.5 Mb region [43, 65, 380]. Further, many of the somatic 22q deletions in schwannomas exhibit heterogeneous breakpoints located outside of the LCR22 sequences suggesting that these LCRs are not directly involved in deletion formation.
In addition to SMARCB1 and LZTR1, other schwannomatosis predisposition genes located on chromosome 22q are likely to exist (Supplementary Table 2). In a proportion of schwannomatosis patients without identifiable germline LZTR1 or SMARCB1 PV, somatic 22q LOH has been detected in schwannomas with or without an identifiable somatic NF2 PV [43]. Patients with this mutational pattern exhibit 22q-related SWN [66] (Table 1). The targeted sequencing of specifically chromosome 22q in 31 patients with 22q-related SWN has indicated five genes on chromosome 22 that might qualify as additional schwannomatosis predisposition genes (Supplementary Table 2), but further verification is required by the analysis of additional patients [43].
Molecular signature of SWN-schwannomas
Biallelic loss-of function of the NF2 gene is the initiating tumorigenic event of schwannomas in patients with NF2-related SWN and also in a large proportion of sporadic schwannomas [reviewed by 384]. Likewise, schwannomas of patients with SMARCB1- and LZTR1-related SWN exhibit biallelic NF2 gene inactivation as mentioned in the previous section. Thus, loss of NF2 gene function is a crucial event initiating tumorigenesis in most schwannomas. Furthermore, schwannomas of patients with SMARCB1- and LZTR1-related SWN are phenotypically and histopathologically indistinguishable from schwannomas of patients with NF2-related SWN and sporadic schwannomas. Nevertheless, it is plausible that the events which drive schwannoma growth differ depending upon the presence or absence of a germline PV in one of the three schwannoma predisposition genes (NF2, SMARCB1 and LZTR1). In view of the genetic heterogeneity, it might be expected that the molecular signature including changes in DNA methylation and gene expression associated with alterations in signalling pathways varies comparing sporadic schwannomas and those of patients with either NF2-, LZTR1- or SMARCB1-related SWN. However, the vast majority of previous studies that addressed the genomic, gene expression and DNA methylation profiles of schwannomas have been focussed on sporadic schwannomas or those of patients with NF2-related SWN [384-387]. By contrast, the molecular signature of schwannomas from patients with genetically confirmed LZTR1- or SMARCB1-related SWN has been analysed so far only in a single study [111]. Perhaps suprisingly, DNA methylation profiling did not indicate clear differences between schwannomatosis-associated schwannomas (SWN-schwannomas) and sporadic schwannomas [111]. Furthermore, no significant differences in the DNA methylation profiles of SWN-schwannomas were detected when comparing tumours from patients with germline PVs in either LZTR1 or SMARCB1 [111]. However, four different DNA methylation subgroups were identified in SWN-associated schwannomas, which were specifically associated with the anatomic location of the tumours. Multiple schwannomas resected from different anatomic areas of the same patient resolved into different methylation clusters. This finding suggests that Schwann cells derived from different regions of the body exhibit different DNA methylation profiles [111]. Moreover, each methylation cluster exhibited a distinct transcriptome profile with upregulated expression of specific pathways including cAMP, NFkB, RB and PIGF. These findings indicate the putative existence of four subtypes of SWN-schwannomas [111]. Further, pathway analysis indicated the upregulation of VEGF, SHH and MEK pathways, in addition to mismatch-repair and DNA repair-related genes in SWN-schwannomas as compared to sporadic schwannomas [111]. Remarkably, SWN-schwannomas exhibited a significantly elevated number of chromosomal copy number variants and higher rates of chromosome 22q loss-of-heterozygosity as compared to sporadic schwannomas [111]. Taken together, these findings suggest that substantial differences may exist in the pathogenesis of sporadic versus SWN-schwannomas. However, significant differences in the molecular profiles of schwannomas derived from patients with either LZTR1- or SMARCB1-related SWN were not obvious. Mansouri et al. [111] investigated 25 schwannomas from 10 patients with SMARCB1-related SWN and 69 schwannomas from 26 patients with LZTR1-related SWN. Most likely, larger numbers of SWN-schwannomas should be analysed comparatively in order to identify any molecular differences between schwannomas derived from patients with either LZTR1- or SMARCB1-related SWN.
Single-cell RNA sequencing (scRNA-seq) of 22 schwannomas including sporadic tumours and those from patients with NF2-related SWN and non-NF2-related SWN indicated intra- and inter-tumoral transcriptional heterogeneity of schwannomas [388]. Nevertheless, six recurring distinct transcriptional programs (meta-programs) have been identified with gene signatures related to stress, myelin production, antigen presentation, interferon signalling, glycolysis and extracellular matrix [388]. The advantage of scRNA-seq is that intra-tumoral heterogeneity (ITH), which is a property of many tumours driven by genetic, epigenetic and microenvironmental influences, can be assessed very efficiently. Numerous scRNA-seq analyses of different tumour types indicated that ITH is often associated with “expression programs” comprising dozens of genes with coordinated variability in their expression across malignant cells within a given tumour. Similar ITH programs have been identified across tumours of the same cancer type, and in some instances even across different tumour types suggesting that ITH expression programs reflect basic principles of tumour biology. The consensus among related ITH programs from different tumours has been designated as meta-programs [389]. In schwannomas, six distinct gene expression meta-programs were identified [388]. Remarkably, these meta-programs were observed in schwannomas of different genetic backgrounds and from different anatomical locations. However, a clear clustering of schwannomas according to their genetic background and anatomical location was not possible suggesting that the schwannomas exhibit similar overall expression profiles. Unfortunately, no distinction was made between SMARCB1- or LZTR1-related SWN in this study [388]. Additional scRNA-seq analyses of schwannomas from patients with characterized germline variants in either SMARCB1 or LZTR1 would be instructive in order to elaborate any transcriptome differences that remain to be identified.
Recurrent SH3PXD2A-HTRA1 fusion in SWN-schwannomas
Remarkably, a recurrent somatic fusion gene has been identified in 10% of sporadic schwannomas and in schwannomas of patients with NF2-related SWN [390]. The in-frame fusion involves the SH3 and PX domains-containing protein 2 A gene (SH3PXD2A; MIM #619455) and the high temperature requirement A serine peptidase 1 gene (HTRA1; MIM #602194), both located on chromosome 10q. The fusion results from a balanced 19-Mb inversion within chromosome 10q [390]. In vitro transfection assays indicated that the overexpression of the SH3PXD2A-HTRA1 fusion protein caused increased levels of phosphorylated extracellular signal-regulated kinase (ERK) indicative of an activated mitogen-activated protein kinase (MAPK) pathway. Further, the SH3PXD2A-HTRA1 fusion has been shown to increase proliferation and invasive growth [390]. The SH3PXD2A-HTRA1 fusion gene is also present in a subset of schwannomas from patients with schwannomatosis [111]. The SH3PXD2A-HTRA1 fusion gene was detected in 2/24 (8.3%) of schwannomas from patients with SMARCB1-related SWN, and in 13/64 (20.3%) of schwannomas from patients with LZTR1-related SWN which is not significantly different [111]. In schwannomas of patients with LZTR1-related SWN, the fusion gene was significantly more frequent in painful schwannomas as compared to pain-free schwannomas. Furthermore, upregulation of the RAS/MAPK pathway was observed in schwannomas of patients with LZTR1-related SWN [111]. The RAS/MAPK pathway is also activated in sporadic schwannomas [390]. However, it is unknown if RAS/MAPK pathway activation also plays an important role in schwannomas of patients with SMARCB1-related SWN. Further studies, including higher numbers of schwannomas from patients with SMARCB1-related SWN, will be necessary to identify the full spectrum of pathways altered in these tumours.
Molecular pathogenesis of neurodevelopmental disorders caused by germline SMARCB1 PVs
In addition to its role as a tumour suppressor, the SMARCB1 protein is also an important regulator during development. Smarcb1 is required for early embryonic survival since homozygous Smarcb1-null mouse embryos die between embryonic days 3.5 and 5.5 post coitum [357, 377, 378]. Human induced pluripotent stem cells and organoid models indicated that SMARCB1 loss during neuronal differentiation leads to a lack of stability among neural progenitor cells and a failure in maturation [319, 320]. Furthermore, significant differences in the response of cells to SMARCB1 loss were detected at different stages of neural differentiation, indicating a narrow time window early in neural commitment during which cells are highly vulnerable to SMARCB1 loss-of-function, exhibiting severe defects in the progression of differentiation [319, 320]. This is in accord with the results of inducible Smarcb1 knockout mouse models [178] (Sect. The time window of SMARCB1 inactivation in rhabdoid tumour development). In human embryonic stem cells, SMARCB1 is required for increased accessibility of chromatin regions associated with neural differentiation but dispensable for mesodermal or endodermal differentiation [228, 229]. In similar vein, induced loss of Smarcb1 protein in mouse embryonic stem cells impaired the expression regulation of genes associated with nervous system development [327]. An important role of SMARCB1 during human neurodevelopment may also be deduced from the observation that germline pathogenic SMARCB1 variants cause some of the SWI/SNF-related intellectual disability disorders (SSRIDDs), which result from dysfunction of BAF complexes [reviewed by 211, 44, 216, 391] as reviewed in the following section.
Molecular pathogenesis of Coffin-Siris syndrome (CSS)
So far, 14 different intragenic SMARCB1 PVs affecting single nucleotides and one partial gene deletion of 9-kb encompassing SMARCB1 exons 8 and 9 as well as the 3’ flanking region of SMARCB1, have been identified in 35 patients with CSS [45, 46, 48, 49, 202–209, 282, 392, 394] (Table 4). The most common recurrent CSS-associated SMARCB1 PV is an in-frame deletion of a single lysine, K364del (identified in 14 unrelated CSS patients). This recurrent PV is located in the C-terminal coiled-coil domain (CTD) of SMARCB1 close to other missense PVs (Table 4). The CSS-causing SMARCB1 variants cluster closely together at exons 8 and 9, indicating a specific position effect in the pathogenesis of SMARCB1-related CSS. It should be noted that the PVs in the SMARCB1 CTD are not only found in the germline of patients with CSS but are also observed as somatic variants in different types of cancer [134, 283].
It is intriguing that PVs affecting single amino acids located within the CTD of SMARCB1 cause a severe neurodevelopmental disorder such as CSS, but are not associated with rhabdoid tumour development. This is suggestive of a very specific role for the SMARCB1 CTD which encompasses an alpha helical domain within a region of densely packed basic and positively-charged amino acids. Importantly, this alpha-helical domain directly binds to the acidic patch of the nucleosome [258]. CSS-causing PVs located within the SMARCB1 CTD do not grossly alter the secondary structure of this domain. Instead, they disrupt nucleosome binding and preclude BAF-mediated nucleosome remodelling and DNA accessibility at enhancer regions [258]. The genome-wide localization of the BAF complex is not affected by PVs in the C-terminal alpha-helical domain of SMARCB1. Nevertheless, these complexes are defective in activating critical target genes [258, 259].
Among the CSS-causing PVs located in the SMARCB1 CTD is the missense variant R377H [45, 394]. Re-expression of this variant in the SMARCB1-deficient G401 cell line did not result in any decrease in cell proliferation, in contrast to the nonsense R377* mutant which caused a significant reduction in cell proliferation [273]. These findings imply that the nonsense R377* mutation partially compromises SMARCB1 tumour suppressor function, whereas the R377H missense PV retains this functionality [273]. This finding may explain why patients with CSS are not affected by pediatric AT/RT or other malignant tumours associated with complete loss of SMARCB1 function.
Importantly, heterozygous CSS-causing SMARCB1 PVs located in the CTD result in gene regulatory and morphological changes during induced pluripotent stem cell (iPSC)-neuronal differentiation. Indeed, differentiated neurons derived from iPSCs harbouring the heterozygous SMARCB1 c.1091_1093del (p.K364del) variant showed less neurite outgrowth than wild-type controls [258, 259].
The conclusion that neurite outgrowth deficits may result from C-terminal non-truncating pathogenic SMARCB1 variants has been confirmed by means of the genetically engineered mouse model created by Brugmans et al. [395]. These mice harboured a deletion of a cytosine in exon 9 at position c.1148 of Smarcb1, causing a frameshift of 36 amino acids until a downstream stop codon (c.1148del; p.P383QfsX36) [395]. Adolescent Smarcb11148del/1148del mice exhibited delayed weight gain and hydrocephalus including enlarged lateral ventricles. In their embryonic and neonatal stages, the brains of these mutant mice did not differ anatomically or histologically from the brains of wild-type controls [395]. Transcriptome analysis by single-cell RNA sequencing of brains from newborn mutant mice indicated that a complete brain is formed with all cell types from a normal mouse brain. Nevertheless, neuronal signalling was perturbed in these newborn mutant mice. Drastically lowered expression of the AP-1 transcription factor family was noted to be the cause of reduced expression of essential regulators of neurite outgrowth via growth cones in the mutant mice [395]. These findings are indicative of the important role of SMARCB1 during neurodevelopment. Impaired SMARCB1 function may also disturb neurite outgrowth and synapse formation in humans causing intellectual disability in patients with neurodevelopmental disorders such as CSS. This is in line with other studies showing neurite outgrowth deficits in neurons with pathogenic variants in other BAF complex genes [269, 396]. Taken together, deficiencies of BAF complex subunits play an important role in the pathogenesis of a subgroup of neurodevelopmental disorders. Indeed, BAF complex genes are the most frequently mutated genes among those involved in chromatin regulation in the context of neurodevelopmental disorders [259].
Importantly, BAF complex subunits are expressed in a temporal and cell-type specific manner during neurodevelopment. The BAF complex begins to switch subunits to those unique to neural progenitors, followed by subunits specific to neurons during differentiation from embryonic stem cells into neurons [263, 270]. Thus, the timely expression of these BAF subunits is essential for regulating cell fate during neurodevelopment. The combinatorial assembly of subunits determines cell lineage specification by creating specific patterns of chromatin states at different developmental stages, which are essential for normal neurodevelopment [reviewed by 397]. The pathogenesis underlying the SWI/SNF-related intellectual disability disorders (SSRIDDs) including CSS indicates that dysfunction of any of these subunits disturbs neural development and results in the overlapping clinical phenotypes of SSIDRs [reviewed by 211, 216, 391]. Since the clinical phenotype of SSIDRs is in several instances overlapping, genetic testing has become necessary in order to arrive at a correct differential diagnosis. For example, patients initially clinically diagnosed with Aicardi syndrome or Nicolaides–Baraitser syndrome have been reclassified as CSS cases after the identification of SMARCB1 PVs in C-terminal domain which have been previously identified as recurrent mutations in patients with classical CSS [49, 393].
The important role of SMARCB1 during nervous system development also became obvious through the conditional knockout mouse models generated by Vitte et al. [179]. In these mice, Smarcb1 deletion was induced by the Cre promoters DHH and mGFAP later during Schwann cell development (beginning at E12.5 or E13.5, respectively). These DHH-CreC; Smarcb flox/flox, mGFAP-Cre; Smarcb1flox/flox and mGFAP-Cre; Smarcb1del/flox mice survived only a few weeks and developed progressive hindlimb paralysis. Their sciatic nerves were thinner and more transparent than those in control mice. Histological analysis of the nerve fibres indicated severe disturbances of structure and organization caused by Smarcb1 loss [179]. These mice did not develop tumours indicating that Smarcb1 loss during later stages of development is on its own not tumorigenic (see Sect. The time window of SMARCB1 inactivation in rhabdoid tumour development). The severe neurological phenotype of these mice implies that SMARCB1 protein deficiency during early development of the human nervous system may well be responsible for the neurological deficits observed in patients with pathogenic SMARCB1 variants and CSS [213, 214].
Patients with CSS and schwannomatosis
Remarkably, two patients have been identified who had both CSS and schwannomatosis [282, 392]. The 28 year-old female patient reported by Gallagher et al. [392] had multiple intra-thoracic schwannomas and a large painful schwannoma of the left upper arm as well as a severe clinical manifestation of CSS. Sequence analysis indicated a de novo germline in-frame deletion in SMARCB1, c.1091_1093del (p.K364del), which represents the most common recurrent SMARCB1 PV in patients with CSS (Table 4). As mentioned in Sect. Molecular pathogenesis of Coffin-Siris syndrome (CSS), this pathogenic SMARCB1 variant is located in the C-terminal domain. It has been shown that this PV impairs the nucleosomal binding of SMARCB1 and leads to changes in gene expression as well as cellular morphology during induced IPSC differentiation which showed less neurite outgrowth than wild-type controls [258, 259].
The patient reported by Gossai et al. [282] harboured the recurrent pathogenic SMARCB1 variant c.1121G > A (p.Arg374Gln) which also prevents the nucleosomal binding of SMARCB1 [258, 259]. The analysis of a schwannoma in this patient indicated somatic loss of the wild-type SMARCB1 allele, in combination with NF2 loss [282]. The patient had a very severe form of CSS and also schwannomatosis with multiple spinal schwannomas and bilateral cranial nerve involvement [282]. Only 8% of patients with schwannomatosis exhibit single non-vestibular cranial schwannomas [29]. The severe phenotype of both patients suggests that at least some patients with CSS and SMARCB1 PVs are at risk of developing schwannomas and should be investigated by MRI to prevent severe problems caused by a delayed diagnosis of the tumours.
Molecular pathogenesis of intellectual disability with choroid plexus hyperplasia (ID-CPH)
The pathogenic de novo missense SMARCB1 variant (c.110G > A; p.Arg37His) identified in four patients with severe intellectual disability, choroid plexus hyperplasia and resultant hydrocephalus termed ID-CPH is located within exon 2 of SMARCB1, encoding the winged-helix DNA-binding domain (WHD) [217]. It remains to be determined how pathogenic variants in different parts of SMARCB1 can lead to clinically different disorders of neurological development associated with severe intellectual deficits such as CSS and ID-CPH. The R37H missense variant located within the N-terminal SMARCB1 WHD in patients with ID-CPH does not impair the ability of the SMARCB1 protein to bind to nucleosomes, as it has been shown for PVs located in the C-terminal domain of SMARCB1 in patients with CSS [258]. Further, the SMARCB1 R37H missense mutation does not impact BAF nucleosome remodelling activity in vitro [258, 259]. Of note, the WHD is isolated from the SMARCB1 C-terminus in the canonical BAF complex but has been predicted to be repositioned closer to the nucleosome binding lobe in the C-terminal domain in the PBAF complex [260, 272]. This is suggestive of a different functional impact for the SMARCB1 R37H mutation in the different BAF complexes (BAF vs. PBAF).
Reduced Schwannoma risk in patients with 22q11.2 deletions
Of the clinical syndromes associated with germline deletions at 22q11.2, the best characterized is the proximal chromosome 22q11.2 microdeletion syndrome, leading to DiGeorge syndrome/velocardiofacial syndrome (DGS) [MIM #188400 ] most often caused by deletions of ∼3 Mb spanning LCR22 A–D [reviewed by 195] (Fig. 2). Of note, these 3-Mb deletions include the LZTR1 gene but not SMARCB1. Remarkably, patients with these 3-Mb deletions have a lower risk for developing schwannomas as compared to the general population [398]. This is most likely due to the observation that in patients with germline whole-gene deletions of a tumour suppressor gene, the second tumour-specific mutational event is never loss-of-heterozygosity (LOH) of the wild-type allele of this tumour suppressor gene but rather invariably a subtle intragenic mutation [399]. In patients with 3-Mb deletions and DGS, LOH leading to loss of LZTR1 and NF2 as well as flanking regions or larger parts of 22q would appear to be impaired. However, as outlined in Sect. Co-involvement of several tumour suppressor genes in schwannoma development, NF2 loss is indispensable for schwannoma growth. Hence, the 3-Mb deletion in proximal 22q11.2 causing DGS appears to confer a reduced risk for schwannoma development [398]. Importantly, schwannomas have not so far been reported in patients with distal 22q11.2 deletions and increased risk for MRTs. Most likely, somatic LOH caused by mitotic recombination leading to the loss of large parts of 22q, including the NF2 gene, is impaired not only in patients with proximal 22q11.2 deletions but also in those with distal 22q11.2 deletions that encompass the SMARCB1 gene.
Conclusion
The important role of the BAF complex during cellular and tissue differentiation, in particular nervous system development, provides a link between tumour suppression and neurodevelopment. Chromatin remodelling is essential for the differentiation of the neural crest. Impairment of proper chromatin remodelling may give rise to neural crest-derived tumours or neurodevelopmental disorders. The exit of neural crest cells from pluripotency towards lineage-specific differentiation would appear to be particularly vulnerable to BAF complex dysfunction, including SMARCB1 loss-of-function in a dosage-dependent manner.
The functional impairment of the SMARCB1 protein or other BAF complex subunits during neural differentiation may either lead to aberrant proliferation and tumorigenesis or intellectual disability and developmental delay. Germline SMARCB1 PVs are associated with RTPS1 and the development of malignant rhabdoid tumours, schwannomatosis or neurodevelopmental disorders such as CSS and ID-CPH. Several factors appear to determine the type of pathology associated with germline SMARCB1 PVs. First, the timing of SMARCB1 inactivation, either during early embryonic development or during later stages in specific progenitor cells, is an important determinant. Several model systems have indicated a very early developmental window for the origin of pediatric MRTs caused by a specific vulnerability to biallelic SMARCB1 inactivation in early neural crest cells. By contrast, schwannomas are likely to result from more differentiated cells such as Schwann cell precursors, which are migrated multipotent progenitors causing schwannoma growth in different body locations during later life.
Second, the different pathologies caused by SMARCB1 mutations are strongly influenced by the type of the germline SMARCB1 pathogenic variant leading to either complete loss of SMARCB1 function in MRTs or a semi-functional SMARCB1 protein resulting from a pathogenic but hypomorphic SMARCB1 variant in patients with schwannomatosis. The type and position of the germline PV within SMARCB1 would also appear to play an important role in the context of SMARCB1-associated neurodevelopmental disorders such as CSS. The majority of heterozygous SMARCB1 PVs causing CSS are single residue alterations located in the C-terminal domain of SMARCB1, which serve to impair the interaction of the BAF complex with the nucleosome. However, these PVs do not interfere with the tumour suppressor functions of SMARCB1. Thus, the CSS-causing SMARCB1 PVs may specifically affect central nervous system development but do not cause malignancy in patients with CSS.
A third determinant of the different pathologies caused by SMARCB1 PVs is represented by additional genomic and epigenetic changes. The mutation type associated with the loss of the second SMARCB1 allele (intragenic SMARCB1 PV, large deletion, complete loss of chromosome 22q) may also influence tumorigenesis. Whilst biallelic NF2 gene inactivation is an absolute requirement for schwannoma growth in addition to biallelic SMARCB1 mutation, complete NF2 gene inactivation is dispensable for MRT development. Malignancy in MRTs is driven by massive changes in the epigenome due to SMARCB1 loss accompanied by changes in the expression of hundreds of genes leading to an undifferentiated tumour phenotype with a very poor prognosis. The continuing analysis of the multifaceted roles of SMARCB1 in cell cycle regulation, DNA repair, gene activation and repression, neurogenesis and nervous system development promises to identify further determinants of SMARCB1-associated pathologies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
H.K.-S. and D.N.C. wrote the review and prepared the figures. Both authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP (1994) Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002–2006. 10.1126/science.7801128 [DOI] [PubMed] [Google Scholar]
- 2.Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O (1998) Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203–206. 10.1038/28212 [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B (1999) Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res 59:74–79 [PubMed] [Google Scholar]
- 4.Sévenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, Jeanpierre C, Jouvet A, Delattre O (1999a) Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet 8:2359–2368. 10.1093/hmg/8.13.2359 [DOI] [PubMed] [Google Scholar]
- 5.Sévenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O (1999b) Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet 65:1342–1348. 10.1086/302639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biegel JA, Fogelgren B, Wainwright LM, Zhou JY, Bevan H, Rorke LB (2000) Germline INI1 mutation in a patient with a central nervous system atypical teratoid tumor and renal rhabdoid tumor. Genes Chromosomes Cancer 28:31–37. [DOI] [PubMed] [Google Scholar]
- 7.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB (2002) Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res 8:3461–3467 [PubMed] [Google Scholar]
- 8.Taylor MD, Gokgoz N, Andrulis IL, Mainprize TG, Drake JM, Rutka JT (2000) Familial posterior fossa brain tumors of infancy secondary to germline mutation of the hSNF5 gene. Am J Hum Genet 66:1403–1406. 10.1086/302833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biegel JA (2006) Molecular genetics of atypical teratoid/rhabdoid tumor. Neurosurg Focus 20:E11. 10.3171/foc.2006.20.1.12 [DOI] [PubMed] [Google Scholar]
- 10.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, Biegel JA, Getz G, Roberts CW (2012) A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122:2983–2988. 10.1172/JCI6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nesvick CL, Lafay-Cousin L, Raghunathan A, Bouffet E, Huang AA, Daniels DJ (2020) Atypical teratoid rhabdoid tumor: molecular insights and translation to novel therapeutics. J Neurooncol 150:47–56. 10.1007/s11060-020-03639-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourdeaut F, Lequin D, Brugières L et al (2011) Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res 17:31–38. 10.1158/1078-0432.CCR-10-1795 [DOI] [PubMed] [Google Scholar]
- 13.Eaton KW, Tooke LS, Wainwright LM, Judkins AR, Biegel JA (2011) Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer 56:7–15. 10.1002/pbc.22831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farouk Sait S, Walsh MF, Karajannis MA (2021) Genetic syndromes predisposing to pediatric brain tumors. Neurooncol Pract 8:375–390. 10.1093/nop/npab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frühwald MC, Hasselblatt M, Nemes K et al (2020) Age and DNA methylation subgroup as potential independent risk factors for treatment stratification in children with atypical teratoid/rhabdoid tumors. Neuro Oncol 22:1006–1017. 10.1093/neuonc/noz244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker NA, Al-Obaidi A, Deutsch JM (2020) SMARCB1/INI1-deficient tumors of adulthood. F1000Res 9:662. 10.12688/f1000research.24808.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margol AS, Judkins AR (2014) Pathology and diagnosis of SMARCB1-deficient tumors. Cancer Genet 207:358–364. 10.1016/j.cancergen.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 18.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45:592–601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohashi K, Oda Y (2017) Oncogenic roles of SMARCB1/INI1 and its deficient tumors. Cancer Sci 108:547–552. 10.1111/cas.13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper GW, Hong AL (2022) SMARCB1-deficient cancers: novel molecular insights and therapeutic vulnerabilities. Cancers (Basel) 14:3645. 10.3390/cancers14153645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N, Qin Y, Du F, Wang X, Song C (2022) Prevalence of SWI/SNF genomic alterations in cancer and association with the response to immune checkpoint inhibitors: A systematic review and meta-analysis. Gene 834:146638. 10.1016/j.gene.2022.146638 [DOI] [PubMed] [Google Scholar]
- 22.Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR (2017) Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 49:213–222. 10.1038/ng.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chun HE, Lim EL, Heravi-Moussavi A et al (2016) Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell 29:394–406. 10.1016/j.ccell.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johann PD, Erkek S, Zapatka M et al (2016) Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 29:379–393. 10.1016/j.ccell.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 25.Torchia J, Picard D, Lafay-Cousin L et al (2015) Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. Lancet Oncol 16:569–582. 10.1016/S1470-2045(15)70114-2 [DOI] [PubMed] [Google Scholar]
- 26.Torchia J, Golbourn B, Feng S et al (2016) Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell 30:891–908. 10.1016/j.ccell.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao F, Zhang Y, Hou J, Zhao B (2025) Chromatin remodeling and cancer: the critical influence of the SWI/SNF complex. Epigenetics Chromatin 18:22. 10.1186/s13072-025-00590-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P (2007) Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet 80:805–810. 10.1086/513207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merker VL, Esparza S, Smith MJ, Stemmer-Rachamimov A, Plotkin SR (2012) Clinical features of schwannomatosis: a retrospective analysis of 87 patients. Oncologist 17:1317–1322. 10.1634/theoncologist.2012-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacCollin M, Woodfin W, Kronn D, Short MP (1996) Schwannomatosis: a clinical and pathologic study. Neurology 46:1072–1079. 10.1212/wnl.46.4.1072 [DOI] [PubMed] [Google Scholar]
- 31.MacCollin M, Chiocca EA, Evans DG, Friedman JM, Horvitz R, Jaramillo D, Lev M, Mautner VF, Niimura M, Plotkin SR, Sang CN, Stemmer-Rachamimov A, Roach ES (2005) Diagnostic criteria for schwannomatosis. Neurology 64:1838–1845. 10.1212/01.WNL.0000163982.78900.AD [DOI] [PubMed] [Google Scholar]
- 32.Evans DG, Bowers NL, Tobi S et al (2018) Schwannomatosis: a genetic and epidemiological study. J Neurol Neurosurg Psychiatry 89:1215–1219. 10.1136/jnnp-2018-318538 [DOI] [PubMed] [Google Scholar]
- 33.Boyd C, Smith MJ, Kluwe L, Balogh A, Maccollin M, Plotkin SR (2008) Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clin Genet 74:358–366. 10.1111/j.1399-0004.2008.01060.x [DOI] [PubMed] [Google Scholar]
- 34.Hadfield KD, Newman WG, Bowers NL, Wallace A, Bolger C, Colley A, McCann E, Trump D, Prescott T, Evans DG (2008) Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet 45:332–339. 10.1136/jmg.2007.056499 [DOI] [PubMed] [Google Scholar]
- 35.Sestini R, Bacci C, Provenzano A, Genuardi M, Papi L (2008) Evidence of a four-hit mechanism involving SMARCB1 and NF2 in schwannomatosis-associated schwannomas. Hum Mutat 29:227–231. 10.1002/humu.20679 [DOI] [PubMed] [Google Scholar]
- 36.Rousseau G, Noguchi T, Bourdon V, Sobol H, Olschwang S (2011) SMARCB1/INI1 germline mutations contribute to 10% of sporadic schwannomatosis. BMC Neurol 11:9. 10.1186/1471-2377-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melean G, Velasco A, Hernández-Imaz E, Rodríguez-Álvarez FJ, Martín Y, Valero A, Hernández-Chico C (2012) RNA-based analysis of two SMARCB1 mutations associated with familial schwannomatosis with meningiomas. Neurogenetics 13:267–274. 10.1007/s10048-012-0335-8 [DOI] [PubMed] [Google Scholar]
- 38.Smith MJ, Wallace AJ, Bowers NL, Rustad CF, Woods CG, Leschziner GD, Ferner RE, Evans DG (2012) Frequency of SMARCB1 mutations in familial and sporadic schwannomatosis. Neurogenetics 13:141–145. 10.1007/s10048-012-0319-8 [DOI] [PubMed] [Google Scholar]
- 39.Smith MJ, Boyd CD, MacCollin MM, Plotkin SR (2009) Identity analysis of schwannomatosis kindreds with recurrent constitutional SMARCB1 (INI1) alterations. Clin Genet 75:501–502. 10.1111/j.1399-0004.2009.01156.x [DOI] [PubMed] [Google Scholar]
- 40.Smith MJ, Wallace AJ, Bowers NL, Eaton H, Evans DG (2014) SMARCB1 mutations in schwannomatosis and genotype correlations with rhabdoid tumors. Cancer Genet 207:373–378. 10.1016/j.cancergen.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 41.Holsten T, Bens S, Oyen F, Nemes K, Hasselblatt M, Kordes U, Siebert R, Frühwald MC, Schneppenheim R, Schüller U (2018) Germline variants in SMARCB1 and other members of the BAF chromatin-remodeling complex across human disease entities: a meta-analysis. Eur J Hum Genet 26:1083–1093. 10.1038/s41431-018-0143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louvrier C, Pasmant E, Briand-Suleau A, Cohen J, Nitschké P, Nectoux J, Orhant L, Zordan C, Goizet C, Goutagny S, Lallemand D, Vidaud M, Vidaud D, Kalamarides M, Parfait B (2018) Targeted next-generation sequencing for differential diagnosis of neurofibromatosis type 2, schwannomatosis, and meningiomatosis. Neuro Oncol 20:917–929. 10.1093/neuonc/noy009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piotrowski A, Koczkowska M, Poplawski AB et al (2022) Targeted massively parallel sequencing of candidate regions on chromosome 22q predisposing to multiple schwannomas: an analysis of 51 individuals in a single-center experience. Hum Mutat 43:74–84. 10.1002/humu.24294 [DOI] [PubMed] [Google Scholar]
- 44.Chmykhalo VK, Deev RV, Tokarev AT, Polunina YA, Xue L, Shidlovskii YV (2025) SWI/SNF complex connects signaling and epigenetic state in cells of nervous system. Mol Neurobiol 62:1536–1557. 10.1007/s12035-024-04355-6 [DOI] [PubMed] [Google Scholar]
- 45.Tsurusaki Y, Okamoto N, Ohashi H et al (2012) Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet 44:376–378. 10.1038/ng.2219 [DOI] [PubMed] [Google Scholar]
- 46.Tsurusaki Y, Okamoto N, Ohashi H et al (2014) Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin Genet 85:548–554. 10.1111/cge.12225 [DOI] [PubMed] [Google Scholar]
- 47.Kosho T, Okamoto N, Ohashi H et al (2013) Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature. Am J Med Genet A 161A:1221–1237. 10.1002/ajmg.a.35933 [DOI] [PubMed] [Google Scholar]
- 48.Santen GW, Aten E, Vulto-van Silfhout AT et al (2013) Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat 34:1519–1528. 10.1002/humu.22394 [DOI] [PubMed] [Google Scholar]
- 49.Wieczorek D, Bögershausen N, Beleggia F et al (2013) A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet 22:5121–5135. 10.1093/hmg/ddt366 [DOI] [PubMed] [Google Scholar]
- 50.Janson K, Nedzi LA, David O, Schorin M, Walsh JW, Bhattacharjee M, Pridjian G, Tan L, Judkins AR, Biegel JA (2006) Predisposition to atypical teratoid/rhabdoid tumor due to an inherited INI1 mutation. Pediatr Blood Cancer 47:279–284. 10.1002/pbc.20622 [DOI] [PubMed] [Google Scholar]
- 51.Ammerlaan AC, Ararou A, Houben MP, Baas F, Tijssen CC, Teepen JL, Wesseling P, Hulsebos TJ (2008) Long-term survival and transmission of INI1-mutation via nonpenetrant males in a family with rhabdoid tumour predisposition syndrome. Br J Cancer 98:474–479. 10.1038/sj.bjc.6604156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swensen JJ, Keyser J, Coffin CM, Biegel JA, Viskochil DH, Williams MS (2009) Familial occurrence of schwannomas and malignant rhabdoid tumour associated with a duplication in SMARCB1. J Med Genet 46:68–72. 10.1136/jmg.2008.060152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruggers CS, Bleyl SB, Pysher T, Barnette P, Afify Z, Walker M, Biegel JA (2011) Clinicopathologic comparison of familial versus sporadic atypical teratoid/rhabdoid tumors (AT/RT) of the central nervous system. Pediatr Blood Cancer 56:1026–1031. 10.1002/pbc.22757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carter JM, O’Hara C, Dundas G, Gilchrist D, Collins MS, Eaton K, Judkins AR, Biegel JA, Folpe AL (2012) Epithelioid malignant peripheral nerve sheath tumor arising in a schwannoma, in a patient with neuroblastoma-like schwannomatosis and a novel germline SMARCB1 mutation. Am J Surg Pathol 36:154–160. 10.1097/PAS.0b013e3182380802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Munckhof P, Christiaans I, Kenter SB, Baas F, Hulsebos TJ (2012) Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the Falx cerebri. Neurogenetics 13:1–7. 10.1007/s10048-011-0300-y [DOI] [PubMed] [Google Scholar]
- 56.Kordes U, Bartelheim K, Modena P, Massimino M, Biassoni V, Reinhard H, Hasselblatt M, Schneppenheim R, Frühwald MC (2014) Favorable outcome of patients affected by rhabdoid tumors due to rhabdoid tumor predisposition syndrome (RTPS). Pediatr Blood Cancer 61:919–921. 10.1002/pbc.24793 [DOI] [PubMed] [Google Scholar]
- 57.Robson DK, Ironside JW (1990) Malignant peripheral nerve sheath tumour arising in a schwannoma. Histopathology 16:295–297. 10.1111/j.1365-2559.1990.tb01118.x [DOI] [PubMed] [Google Scholar]
- 58.Woodruff JM, Selig AM, Crowley K, Allen PW (1994) Schwannoma (neurilemoma) with malignant transformation. A rare, distinctive peripheral nerve tumor. Am J Surg Pathol 18:882–895. 10.1097/00000478-199409000-00003 [DOI] [PubMed] [Google Scholar]
- 59.Nayler SJ, Leiman G, Omar T, Cooper K (1996) Malignant transformation in a schwannoma. Histopathology 29:189–192. 10.1046/j.1365-2559.1996.d01-491.x [DOI] [PubMed] [Google Scholar]
- 60.McMenamin ME, Fletcher CD (2001) Expanding the spectrum of malignant change in schwannomas: epithelioid malignant change, epithelioid malignant peripheral nerve sheath tumor, and epithelioid angiosarcoma: a study of 17 cases. Am J Surg Pathol 25:13–25. 10.1097/00000478-200101000-00002 [DOI] [PubMed] [Google Scholar]
- 61.Hilton DA, Hanemann CO (2014) Schwannomas and their pathogenesis. Brain Pathol 24:205–220. 10.1111/bpa.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacCollin M, Willett C, Heinrich B, Jacoby LB, Acierno JS Jr, Perry A, Louis DN (2003) Familial schwannomatosis: exclusion of the NF2 locus as the germline event. Neurology 60:1968–1974. 10.1212/01.wnl.0000070184.08740.e0 [DOI] [PubMed] [Google Scholar]
- 63.Jacoby LB, Jones D, Davis K, Kronn D, Short MP, Gusella J, MacCollin M (1997) Molecular analysis of the NF2 tumor-suppressor gene in schwannomatosis. Am J Hum Genet 61:1293–1302. 10.1086/301633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baser ME, Friedman JM, Evans DG (2006) Increasing the specificity of diagnostic criteria for schwannomatosis. Neurology 66:730–732. 10.1212/01.wnl.0000201190.89751.41 [DOI] [PubMed] [Google Scholar]
- 65.Piotrowski A, Xie J, Liu YF et al (2014) Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nat Genet 46:182–187. 10.1038/ng.2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plotkin SR, Messiaen L, Legius E et al (2022) Updated diagnostic criteria and nomenclature for neurofibromatosis type 2 and schwannomatosis: an international consensus recommendation. Genet Med 24:1967–1977. 10.1016/j.gim.2022.05.007 [DOI] [PubMed] [Google Scholar]
- 67.Hutter S, Piro RM, Reuss DE et al (2014) (2014) Whole exome sequencing reveals that the majority of schwannomatosis cases remain unexplained after excluding SMARCB1 and LZTR1 germline variants. Acta Neuropathol 128:449–452. 10.1007/s00401-014-1311-1 [DOI] [PubMed]
- 68.Smith MJ, Isidor B, Beetz C et al (2015) Mutations in LZTR1 add to the complex heterogeneity of schwannomatosis. Neurology 84:141–147. 10.1212/WNL.0000000000001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deng F, Evans DG, Smith MJ (2022) Comparison of the frequency of loss-of-function LZTR1 variants between schwannomatosis patients and the general population. Hum Mutat 43:919–927. 10.1002/humu.24376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang K, Lin JW, Wang J, Wu X, Gao H, Hsieh YC, Hwu P, Liu YR, Su L, Chiou HY, Wang D, Yuan YC, Whang-Peng J, Chiu WT, Yen Y (2014) A germline missense mutation in COQ6 is associated with susceptibility to Familial schwannomatosis. Genet Med 16:787–792. 10.1038/gim.2014.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trevisson E, Clementi M, Salviati L (2015) Is there a link between COQ6 and schwannomatosis? Genet Med 17:312–313. 10.1038/gim.2014.211 [DOI] [PubMed] [Google Scholar]
- 72.Min BJ, Kang YK, Chung YG, Seo ME, Chang KB, Joo MW (2020) Germline mutations for novel candidate predisposition genes in sporadic schwannomatosis. Clin Orthop Relat Res 478:2442–2450. 10.1097/CORR.0000000000001239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivera B, Nadaf J, Fahiminiya S et al (2020) DGCR8 microprocessor defect characterizes familial multinodular goiter with schwannomatosis. J Clin Invest 130:1479–1490. 10.1172/JCI130206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nogué C, Chong AS, Grau E, Han H, Dorca E, Roca C, Mosquera JL, Lázaro C, Foulkes WD, Brunet J, Rivera B (2022) DGCR8 and the six hit, three-step model of schwannomatosis. Acta Neuropathol 143:115–117. 10.1007/s00401-021-02387-z [DOI] [PubMed] [Google Scholar]
- 75.Perez-Becerril C, Wallace AJ, Schlecht H, Bowers NL, Smith PT, Gokhale C, Eaton H, Charlton C, Robinson R, Charlton RS, Evans DG, Smith MJ (2022) Screening of potential novel candidate genes in schwannomatosis patients. Hum Mutat 43:1368–1376. 10.1002/humu.24424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terry M, Gupta R, Ravindranathan A, Wu J, Chan E, Bollen AW, Chang SM, Berger MS, Jacques L, Solomon DA (2023) Somatic mosaic SOX10 indel mutations underlie a form of segmental schwannomatosis. Acta Neuropathol 146:857–860. 10.1007/s00401-023-02641-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams EA, Ravindranathan A, Gupta R et al (2023) Novel SOX10 indel mutations drive schwannomas through impaired transactivation of myelination gene programs. Neuro Oncol 25:2221–2236. 10.1093/neuonc/noad121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee PH, Huang SC, Lee JC, Li SC, Tsai JW, Chang YM, Kao YC, Fan WL, Chang CD, Chen HC, Li CH, Hu CF, Liu TT, Wu PS, Nam MH, Yu SC, Wang JC, Huang HY (2025) In-frame insertions of SOX10 are highly enriched and characterize a distinct transcriptomic profile in gastrointestinal schwannomas. J Pathol 25 Apr 24. Epub ahead of print. PMID: 40272443 10.1002/path.6426 [DOI] [PubMed]
- 79.Forde C, Smith MJ, Burghel GJ et al (2024) NF2-related schwannomatosis and other schwannomatosis: an updated genetic and epidemiological study. J Med Genet 61:856–860. 10.1136/jmg-2024-110065 [DOI] [PubMed] [Google Scholar]
- 80.Halliday D, Emmanouil B, Evans DGR (2023) Updated protocol for genetic testing, screening and clinical management of individuals at risk of NF2-related schwannomatosis. Clin Genet 103:540–552. 10.1111/cge.14310 [DOI] [PubMed] [Google Scholar]
- 81.Smith MJ, Perez-Becerril C, van der Meer M et al (2024) Genetic findings in people with schwannomas who do not meet clinical diagnostic criteria for NF2-related schwannomatosis. J Med Genet 61:1011–1015. 10.1136/jmg-2024-110217 [DOI] [PubMed] [Google Scholar]
- 82.Merker VL, Slobogean B, Jordan JT, Langmead S, Meterko M, Charns MP, Elwy AR, Blakeley JO, Plotkin SR (2022) Understanding barriers to diagnosis in a rare, genetic disease: delays and errors in diagnosing schwannomatosis. Am J Med Genet A 188:2672–2683. 10.1002/ajmg.a.62860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith MJ, Bowers NL, Bulman M, Gokhale C, Wallace AJ, King AT, Lloyd SK, Rutherford SA, Hammerbeck-Ward CL, Freeman SR, Evans DG (2017) Revisiting neurofibromatosis type 2 diagnostic criteria to exclude LZTR1-related schwannomatosis. Neurology 88:87–92. 10.1212/WNL.0000000000003418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kehrer-Sawatzki H, Kluwe L, Friedrich RE, Summerer A, Schäfer E, Wahlländer U, Matthies C, Gugel I, Farschtschi S, Hagel C, Cooper DN, Mautner VF (2018) Phenotypic and genotypic overlap between mosaic NF2 and schwannomatosis in patients with multiple non-intradermal schwannomas. Hum Genet 137:543–552. 10.1007/s00439-018-1909-9 [DOI] [PubMed] [Google Scholar]
- 85.Smith MJ, Burghel GJ, Evans DG (2025) Effects of higher-than-expected control population allele frequency on classification of loss-of-function variants in cancer susceptibility genes. J Med Genet 11:jmg–2025. 10.1136/jmg-2025-110703 [DOI] [PubMed] [Google Scholar]
- 86.Karczewski KJ, Francioli LC, Tiao G et al (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kluwe L, Mautner V, Heinrich B, Dezube R, Jacoby LB, Friedrich RE, MacCollin M (2003) Molecular study of frequency of mosaicism in neurofibromatosis 2 patients with bilateral vestibular schwannomas. J Med Genet 40:109–114. 10.1136/jmg.40.2.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moyhuddin A, Baser ME, Watson C, Purcell S, Ramsden RT, Heiberg A, Wallace AJ, Evans DG (2003) Somatic mosaicism in neurofibromatosis 2: prevalence and risk of disease transmission to offspring. J Med Genet 40:459–463. 10.1136/jmg.40.6.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evans DG, Ramsden RT, Shenton A, Gokhale C, Bowers NL, Huson SM, Pichert G, Wallace A (2007) Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. J Med Genet 44:424–428. 10.1136/jmg.2006.047753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teranishi Y, Miyawaki S, Hongo H, Dofuku S, Okano A, Takayanagi S, Ota T, Yoshimura J, Qu W, Mitsui J, Nakatomi H, Morishita S, Tsuji S, Saito N (2021) Targeted deep sequencing of DNA from multiple tissue types improves the diagnostic rate and reveals a highly diverse phenotype of mosaic neurofibromatosis type 2. J Med Genet 58:701–711. 10.1136/jmedgenet-2020-106973 [DOI] [PubMed] [Google Scholar]
- 91.Murray AJ, Hughes TA, Neal JW, Howard E, Evans DG, Harper PS (2006) A case of multiple cutaneous schwannomas; schwannomatosis or neurofibromatosis type 2? J Neurol Neurosurg Psychiatry 77:269–271. 10.1136/jnnp.2005.067017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Contini E, Paganini I, Sestini R, Candita L, Capone GL, Barbetti L, Falconi S, Frusconi S, Giotti I, Giuliani C, Torricelli F, Benelli M, Papi L (2015) A systematic assessment of accuracy in detecting somatic mosaic variants by deep amplicon sequencing: application to NF2 gene. PLoS ONE 10:e0129099. 10.1371/journal.pone.0129099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sadler KV, Bowers NL, Hartley C et al (2021) Sporadic vestibular schwannoma: a molecular testing summary. J Med Genet 58:227–233. 10.1136/jmedgenet-2020-107022 [DOI] [PubMed] [Google Scholar]
- 94.Plotkin SR, Bredella MA, Cai W, Kassarjian A, Harris GJ, Esparza S, Merker VL, Munn LL, Muzikansky A, Askenazi M, Nguyen R, Wenzel R, Mautner VF (2012) Quantitative assessment of whole-body tumor burden in adult patients with neurofibromatosis. PLoS ONE 7:e35711. 10.1371/journal.pone.0035711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Al-Mistarehi AH, Jiang K, Khalifeh JM, Albert AN, Weber-Levine C, Orlando N, Blanks W, Hersh AM, Romo CG, Blakeley J, Belzberg AJ, Lubelski D (2025) Surgical management of schwannomas in schwannomatosis: a comprehensive analysis of clinical outcomes and determinants of local recurrence. Neurosurg Focus 58:E2. 10.3171/2025.2.FOCUS24751 [DOI] [PubMed] [Google Scholar]
- 96.Pathmanaban ON, Sadler KV, Kamaly-Asl ID, King AT, Rutherford SA, Hammerbeck-Ward C, McCabe MG, Kilday JP, Beetz C, Poplawski NK, Evans DG, Smith MJ (2017) Association of genetic predisposition with solitary schwannoma or meningioma in children and young adults. JAMA Neurol 74:1123–1129. 10.1001/jamaneurol.2017.1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Evans DGR, Salvador H, Chang VY, Erez A, Voss SD, Druker H, Scott HS, Tabori U (2017) Cancer and central nervous system tumor surveillance in pediatric neurofibromatosis 2 and related disorders. Clin Cancer Res 23:e54–e61. 10.1158/1078-0432.CCR-17-0590 [DOI] [PubMed] [Google Scholar]
- 98.Baser ME, Friedman JM, Aeschliman D, Joe H, Wallace AJ, Ramsden RT, Evans DG (2002) Predictors of the risk of mortality in neurofibromatosis 2. Am J Hum Genet 71:715–723. 10.1086/342716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hexter A, Jones A, Joe H, Heap L, Smith MJ, Wallace AJ, Halliday D, Parry A, Taylor A, Raymond L, Shaw A, Afridi S, Obholzer R, Axon P, King AT, English Specialist NF2 Research Group, Friedman JM, Evans DG (2015) Clinical and molecular predictors of mortality in neurofibromatosis 2: a UK National analysis of 1192 patients. J Med Genet 52:699–705. 10.1136/jmedgenet-2015-103290 [DOI] [PubMed] [Google Scholar]
- 100.Halliday D, Emmanouil B, Pretorius P, MacKeith S, Painter S, Tomkins H, Evans DG, Parry A (2017) Genetic severity score predicts clinical phenotype in NF2. J Med Genet 54:657–664. 10.1136/jmedgenet-2017-104519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aboukais R, Zairi F, Bonne NX, Baroncini M, Schapira S, Vincent C, Lejeune JP (2015) Causes of mortality in neurofibromatosis type 2. Br J Neurosurg 29:37–40. 10.3109/02688697.2014.952266 [DOI] [PubMed] [Google Scholar]
- 102.Evans DG, Huson SM, Birch JM (2012) Malignant peripheral nerve sheath tumours in inherited disease. Clin Sarcoma Res 2:17. 10.1186/2045-3329-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Evans DG, Mostaccioli S, Pang D, Fadzil O, Connor M, Pittara M, Champollion N, Wolkenstein P, Thomas N, Ferner RE, Kalamarides M, Peyre M, Papi L, Legius E, Becerra JL, King A, Duff C, Stivaros S, Blanco I (2022) ERN GENTURIS clinical practice guidelines for the diagnosis, treatment, management and surveillance of people with schwannomatosis. Eur J Hum Genet 30:812–817. 10.1038/s41431-022-01086-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jordan JT, Smith MJ, Walker JA, Erdin S, Talkowski ME, Merker VL, Ramesh V, Cai W, Harris GJ, Bredella MA, Seijo M, Suuberg A, Gusella JF, Plotkin SR (2018) Pain correlates with germline mutation in schwannomatosis. Med (Baltim) 97:e9717. 10.1097/MD.0000000000009717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peyre M, Addi A, Parfait B, Fertitta L, Tran S, Wolkenstein P, Kalamarides M (2025) Surgical management of peripheral nerve schwannomas in non-neurofibromatosis type 2 schwannomatosis. Oper neurosurg (Hagerstown) 25 Apr 28. 10.1227/ons.0000000000001595 [DOI] [PubMed]
- 106.Merker VL, Bredella MA, Cai W, Kassarjian A, Harris GJ, Muzikansky A, Nguyen R, Mautner VF, Plotkin SR (2014) Relationship between whole-body tumor burden, clinical phenotype, and quality of life in patients with neurofibromatosis. Am J Med Genet A 164A:1431–1437. 10.1002/ajmg.a.36466 [DOI] [PubMed] [Google Scholar]
- 107.Meller E, Amann A, Malik RA, Kampik D, Matthies C, Üçeyler N (2025) Clinical characterization of neuropathic pain and small fiber impairment in neurofibromatosis. Pain Rep 10:e1285. 10.1097/PR9.0000000000001285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hino U, Tamura R, Toda M (2025) Optimal delivery of pain management in schwannomatosis: a literature review. Ther Clin Risk Manag 21:61–68. 10.2147/TCRM.S362794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Farschtschi SC, Kluwe L, Schön G, Friedrich RE, Matschke J, Glatzel M, Weis J, Hagel C, Mautner VF (2020) Distinctive low epidermal nerve fiber density in schwannomatosis patients provides a major parameter for diagnosis and differential diagnosis. Brain Pathol 30:386–391. 10.1111/bpa.12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Misra K, Ślęczkowska M, Santoro S, Gerrits MM, Mascia E, Marchi M, Salvi E, Smeets HJM, Hoeijmakers JGJ, Martinelli Boneschi FG, Filippi M, Lauria Pinter G, Faber CG, Esposito F (2024) Broadening the genetic spectrum of painful small-fiber neuropathy through whole-exome study in early-onset cases. Int J Mol Sci 25:7248. 10.3390/ijms25137248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mansouri S, Suppiah S, Mamatjan Y et al (2021) Epigenomic, genomic, and transcriptomic landscape of schwannomatosis. Acta Neuropathol 141:101–116. 10.1007/s00401-020-02230-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abe T, Umeki I, Kanno SI, Inoue SI, Niihori T, Aoki Y (2020) LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ 27:1023–1035. 10.1038/s41418-019-0395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H, Cao X, Wang J, Li Q, Zhao Y, Jin X (2021) LZTR1: A promising adaptor of the CUL3 family. Oncol Lett 22:564. 10.3892/ol.2021.12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta K, Harvima IT (2018) Mast cell-neural interactions contribute to pain and itch. Immunol Rev 282:168–187. 10.1111/imr.12622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Plum T, Feyerabend TB, Rodewald HR (2024) Beyond classical immunity: mast cells as signal converters between tissues and neurons. Immunity 57:2723–2736. 10.1016/j.immuni.2024.11.016 [DOI] [PubMed] [Google Scholar]
- 116.Ostrow KL, Donaldson KJ, Caterina MJ, Belzberg A, Hoke A (2019) The secretomes of painful versus nonpainful human schwannomatosis tumor cells differentially influence sensory neuron gene expression and sensitivity. Sci Rep 9:13098. 10.1038/s41598-019-49705-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ostrow KL, Donaldson K, Blakeley J, Belzberg A, Hoke A (2015) Immortalized human Schwann cell lines derived from tumors of schwannomatosis patients. PLoS ONE 10:e0144620. 10.1371/journal.pone.0144620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rubright R, Caterina MJ, Belzberg A, Ostrow KL (2025) Conditioned medium from painful non-NF2 schwannomatosis tumors increases pain behaviors in mice. Sci Rep 15:15851. 10.1038/s41598-025-99820-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perrino MR, Jongmans MCJ, Tomlinson GE et al (2025) Update on cancer and central nervous system tumor purveillance in pediatric NF2-, SMARCB1-, and LZTR1-related schwannomatosis. Clin Cancer Res 31:1400–1406. 10.1158/1078-0432.CCR-24-3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Evans DG, King AT, Bowers NL et al (2019) English specialist NF2 research group. Identifying the deficiencies of current diagnostic criteria for neurofibromatosis 2 using databases of 2777 individuals with molecular testing. Genet Med 21:1525–1533. 10.1038/s41436-018-0384-y [DOI] [PubMed] [Google Scholar]
- 121.Coy S, Rashid R, Stemmer-Rachamimov A, Santagata S (2020) An update on the CNS manifestations of neurofibromatosis type 2. Acta Neuropathol 139:643–665. 10.1007/s00401-019-02029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith MJ, Kulkarni A, Rustad C, Bowers NL, Wallace AJ, Holder SE, Heiberg A, Ramsden RT, Evans DG (2012) Vestibular schwannomas occur in schwannomatosis and should not be considered an exclusion criterion for clinical diagnosis. Am J Med Genet A 158A:215–219. 10.1002/ajmg.a.34376 [DOI] [PubMed] [Google Scholar]
- 123.Wu J, Kong M, Bi Q (2015) Identification of a novel germline SMARCB1 nonsense mutation in a family manifesting both schwannomatosis and unilateral vestibular schwannoma. J Neurooncol 125:439–441. 10.1007/s11060-015-1918-7 [DOI] [PubMed] [Google Scholar]
- 124.Alaidarous A, Parfait B, Ferkal S, Cohen J, Wolkenstein P, Mazereeuw-Hautier J (2019) Segmental schwannomatosis: characteristics in 12 patients. Orphanet J Rare Dis 14:207. 10.1186/s13023-019-1176-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Farschtschi S, Mautner VF, Pham M, Nguyen R, Kehrer-Sawatzki H, Hutter S, Friedrich RE, Schulz A, Morrison H, Jones DT, Bendszus M, Bäumer P (2016) Multifocal nerve lesions and LZTR1 germline mutations in segmental schwannomatosis. Ann Neurol 80:625–628. 10.1002/ana.24753 [DOI] [PubMed] [Google Scholar]
- 126.Lee JH, Jeong JS, Chae KJ, Han YH, Kim SR, Lee YC (2022) A rare case of familial schwannomatosis showing intrafamilial variability with identification of a shared novel germline SMARCB1 mutation. Med (Kaunas) 58:1592. 10.3390/medicina58111592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bacci C, Sestini R, Provenzano A, Paganini I, Mancini I, Porfirio B, Vivarelli R, Genuardi M, Papi L (2010) Schwannomatosis associated with multiple meningiomas due to a familial SMARCB1 mutation. Neurogenetics 11:73–80. 10.1007/s10048-009-0204-2 [DOI] [PubMed] [Google Scholar]
- 128.Christiaans I, Kenter SB, Brink HC, van Os TA, Baas F, van den Munckhof P, Kidd AM, Hulsebos TJ (2011) Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet 48:93–97. 10.1136/jmg.2010.082420 [DOI] [PubMed] [Google Scholar]
- 129.Prescott TE, Smith MJ, Evans DG (2012) Comment on the Article germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the Falx cerebri by Van Den Munckhof et al. Neurogenetics 13:103–104. 10.1007/s10048-011-0309-2 [DOI] [PubMed] [Google Scholar]
- 130.Smith MJ (2015) Germline and somatic mutations in meningiomas. Cancer Genet 208:107–114. 10.1016/j.cancergen.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 131.Fountain DM, Smith MJ, O’Leary C, Pathmanaban ON, Roncaroli F, Bobola N, King AT, Evans DG (2021) The spatial phenotype of genotypically distinct meningiomas demonstrate potential implications of the embryology of the meninges. Oncogene 40:875–884. 10.1038/s41388-020-01568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hadfield KD, Smith MJ, Trump D, Newman WG, Evans DG (2010) SMARCB1 mutations are not a common cause of multiple meningiomas. J Med Genet 47:567–568. 10.1136/jmg.2009.075721 [DOI] [PubMed] [Google Scholar]
- 133.Smith MJ, Higgs JE, Bowers NL, Halliday D, Paterson J, Gillespie J, Huson SM, Freeman SR, Lloyd S, Rutherford SA, King AT, Wallace AJ, Ramsden RT, Evans DG (2011) Cranial meningiomas in 411 neurofibromatosis type 2 (NF2) patients with proven gene mutations: clear positional effect of mutations, but absence of female severity effect on age at onset. J Med Genet 48:261–265. 10.1136/jmg.2010.085241 [DOI] [PubMed] [Google Scholar]
- 134.Schmitz U, Mueller W, Weber M, Sévenet N, Delattre O, von Deimling A (2001) INI1 mutations in meningiomas at a potential hotspot in exon 9. Br J Cancer 84:199–201. 10.1054/bjoc.2000.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rieske P, Zakrzewska M, Piaskowski S, Jaskólski D, Sikorska B, Papierz W, Zakrzewski K, Liberski PP (2003) Molecular heterogeneity of meningioma with INI1 mutation. Mol Pathol 56:299–301. 10.1136/mp.56.5.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Clark VE, Harmancı AS, Bai H et al (2016) Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet 48:1253–1259. 10.1038/ng.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang AS, Jamshidi AO, Oh N, Sahyouni R, Nowroozizadeh B, Kim R, Hsu FPK, Bota D (2018) Somatic SMARCB1 mutation in sporadic multiple meningiomas: case report. Front Neurol 9:919. 10.3389/fneur.2018.00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hulsebos TJ, Kenter S, Siebers-Renelt U, Hans V, Wesseling P, Flucke U (2014) SMARCB1 involvement in the development of leiomyoma in a patient with schwannomatosis. Am J Surg Pathol 38:421–425. 10.1097/PAS.0000000000000110 [DOI] [PubMed] [Google Scholar]
- 139.Mehine M, Kaasinen E, Heinonen HR, Mäkinen N, Kämpjärvi K, Sarvilinna N, Aavikko M, Vähärautio A, Pasanen A, Bützow R, Heikinheimo O, Sjöberg J, Pitkänen E, Vahteristo P, Aaltonen LA (2016) Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci U S A 113:1315–1320. 10.1073/pnas.1518752113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eelloo JA, Smith MJ, Bowers NL, Ealing J, Hulse P, Wylie JP, Shenjere P, Clarke NW, Soh C, Whitehouse RW, Jones M, Duff C, Freemont A, Gareth Evans D (2019) Multiple primary malignancies associated with a germline SMARCB1 pathogenic variant. Fam Cancer 18:445–449. 10.1007/s10689-019-00138-4 [DOI] [PubMed] [Google Scholar]
- 141.Sørensen SA, Mulvihill JJ, Nielsen A (1986) Long-term follow-up of von Recklinghausen neurofibromatosis. Survival and malignant neoplasms. N Engl J Med 314:1010–1015. 10.1056/NEJM198604173141603 [DOI] [PubMed] [Google Scholar]
- 142.Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A (2002) Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet 39:311–314. 10.1136/jmg.39.5.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Evans DG, Birch JM, Ramsden RT, Sharif S, Baser ME (2006) Malignant transformation and new primary tumours after therapeutic radiation for benign disease: substantial risks in certain tumour prone syndromes. J Med Genet 43:289–294. 10.1136/jmg.2005.036319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hulsebos TJ, Kenter S, Baas F, Nannenberg EA, Bleeker FE, van Minkelen R, van den Ouweland AM, Wesseling P, Flucke U (2016) Type 1 papillary renal cell carcinoma in a patient with schwannomatosis: mosaic versus loss of SMARCB1 expression in respectively schwannoma and renal tumor cells. Genes Chromosomes Cancer 55:350–354. 10.1002/gcc.22338 [DOI] [PubMed] [Google Scholar]
- 145.Paganini I, Sestini R, Cacciatore M, Capone GL, Candita L, Paolello C, Sbaraglia M, Dei Tos AP, Rossi S, Papi L (2015) Broadening the spectrum of SMARCB1-associated malignant tumors: a case of uterine leiomyosarcoma in a patient with schwannomatosis. Hum Pathol 46:1226–1231. 10.1016/j.humpath.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 146.Andres S, Huang K, Shatara M, Abdelbaki MS, Ranalli M, Finlay J, Gupta A (2024) Rhabdoid tumor predisposition syndrome: A historical review of treatments and outcomes for associated pediatric malignancies. Pediatr Blood Cancer 71:e30979. 10.1002/pbc.30979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakano Y, Acker M, Druker H, van Engelen K, Meyn MS, Wasserman JD, Venier RE, Goudie C, Stosic A, Huang A, Greer MC, Malkin D, Villani A, Gallinger B (2024) Late-onset tumors in rhabdoid tumor predisposition syndrome type-1 (RTPS1) and implications for surveillance. Eur J Hum Genet 32:1474–1482. 10.1038/s41431-024-01674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hasselblatt M, Nagel I, Oyen F et al (2014) SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol 128:453–456. 10.1007/s00401-014-1323-x [DOI] [PubMed] [Google Scholar]
- 149.Meyers SP, Khademian ZP, Biegel JA, Chuang SH, Korones DN, Zimmerman RA (2006) Primary intracranial atypical teratoid/rhabdoid tumors of infancy and childhood: MRI features and patient outcomes. AJNR Am J Neuroradiol 27:962–971 [PMC free article] [PubMed] [Google Scholar]
- 150.Frühwald MC, Nemes K, Boztug H et al (2021) Current recommendations for clinical surveillance and genetic testing in rhabdoid tumor predisposition: a report from the SIOPE host genome working group. Fam Cancer 20:305–316. 10.1007/s10689-021-00229-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schaefer IM, Hornick JL (2021) SWI/SNF complex-deficient soft tissue neoplasms: an update. Semin Diagn Pathol 38:222–231. 10.1053/j.semdp.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23:1231–1251. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dho YS, Kim SK, Cheon JE, Park SH, Wang KC, Lee JY, Phi JH (2015) Investigation of the location of atypical teratoid/rhabdoid tumor. Childs Nerv Syst 31:1305–1311. 10.1007/s00381-015-2739-x [DOI] [PubMed] [Google Scholar]
- 154.Chi SN, Zimmerman MA, Yao X et al (2009) Intensive multimodality treatment for children with newly diagnosed CNS atypical teratoid rhabdoid tumor. J Clin Oncol 27:385–389. 10.1200/JCO.2008.18.7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dufour C, Beaugrand A, Le Deley MC, Bourdeaut F, André N, Leblond P, Bertozzi AI, Frappaz D, Rialland X, Fouyssac F, Edan C, Grill J, Quidot M, Varlet P (2012) Clinicopathologic prognostic factors in childhood atypical teratoid and rhabdoid tumor of the central nervous system: a multicenter study. Cancer 118:3812–3821. 10.1002/cncr.26684 [DOI] [PubMed] [Google Scholar]
- 156.Lafay-Cousin L, Hawkins C, Carret AS, Johnston D, Zelcer S, Wilson B, Jabado N, Scheinemann K, Eisenstat D, Fryer C, Fleming A, Mpofu C, Larouche V, Strother D, Bouffet E, Huang A (2012) Central nervous system atypical teratoid rhabdoid tumours: the Canadian paediatric brain tumour consortium experience. Eur J Cancer 48:353–359. 10.1016/j.ejca.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 157.Bartelheim K, Nemes K, Seeringer A et al (2016) Improved 6-year overall survival in AT/RT - results of the registry study Rhabdoid 2007. Cancer Med 5:1765–1775. 10.1002/cam4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fossey M, Li H, Afzal S, Carret AS et al (2017) Atypical teratoid rhabdoid tumor in the first year of life: the Canadian ATRT registry experience and review of the literature. J Neurooncol 132:155–162. 10.1007/s11060-016-2353-0 [DOI] [PubMed] [Google Scholar]
- 159.Gastberger K, Fincke VE, Mucha M, Siebert R, Hasselblatt M, Frühwald MC (2023) Current molecular and clinical landscape of ATRT - the link to future therapies. Cancer Manag Res 15:1369–1393. 10.2147/CMAR.S379451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Z, Yang J, Liu X, Liu W (2025) Clinical characteristics of patients with atypical teratoid/rhabdoid tumors: a monocentric retrospective analysis. Front Pediatr 13:1463510. 10.3389/fped.2025.1463510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Raisanen J, Biegel JA, Hatanpaa KJ, Judkins A, White CL, Perry A (2005) Chromosome 22q deletions in atypical teratoid/rhabdoid tumors in adults. Brain Pathol 15:23–28. 10.1111/j.1750-3639.2005.tb00096.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zarovnaya EL, Pallatroni HF, Hug EB, Ball PA, Cromwell LD, Pipas JM, Fadul CE, Meyer LP, Park JP, Biegel JA, Perry A, Rhodes CH (2007) Atypical teratoid/rhabdoid tumor of the spine in an adult: case report and review of the literature. J Neurooncol 84:49–55. 10.1007/s11060-007-9339-x [DOI] [PubMed] [Google Scholar]
- 163.Makuria AT, Rushing EJ, McGrail KM, Hartmann DP, Azumi N, Ozdemirli M (2008) Atypical teratoid rhabdoid tumor (AT/RT) in adults: review of four cases. J Neurooncol 88:321–330. 10.1007/s11060-008-9571-z [DOI] [PubMed] [Google Scholar]
- 164.Chan V, Marro A, Findlay JM, Schmitt LM, Das S (2018) A systematic review of atypical teratoid rhabdoid tumor in adults. Front Oncol 8:567. 10.3389/fonc.2018.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Johann PD, Bens S, Oyen F et al (2018) Sellar region atypical teratoid/rhabdoid tumors (ATRT) in adults display DNA methylation profiles of the ATRT-MYC subgroup. Am J Surg Pathol 42:506–511. 10.1097/PAS.0000000000001023 [DOI] [PubMed] [Google Scholar]
- 166.Paolini MA, Kipp BR, Sukov WR et al (2018) Sellar region atypical teratoid/rhabdoid tumors in adults: clinicopathological characterization of five cases and review of the literature. J Neuropathol Exp Neurol 77:1115-1121. 10.1093/jnen/nly091 [DOI] [PubMed]
- 167.Voisin MR, Ovenden C, Tsang DS, Gupta AA, Huang A, Gao AF, Diamandis P, Almeida JP, Gentili F (2019) Atypical teratoid/rhabdoid sellar tumor in an adult with a Familial history of a germline SMARCB1 mutation: case report and review of the literature. World Neurosurg 127:336–345. 10.1016/j.wneu.2019.04.083 [DOI] [PubMed] [Google Scholar]
- 168.Duan Z, Yao K, Yang S, Qu Y, Ren M, Zhang Y, Fan T, Zhao H, Gao J, Feng J, Fan X, Qi X (2022) Primary adult sellar SMARCB1/INI1-deficient tumor represents a subtype of atypical teratoid/rhabdoid tumor. Mod Pathol 35:1910–1920. 10.1038/s41379-022-01127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Georgountzos G, Gkalonakis I, Kyriakopoulos G, Doukaki C, Vassiliadi DA, Barkas K (2024) A rare case of atypical teratoid rhabdoid tumor at the sellar region in an adult: case report and review of literature. Brain Spine 4:104138. 10.1016/j.bas.2024.104138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Thangjam N, Dey B, Raphael V, Mishra J, Lynser D, Ghosh T, Kumar S, Bhattacharjee M (2024) Adult primary central nervous system atypical teratoid/rhabdoid tumor metastasizing to the cervical lymph node. Cureus 16:e73742. 10.7759/cureus [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kordes U, Gesk S, Frühwald MC, Graf N, Leuschner I, Hasselblatt M, Jeibmann A, Oyen F, Peters O, Pietsch T, Siebert R, Schneppenheim R (2010) Clinical and molecular features in patients with atypical teratoid rhabdoid tumor or malignant rhabdoid tumor. Genes Chromosomes Cancer 49:176–181. 10.1002/gcc.20729 [DOI] [PubMed] [Google Scholar]
- 172.Upadhyaya SA, Robinson GW, Onar-Thomas A et al (2021) Relevance of molecular groups in children with newly diagnosed atypical teratoid rhabdoid tumor: results from prospective St. Jude multi-institutional trials. Clin Cancer Res 27:2879–2889. 10.1158/1078-0432.CCR-20-4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nemes K, Johann PD, Steinbügl M (2022) Infants and newborns with atypical teratoid rhabdoid tumors (ATRT) and extracranial malignant rhabdoid tumors (eMRT) in the EU-RHAB registry: a unique and challenging population. Cancers (Basel) 14:2185. 10.3390/cancers14092185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Abu Arja MH, Patel P, Shah SH, Auletta JJ, Meyer EK, Conley SE, Aldrink JH, Pindrik JA, AbdelBaki MS (2018) Synchronous central nervous system atypical teratoid/rhabdoid tumor and malignant rhabdoid tumor of the kidney: case report of a long-term survivor and review of the literature. World Neurosurg 111:6–15. 10.1016/j.wneu.2017.11.158 [DOI] [PubMed] [Google Scholar]
- 175.Pinto EM, Hamideh D, Bahrami A, Orr BA, Lin T, Pounds S, Zambetti GP, Pappo AS, Gajjar A, Agnihotri S, Broniscer A (2018) Malignant rhabdoid tumors originating within and outside the central nervous system are clinically and molecularly heterogeneous. Acta Neuropathol 136:315–326. 10.1007/s00401-018-1814-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim T, Phi JH (2025) Rhabdoid tumor predisposition syndrome: a comprehensive review of genetics, clinical manifestations, and management. J Korean Neurosurg Soc 2025 Mar 27. 10.3340/jkns.2025.0014Epub ahead of print [DOI] [PMC free article] [PubMed]
- 177.Sredni ST, Tomita T (2015) Rhabdoid tumor predisposition syndrome. Pediatr Dev Pathol 18:49–58. 10.2350/14-07-1531-MISC.1 [DOI] [PubMed] [Google Scholar]
- 178.Han ZY, Richer W, Fréneaux P et al (2016) The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun 7:10421. 10.1038/ncomms10421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vitte J, Gao F, Coppola G, Judkins AR, Giovannini M (2017) Timing of Smarcb1 and Nf2 inactivation determines schwannoma versus rhabdoid tumor development. Nat Commun 8:300. 10.1038/s41467-017-00346-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fountain DM, Sauka-Spengler T (2023) The SWI/SNF complex in neural crest cell development and disease. Annu Rev Genomics Hum Genet 24:203–223. 10.1146/annurev-genom-011723-082913 [DOI] [PubMed] [Google Scholar]
- 181.Tekautz TM, Fuller CE, Blaney S, Fouladi M, Broniscer A, Merchant TE, Krasin M, Dalton J, Hale G, Kun LE, Wallace D, Gilbertson RJ, Gajjar A (2005) Atypical teratoid/rhabdoid tumors (ATRT): improved survival in children 3 years of age and older with radiation therapy and high-dose alkylator-based chemotherapy. J Clin Oncol 23:1491–1499. 10.1200/JCO.2005.05.187 [DOI] [PubMed] [Google Scholar]
- 182.Squire SE, Chan MD, Marcus KJ (2007) Atypical teratoid/rhabdoid tumor: the controversy behind radiation therapy. J Neurooncol 81:97–111. 10.1007/s11060-006-9196-z [DOI] [PubMed] [Google Scholar]
- 183.Shirai R, Osumi T, Terashima K et al (2020) High prevalence of SMARCB1 constitutional abnormalities including mosaicism in malignant rhabdoid tumors. Eur J Hum Genet 28:1124–1128. 10.1038/s41431-020-0614-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fukushima H, Yamasaki K, Sakaida M, Tsujio N, Okuno T, Ishii N, Okada K, Fujisaki H, Matsusaka Y, Sakamoto H, Yoneda A, Hara J, Inoue T (2021) Rhabdoid tumor predisposition syndrome with renal tumor 10 years after brain tumor. Pathol Int 71:155–160. 10.1111/pin.13056 [DOI] [PubMed] [Google Scholar]
- 185.Baker TG, Lyons MJ, Leddy L, Parham DM, Welsh CT (2021) Epithelioid sarcoma arising in a long-term survivor of an atypical teratoid/rhabdoid tumor in a patient with rhabdoid tumor predisposition syndrome. Pediatr Dev Pathol 24:164–168. 10.1177/1093526620986492 [DOI] [PubMed] [Google Scholar]
- 186.Kehrer-Sawatzki H, Kordes U, Seiffert S, Summerer A, Hagel C, Schüller U, Farschtschi S, Schneppenheim R, Bendszus M, Godel T, Mautner VF (2018) Co-occurrence of schwannomatosis and rhabdoid tumor predisposition syndrome 1. Mol Genet Genomic Med 6:627–637. 10.1002/mgg3.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Forest F, David A, Arrufat S, Pierron G, Ranchere-Vince D, Stephan JL, Clemenson A, Delattre O, Bourdeaut F (2012) Conventional chondrosarcoma in a survivor of rhabdoid tumor: enlarging the spectrum of tumors associated with SMARCB1 germline mutations. Am J Surg Pathol 36:1892–1896. 10.1097/PAS.0b013e31826cbe7a [DOI] [PubMed] [Google Scholar]
- 188.Upadhyaya SA, McGee RB, Wilky BA, Broniscer A (2018) Malignant progression of a peripheral nerve sheath tumor in the setting of rhabdoid tumor predisposition syndrome. Pediatr Blood Cancer 65:e27030. 10.1002/pbc.27030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dunham C (2015) Uncommon pediatric tumors of the posterior fossa: pathologic and molecular features. Childs Nerv Syst 31:1729–1737. 10.1007/s00381-015-2735-1 [DOI] [PubMed] [Google Scholar]
- 190.Johann PD, Hovestadt V, Thomas C et al (2017) Cribriform neuroepithelial tumor: molecular characterization of a SMARCB1-deficient non-rhabdoid tumor with favorable long-term outcome. Brain Pathol 27:411–418. 10.1111/bpa.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tauziède-Espariat A, Guerrini-Rousseau L, Puget S, Masliah-Planchon J, Bourdeaut F, Hasty L, Grill J, Dangouloff-Ros V, Boddaert N, Chrétien F, Lechapt E, Dufour C, Varlet P (2021) A novel case of cribriform neuroepithelial tumor: a potential diagnostic pitfall in the ventricular system. Pediatr Blood Cancer 68:e29037. 10.1002/pbc.29037 [DOI] [PubMed] [Google Scholar]
- 192.Hasselblatt M, Oyen F, Gesk S, Kordes U, Wrede B, Bergmann M, Schmid H, Frühwald MC, Schneppenheim R, Siebert R, Paulus W (2009) Cribriform neuroepithelial tumor (CRINET): a nonrhabdoid ventricular tumor with INI1 loss and relatively favorable prognosis. J Neuropathol Exp Neurol 68:1249–1255. 10.1097/NEN.0b013e3181c06a51 [DOI] [PubMed] [Google Scholar]
- 193.Rigsby RK, Brahmbhatt P, Desai AB, Bathla G, Ebner BA, Gupta V, Vibhute P, Agarwal AK (2023) Newly recognized CNS tumors in the 2021 world health organization classification: imaging overview with histopathologic and genetic correlation. AJNR Am J Neuroradiol 44:367–380. 10.3174/ajnr.A7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cyrta J, Masliah-Planchon J, Hoare O et al (2025) SMARCB1-deficient malignant melanocytic uveal tumours: a new neural crest-derived tumour entity with SMARCB1-related germline predisposition. J Pathol 265:357–371. 10.1002/path.6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Burnside RD (2015) 22q11.21 deletion syndromes: a review of proximal, central, and distal deletions and their associated features. Cytogenet Genome Res 146:89–99. 10.1159/000438708 [DOI] [PubMed] [Google Scholar]
- 196.Wieser R, Fritz B, Ullmann R, Müller I, Galhuber M, Storlazzi CT, Ramaswamy A, Christiansen H, Shimizu N, Rehder H (2005) Novel rearrangement of chromosome band 22q11.2 causing 22q11 microdeletion syndrome-like phenotype and rhabdoid tumor of the kidney. Hum Mutat 26:78–83. 10.1002/humu.20195. PMID: 15957176 [DOI] [PubMed]
- 197.Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, Nikkel SM, Zuppan CW, Wainwright LM, Zhang F, Biegel JA (2007) High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet 122:117–127. 10.1007/s00439-007-0386-3 [DOI] [PubMed] [Google Scholar]
- 198.Beddow RA, Smith M, Kidd A, Corbett R, Hunter AG (2011) Diagnosis of distal 22q11.2 deletion syndrome in a patient with a teratoid/rhabdoid tumour. Eur J Med Genet 54:295–298. 10.1016/j.ejmg.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 199.Toth G, Zraly CB, Thomson TL, Jones C, Lapetino S, Muraskas J, Zhang J, Dingwall AK (2011) Congenital anomalies and rhabdoid tumor associated with 22q11 germline deletion and somatic inactivation of the SMARCB1 tumor suppressor. Genes Chromosomes Cancer 50:379–388. 10.1002/gcc.20862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chakrapani AL, White CR, Korcheva V, White K, Lofgren S, Zonana J, Moore S, Krol A, Mansoor A (2012) Congenital extrarenal malignant rhabdoid tumor in an infant with distal 22q11.2 deletion syndrome: the importance of SMARCB1. Am J Dermatopathol 34:e77–80. 10.1097/DAD.0b013e31825793c3 [DOI] [PubMed] [Google Scholar]
- 201.Bosse KR, Shukla AR, Pawel B, Chikwava KR, Santi M, Tooke L, Castagna K, Biegel JA, Bagatell R (2014) Malignant rhabdoid tumor of the bladder and ganglioglioma in a 14 year-old male with a germline 22q11.2 deletion. Cancer Genet 207:415–419. 10.1016/j.cancergen.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mari F, Marozza A, Mencarelli MA et al (2015) Coffin-Siris and Nicolaides-Baraitser syndromes are a common well recognizable cause of intellectual disability. Brain Dev 37:527–536. 10.1016/j.braindev.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 203.Monroe GR, Frederix GW, Savelberg SM et al (2016) Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med 18:949–956. 10.1038/gim.2015.200 [DOI] [PubMed] [Google Scholar]
- 204.Han JY, Jang JH, Park J, Lee IG (2018) Targeted next-generation sequencing of Korean patients with developmental delay and/or intellectual disability. Front Pediatr 6:391. 10.3389/fped.2018.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sekiguchi F, Tsurusaki Y, Okamoto N et al (2019) Genetic abnormalities in a large cohort of Coffin-Siris syndrome patients. J Hum Genet 64:1173–1186. 10.1038/s10038-019-0667-4 [DOI] [PubMed] [Google Scholar]
- 206.Cheng SSW, Luk HM, Mok MT, Leung SS, Lo IFM (2021) Genotype and phenotype in 18 Chinese patients with Coffin-Siris syndrome. Am J Med Genet A 185:2250–2261. 10.1002/ajmg.a.62187 [DOI] [PubMed] [Google Scholar]
- 207.Guo Z, Bai J, Liu Y, Zhang X, Yang W, Wang J, Zhang Y, Xiao H, Hao B, Liao S (2024) A novel mutation in SMARCB1 associated with adult Coffin-Siris syndrome and meningioma. Acta Biochim Biophys Sin (Shanghai) 13. 10.3724/abbs.2024204 [DOI] [PMC free article] [PubMed]
- 208.Keskinen S, Paakkola T, Mattila M, Hietala M, Koillinen H, Laine J, Haanpää MK (2024) Prenatal Coffin-Siris syndrome: expanding the phenotypic and genotypic spectrum of the disease. Pediatr Dev Pathol 27:181–186. 10.1177/10935266231210155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schmetz A, Lüdecke HJ, Surowy H (2024) Delineation of the adult phenotype of Coffin-Siris syndrome in 35 individuals. Hum Genet 143:71–84. 10.1007/s00439-023-02622-5 [DOI] [PubMed] [Google Scholar]
- 210.Bramswig NC, Lüdecke HJ, Alanay Y et al (2015) Exome sequencing unravels unexpected differential diagnoses in individuals with the tentative diagnosis of Coffin-Siris and Nicolaides-Baraitser syndromes. Hum Genet 134:553–568. 10.1007/s00439-015-1535-8 [DOI] [PubMed] [Google Scholar]
- 211.Santen GW, Aten E, Sun Y et al (2012) Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet 44:379–380. 10.1038/ng.2217 [DOI] [PubMed] [Google Scholar]
- 212.Miyake N, Tsurusaki Y, Matsumoto N (2014) Numerous BAF complex genes are mutated in Coffin-Siris syndrome. Am J Med Genet C Semin Med Genet 166 C:257–261. 10.1002/ajmg.c.31406 [DOI] [PubMed] [Google Scholar]
- 213.Kosho T, Miyake N, Carey JC (2014) Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am J Med Genet C Semin Med Genet 166 C:241–251. 10.1002/ajmg.c.31415 [DOI] [PubMed] [Google Scholar]
- 214.Kosho T, Okamoto N (2014) Coffin-Siris Syndrome International Collaborators. Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am J Med Genet C Semin Med Genet 166 C:262–275. 10.1002/ajmg.c.31407 [DOI] [PubMed]
- 215.Filatova A, Rey LK, Lechler MB, Schaper J, Hempel M, Posmyk R, Szczaluba K, Santen GWE, Wieczorek D, Nuber UA (2019) Mutations in SMARCB1 and in other Coffin-Siris syndrome genes lead to various brain midline defects. Nat Commun 10:2966. 10.1038/s41467-019-10849-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bögershausen N, Wollnik B (2018) Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front Mol Neurosci 11:252. 10.3389/fnmol.2018.00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Diets IJ, Prescott T, Champaigne NL, Mancini GMS, Krossnes B, Frič R, Kocsis K, Jongmans MCJ, Kleefstra T (2019) A recurrent de novo missense pathogenic variant in SMARCB1 causes severe intellectual disability and choroid plexus hyperplasia with resultant hydrocephalus. Genet Med 21:572–579. 10.1038/s41436-018-0079-4 [DOI] [PubMed] [Google Scholar]
- 218.Kleefstra T, Kramer JM, Neveling K et al (2012) Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet 91:73–82. 10.1016/j.ajhg.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Campeau PM, Kasperaviciute D, Lu JT et al (2014) The genetic basis of DOORS syndrome: an exome-sequencing study. Lancet Neurol 13:44–58. 10.1016/S1474-4422(13)70265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Shaikh TH, Kurahashi H, Emanuel BS (2001) Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: an update and literature review. Genet Med 3:6–13. 10.1097/00125817-200101000-00003 [DOI] [PubMed] [Google Scholar]
- 221.Lafay-Cousin L, Payne E, Strother D, Chernos J, Chan M, Bernier FP (2009) Goldenhar phenotype in a child with distal 22q11.2 deletion and intracranial atypical teratoid rhabdoid tumor. Am J Med Genet A 149A:2855–2859. 10.1002/ajmg.a.33119 [DOI] [PubMed] [Google Scholar]
- 222.Tan TY, Collins A, James PA, McGillivray G, Stark Z, Gordon CT, Leventer RJ, Pope K, Forbes R, Crolla JA, Ganesamoorthy D, Burgess T, Bruno DL, Slater HR, Farlie PG, Amor DJ (2011) Phenotypic variability of distal 22q11.2 copy number abnormalities. Am J Med Genet A 155A:1623–1633. 10.1002/ajmg.a.34051 [DOI] [PubMed] [Google Scholar]
- 223.Lee JC, Tran QT, McGee RB et al (2024) Atypical teratoid/rhabdoid tumour-TYR subtype arising in the setting of germline ring chromosome 22: an uncommon form of tumour predisposition. Neuropathol Appl Neurobiol 50:e12971. 10.1111/nan.12971 [DOI] [PubMed] [Google Scholar]
- 224.Vries RG, Bezrookove V, Zuijderduijn LM, Kia SK, Houweling A, Oruetxebarria I, Raap AK, Verrijzer CP (2005) Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev 19:665–670. 10.1101/gad.3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13:339–346. 10.1038/nsmb1071 [DOI] [PubMed] [Google Scholar]
- 226.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR (2009) An embryonic stem cell chromatin remodeling complex, esbaf, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A 106:5187–5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, Zhu J, Fan G, Archer TK, Wang Y (2014) Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Rep 3:460–474. 10.1016/j.stemcr.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Langer LF, Ward JM, Archer TK (2019) Tumor suppressor SMARCB1 suppresses super-enhancers to govern hESC lineage determination. Elife 8:e45672. 10.7554/eLife.45672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Carmel-Gross I, Levy E, Armon L, Yaron O, Waldman Ben-Asher H, Urbach A (2020) Human pluripotent stem cell fate regulation by SMARCB1. Stem Cell Rep 15:1037–1046. 10.1016/j.stemcr.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Saha D, Animireddy S, Bartholomew B (2024) The SWI/SNF ATP-dependent chromatin remodeling complex in cell lineage priming and early development. Biochem Soc Trans 52:603–616. 10.1042/BST20230416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE (2002) Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 21:5193–5203. 10.1038/sj.onc.1205706 [DOI] [PubMed] [Google Scholar]
- 232.Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, Kalpana GV (2002) Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol 22:5975–5988. 10.1128/MCB.22.16.5975-5988.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Versteege I, Medjkane S, Rouillard D, Delattre O (2002) A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene 21:6403–6412. 10.1038/sj.onc.1205841 [DOI] [PubMed] [Google Scholar]
- 234.Medjkane S, Novikov E, Versteege I, Delattre O (2004) The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res 64:3406–3413. 10.1158/0008-5472.CAN-03-3004 [DOI] [PubMed] [Google Scholar]
- 235.Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE (2010) Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res 70:1854–1865. 10.1158/0008-5472.CAN-09-1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nguyen LT, Hains AE, Aziz-Zanjani MO, Dalsass M, Farooqee SBUD, Lu Y, Jackson PK, Van Rechem C (2024) Absence of SMARCB1 in rhabdoid tumor cells increases sensitivity to translation Inhibition and alters translation efficiency of specific mRNAs. J Biol Chem 300:107988. 10.1016/j.jbc.2024.107988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hollmann TJ, Hornick JL (2011) INI1-deficient tumors: diagnostic features and molecular genetics. Am J Surg Pathol 35:e47–63. 10.1097/PAS.0b013e31822b325b [DOI] [PubMed] [Google Scholar]
- 238.Wang L, Leite de Oliveira R, Wang C, Fernandes Neto JM, Mainardi S, Evers B, Lieftink C, Morris B, Jochems F, Willemsen L, Beijersbergen RL, Bernards R (2017) High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep 21:773–783. 10.1016/j.celrep.2017.09.085 [DOI] [PubMed] [Google Scholar]
- 239.Hong SH, Son KH, Ha SY, Wee TI, Choi SK, Won JE, Han HD, Ro Y, Park YM, Eun JW, Nam SW, Han JW, Kang K, You JS (2021) Nucleoporin 210 serves a key scaffold for SMARCB1 in liver cancer. Cancer Res 81:356–370. 10.1158/0008-5472 [DOI] [PubMed] [Google Scholar]
- 240.Favre M, Butticaz C, Stevenson B, Jongeneel CV, Telenti A (2003) High frequency of alternative splicing of human genes participating in the HIV-1 life cycle: a model using TSG101, betatrcp, PPIA, INI1, NAF1, and PML. J Acquir Immune Defic Syndr 34:127–133. 10.1097/00126334-200310010-00002 [DOI] [PubMed] [Google Scholar]
- 241.Bruder CE, Dumanski JP, Kedra D (1999) The mouse ortholog of the human SMARCB1 gene encodes two splice forms. Biochem Biophys Res Commun 257:886–890. 10.1006/bbrc.1999.0563 [DOI] [PubMed] [Google Scholar]
- 242.Xu Y, Yan W, Chen X (2010) SNF5, a core component of the SWI/SNF complex, is necessary for p53 expression and cell survival, in part through eIF4E. Oncogene 29:4090–4100. 10.1038/onc.2010.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Reincke BS, Rosson GB, Oswald BW, Wright CF (2003) INI1 expression induces cell cycle arrest and markers of senescence in malignant rhabdoid tumor cells. J Cell Physiol 194:303–313. 10.1002/jcp.10201 [DOI] [PubMed] [Google Scholar]
- 244.Pyeon D, Price L, Park IW (2015) Comparative molecular genetic analysis of simian and human HIV-1 integrase interactor INI1/SMARCB1/SNF5. Arch Virol 160:3085–3091. 10.1007/s00705-015-2585-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CW (2017) The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 8:14648. 10.1038/ncomms14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Nakayama RT, Pulice JL, Valencia AM et al (2017) SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49:1613–1623. 10.1038/ng.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang X, Lee RS, Alver BH et al (2017) SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49:289–295. 10.1038/ng.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wang W, Côté J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR (1996) Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15:5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 249.Lemon B, Inouye C, King DS, Tjian R (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924–928. 10.1038/414924a [DOI] [PubMed] [Google Scholar]
- 250.Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, Valencia AM, Poynter SJ, Cassel SH, Ranish JA, Kadoch C (2018) Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175:1272–1288e20. 10.1016/j.cell.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Michel BC, D’Avino AR, Cassel SH et al (2018) A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol 20:1410–1420. 10.1038/s41556-018-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang X, Wang S, Troisi EC, Howard TP, Haswell JR, Wolf BK, Hawk WH, Ramos P, Oberlick EM, Tzvetkov EP, Ross A, Vazquez F, Hahn WC, Park PJ, Roberts CWM (2019) BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat Commun 10:1881. 10.1038/s41467-019-09891-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kadoch C, Crabtree GR (2015) Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 1:e1500447. 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sokpor G, Xie Y, Rosenbusch J, Tuoc T (2017) Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front Mol Neurosci 10:243. 10.3389/fnmol.2017.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B (2010) SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell 38:590–602. 10.1016/j.molcel.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu N, Balliano A, Hayes JJ (2011) Mechanism(s) of SWI/SNF-induced nucleosome mobilization. ChemBioChem 12:196–204. 10.1002/cbic.201000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Morrison EA, Sanchez JC, Ronan JL, Farrell DP, Varzavand K, Johnson JK, Gu BX, Crabtree GR, Musselman CA (2017) DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun 8:16080. 10.1038/ncomms16080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Valencia AM, Collings CK, Dao HT et al (2019) Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 179:1342–1356e23. 10.1016/j.cell.2019.10.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Valencia AM, Sankar A, van der Sluijs PJ, Satterstrom FK, Fu J, Talkowski ME, Vergano SAS, Santen GWE, Kadoch C (2023) Landscape of mSWI/SNF chromatin remodeling complex perturbations in neurodevelopmental disorders. Nat Genet 55:1400–1412. 10.1038/s41588-023-01451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y (2020) Structure of nucleosome-bound human BAF complex. Science 367:875–881. 10.1126/science.aaz9761 [DOI] [PubMed] [Google Scholar]
- 261.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW (2010) Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18:316–328. 10.1016/j.ccr.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhu Z, Chen X, Guo A, Manzano T, Walsh PJ, Wills KM, Halliburton R, Radko-Juettner S, Carter RD, Partridge JF, Green DR, Zhang J, Roberts CWM (2023) Mitotic bookmarking by SWI/SNF subunits. Nature 618:180–187. 10.1038/s41586-023-06085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55:201–215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Liao J, Ho J, Burns M, Dykhuizen EC, Hargreaves DC (2024) Collaboration between distinct SWI/SNF chromatin remodeling complexes directs enhancer selection and activation of macrophage inflammatory genes. Immunity 57:1780–1795e6. 10.1016/j.immuni.2024.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, Löbrich M, Jeggo PA, Downs JA (2014) Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell 55:723–732. 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Niimi A, Hopkins SR, Downs JA, Masutani C (2015) The BAH domain of BAF180 is required for PCNA ubiquitination. Mutat Res 779:16–23. 10.1016/j.mrfmmm.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 267.Carcamo S, Nguyen CB, Grossi E, Filipescu D, Alpsoy A, Dhiman A, Sun D, Narang S, Imig J, Martin TC, Parsons R, Aifantis I, Tsirigos A, Aguirre-Ghiso JA, Dykhuizen EC, Hasson D, Bernstein E (2022) Altered BAF occupancy and transcription factor dynamics in PBAF-deficient melanoma. Cell Rep 39:110637. 10.1016/j.celrep.2022.110637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Alfert A, Moreno N, Kerl K (2019) The BAF complex in development and disease. Epigenetics Chromatin 12:19. 10.1186/s13072-019-0264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR (2007) Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56:94–108. 10.1016/j.neuron.2007.08.021 [DOI] [PubMed] [Google Scholar]
- 270.Bachmann C, Nguyen H, Rosenbusch J, Pham L, Rabe T, Patwa M, Sokpor G, Seong RH, Ashery-Padan R, Mansouri A, Stoykova A, Staiger JF, Tuoc T (2016) mSWI/SNF (BAF) complexes are indispensable for the neurogenesis and development of embryonic olfactory epithelium. PLoS Genet 12:e1006274. 10.1371/journal.pgen.1006274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Allen MD, Freund SM, Zinzalla G, Bycroft M (2015) The SWI/SNF subunit INI1 contains an N-terminal winged helix DNA binding domain that is a target for mutations in schwannomatosis. Structure 23:1344–1349. 10.1016/j.str.2015.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yuan J, Chen K, Zhang W, Chen Z (2022) Structure of human chromatin-remodelling PBAF complex bound to a nucleosome. Nature 605:166–171. 10.1038/s41586-022-04658-5 [DOI] [PubMed] [Google Scholar]
- 273.Cooper GW, Lee BP, Kim WJ et al (2025) SMARCB1 missense mutants disrupt SWI/SNF complex stability and remodeling activity. Res Sq [Preprint] Mar 26:rs.3.rs-6018128.10.21203/rs.3.rs-6018128/v1
- 274.van der Lee R, Buljan M, Lang B et (2014) Classification of intrinsically disordered regions and proteins. Chem Rev 114:6589–6631. 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen G, Zhou H, Liu B, Wang Y, Zhao J, Giancotti FG, Long J (2020) A heterotrimeric SMARCB1-SMARCC2 subcomplex is required for the assembly and tumor suppression function of the BAF chromatin-remodeling complex. Cell Discov 6:66. 10.1038/s41421-020-00196-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV (1999) c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet 22:102–105. 10.1038/8811 [DOI] [PubMed] [Google Scholar]
- 277.Takayama MA, Taira T, Tamai K, Iguchi-Ariga SM, Ariga H (2000) ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells5:481– 90. 10.1046/j.1365-2443.2000.00338.x [DOI] [PubMed]
- 278.Craig E, Zhang ZK, Davies KP, Kalpana GV (2002) A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: implications for tumorigenesis. EMBO J 21:31–42. 10.1093/emboj/21.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Pathak R, Zin F, Thomas C et al (2021) Inhibition of nuclear export restores nuclear localization and residual tumor suppressor function of truncated SMARCB1/INI1 protein in a molecular subset of atypical teratoid/rhabdoid tumors. Acta Neuropathol 142:361–374. 10.1007/s00401-021-02328-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Judkins AR, Mauger J, Ht A, Rorke LB, Biegel JA (2004) Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol 28:644–650. 10.1097/00000478-200405000-00013 [DOI] [PubMed] [Google Scholar]
- 281.Ho B, Johann PD, Grabovska Y, De Dieu Andrianteranagna MJ, Yao F, Frühwald M, Hasselblatt M, Bourdeaut F, Williamson D, Huang A, Kool M (2020) Molecular subgrouping of atypical teratoid/rhabdoid tumors-a reinvestigation and current consensus. Neuro Onco 22:613–624. 10.1093/neuonc/noz235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Gossai N, Biegel JA, Messiaen L, Berry SA, Moertel CL (2015) Report of a patient with a constitutional missense mutation in SMARCB1, Coffin-Siris phenotype, and schwannomatosis. Am J Med Genet A 167A:3186–3191. 10.1002/ajmg.a.37356 [DOI] [PubMed] [Google Scholar]
- 283.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45:D777–D783. 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Erkek S, Johann PD, Finetti MA et al (2019) Comprehensive analysis of chromatin states in atypical teratoid/rhabdoid tumor identifies diverging roles for SWI/SNF and polycomb in gene regulation. Cancer Cell 35:95–110e8. 10.1016/j.ccell.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wei D, Goldfarb D, Song S, Cannon C, Yan F, Sakellariou-Thompson D, Emanuele M, Major MB, Weissman BE, Kuwahara Y (2014) SNF5/INI1 deficiency redefines chromatin remodeling complex composition during tumor development. Mol Cancer Res 12:1574–1585. 10.1158/1541-7786.MCR-14-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA (2013) Super-enhancers in the control of cell identity and disease. Cell 155:934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Iurlaro M, Stadler MB, Masoni F, Jagani Z, Galli GG, Schübeler D (2021) Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet 53:279–287. 10.1038/s41588-020-00768-w [DOI] [PubMed] [Google Scholar]
- 289.Zaret KS (2020) Pioneer transcription factors initiating gene network changes. Annu Rev Genet 54:367–385. 10.1146/annurev-genet-030220-015007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Brahma S, Henikoff S (2024) The BAF chromatin remodeler synergizes with RNA polymerase II and transcription factors to evict nucleosomes. Nat Genet 56:100–111. 10.1038/s41588-023-01603-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Gourisankar S, Krokhotin A, Wenderski W, Crabtree GR (2024) Context-specific functions of chromatin remodellers in development and disease. Nat Rev Genet 25:340–361. 10.1038/s41576-023-00666-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Phanstiel DH, Van Bortle K, Spacek D, Hess GT, Shamim MS, Machol I, Love MI, Aiden EL, Bassik MC, Snyder MP (2017) Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol Cell 67:1037–1048e6. 10.1016/j.molcel.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME (2017) AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol Cell 68:1067–1082e12. 10.1016/j.molcel.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Nesvick CL, Zhang L, Yan Y, Wixom AQ, Hamdan FH, Ge J, Anderson JB, Gaspar-Maia A, Johnsen SA, Daniels DJ (2025) SWI/SNF complexes govern ontology-specific transcription factor function in MYC-subtype atypical teratoid rhabdoid tumor. Neuro Oncol Mar 23:noaf081. 10.1093/neuonc/noaf081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Comandante-Lou N, Baumann DG, Fallahi-Sichani M (2022) AP-1 transcription factor network explains diverse patterns of cellular plasticity in melanoma cells. Cell Rep 40:111147. 10.1016/j.celrep.2022.111147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Grossi E, Nguyen CB, Carcamo S, Kirigin Callaú V, Moran S, Filipescu D, Tagore S, Firestone TM, Keogh MC, Sun L, Izar B, Hasson D, Bernstein E (2025) The SWI/SNF PBAF complex facilitates REST occupancy at repressive chromatin. Mol Cell 85:1714–1729e7. 10.1016/j.molcel.2025.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hasselblatt M, Isken S, Linge A, Eikmeier K, Jeibmann A, Oyen F, Nagel I, Richter J, Bartelheim K, Kordes U, Schneppenheim R, Frühwald M, Siebert R, Paulus W (2013) High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer 52:185–190. 10.1002/gcc.22018 [DOI] [PubMed] [Google Scholar]
- 298.Kieran MW, Roberts CW, Chi SN, Ligon KL, Rich BE, Macconaill LE, Garraway LA, Biegel JA (2012) Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer 59:1155–1157. 10.1002/pbc.24315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kakkar A, Biswas A, Goyal N, Suri V, Sharma MC, Gupta D, Julka PK, Sarkar C (2016) The expression of Cyclin D1, VEGF, EZH2, and H3K27me3 in atypical teratoid/rhabdoid tumors of the CNS: A possible role in targeted therapy. Appl Immunohistochem Mol Morphol 24:729–737. 10.1097/PAI.0000000000000247 [DOI] [PubMed] [Google Scholar]
- 301.Hasselblatt M, Johann PD, Kool M, Frühwald MC (2017) Reduced histone H3 K27 trimethylation is encountered in about 50% of atypical teratoid/rhabdoid tumors (AT/RT) but is not associated with molecular subgroup status and outcome. Acta Neuropathol 134:817–818. 10.1007/s00401-017-1766-y [DOI] [PubMed] [Google Scholar]
- 302.Birks DK, Donson AM, Patel PR, Sufit A, Algar EM, Dunham C, Kleinschmidt-DeMasters BK, Handler MH, Vibhakar R, Foreman NK (2013) Pediatric rhabdoid tumors of kidney and brain show many differences in gene expression but share dysregulation of cell cycle and epigenetic effector genes. Pediatr Blood Cancer 60:1095–1102. 10.1002/pbc.24481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Capper D, Jones DTW, Sill M et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Chun HE, Johann PD, Milne K (2019) Identification and analyses of extra-cranial and cranial rhabdoid tumor molecular subgroups reveal tumors with cytotoxic T cell infiltration. Cell Rep 29:2338–2354e7. 10.1016/j.celrep.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Huhtala L, Karabiyik G, Rautajoki KJ (2024) Development and epigenetic regulation of atypical teratoid/rhabdoid tumors in the context of cell-of-origin and halted cell differentiation. Neurooncol Adv 6:vdae162. 10.1093/noajnl/vdae162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hasselblatt M, Thomas C, Nemes K et al (2020) Tyrosinase immunohistochemistry can be employed for the diagnosis of atypical teratoid/rhabdoid tumours of the tyrosinase subgroup (ATRT-TYR). Neuropathol Appl Neurobiol 46:186–189. 10.1111/nan.12560 [DOI] [PubMed] [Google Scholar]
- 307.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW (2016) Targeting the Sonic Hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel) 8:22. 10.3390/cancers8020022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sachan N, Sharma V, Mutsuddi M, Mukherjee A (2024) Notch signalling: multifaceted role in development and disease. FEBS J 291:3030–3059. 10.1111/febs.16815 [DOI] [PubMed] [Google Scholar]
- 309.Păun O, Tan YX, Patel H, Strohbuecker S, Ghanate A, Cobolli-Gigli C, Llorian Sopena M, Gerontogianni L, Goldstone R, Ang SL, Guillemot F, Dias C (2023) Pioneer factor ASCL1 cooperates with the mSWI/SNF complex at distal regulatory elements to regulate human neural differentiation. Genes Dev 37:218–242. 10.1101/gad.350269.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Nowak J, Nemes K, Hohm A, Vandergrift LA, Hasselblatt M, Johann PD, Kool M, Frühwald MC, Warmuth-Metz M (2018) Magnetic resonance imaging surrogates of molecular subgroups in atypical teratoid/rhabdoid tumor. Neuro Oncol 20:1672–1679. 10.1093/neuonc/noy111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Leruste A, Tosello J, Ramos RN et al (2019) Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell 36:597–612e8. 10.1016/j.ccell.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 312.Grabovska Y, Mackay A, O’Hare P et al (2020) Pediatric pan-central nervous system tumor analysis of immune-cell infiltration identifies correlates of antitumor immunity. Nat Commun 11:4324. 10.1038/s41467-020-18070-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Jessa S, Blanchet-Cohen A, Krug B et al (2019) Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 51:1702–1713. 10.1038/s41588-019-0531-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lobón-Iglesias MJ, Andrianteranagna M, Han ZY et al (2023) Imaging and multi-omics datasets converge to define different neural progenitor origins for ATRT-SHH subgroups. Nat Commun 14:6669. 10.1038/s41467-023-42371-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Custers L, Khabirova E, Coorens THH et al (2021) Somatic mutations and single-cell transcriptomes reveal the root of malignant rhabdoid tumours. Nat Commun 12:1407. 10.1038/s41467-021-21675-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Graf M, Interlandi M, Moreno N, Holdhof D et al (2022) Single-cell transcriptomics identifies potential cells of origin of MYC rhabdoid tumors. Nat Commun 13:1544. 10.1038/s41467-022-29152-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hancock GV, Wamaitha SE, Peretz L, Clark AT (2021) Mammalian primordial germ cell specification. Development 148:dev189217. 10.1242/dev.189217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tief K, Schmidt A, Aguzzi A, Beermann F (1996) Tyrosinase is a new marker for cell populations in the mouse neural tube. Dev Dyn 205:445–456. [DOI] [PubMed] [Google Scholar]
- 319.Parisian AD, Koga T, Miki S, Johann PD, Kool M, Crawford JR, Furnari FB (2020) SMARCB1 loss interacts with neuronal differentiation state to block maturation and impact cell stability. Genes Dev 34:1316–1329. 10.1101/gad.339978.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Terada Y, Jo N, Arakawa Y, Sakakura M, Yamada Y, Ukai T, Kabata M, Mitsunaga K, Mineharu Y, Ohta S, Nakagawa M, Miyamoto S, Yamamoto T, Yamada Y (2019) Human pluripotent stem cell-derived tumor model uncovers the embryonic stem cell signature as a key driver in atypical teratoid/rhabdoid tumor. Cell Rep 26:2608–2621e6. 10.1016/j.celrep.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 321.Pekkarinen M, Nordfors K, Uusi-Mäkelä J et al (2024) Aberrant DNA methylation distorts developmental trajectories in atypical teratoid/rhabdoid tumors. Life Sci Alliance 7:e202302088. 10.26508/lsa.202302088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Smith DK, Yang J, Liu ML, Zhang CL (2016) Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep 7:955–969. 10.1016/j.stemcr.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK (2016) NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35:24–45. 10.15252/embj.201591206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lu C, Garipler G, Dai C, Roush T, Salome-Correa J, Martin A, Liscovitch-Brauer N, Mazzoni EO, Sanjana NE (2023) Essential transcription factors for induced neuron differentiation. Nat Commun 14:8362. 10.1038/s41467-023-43602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12:31–46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 326.Moreno N, Schmidt C, Ahlfeld J, Pöschl J, Dittmar S, Pfister SM, Kool M, Kerl K, Schüller U (2014) Loss of Smarc proteins impairs cerebellar development. J Neurosci 34:13486–13491. 10.1523/JNEUROSCI.2560-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Alfert A, Walter C, Moreno N, Melcher V, Graf M, Hotfilder M, Dugas M, Albert T, Kerl K (2022) Smarcb1 loss results in a deregulation of EsBAF binding and impacts the expression of neurodevelopmental genes. Cells 11:1354. 10.3390/cells11081354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Alimova I, Wang D, DeSisto J, Danis E, Lakshmanachetty S, Prince E, Murdock G, Pierce A, Donson A, Balakrishnan I, Serkova N, Lin H, Foreman NK, Dahl N, Venkataraman S, Vibhakar R (2025) SIRT2 regulates the SMARCB1 loss-driven differentiation block in ATRT. Mol Cancer Res Feb 17. 10.1158/1541-7786.MCR-24-0926Epub ahead of print [DOI] [PMC free article] [PubMed]
- 329.Morin A, Soane C, Pierce A, Sanford B, Jones KL, Crespo M, Zahedi S, Vibhakar R, Mulcahy Levy JM (2020) Proteasome Inhibition as a therapeutic approach in atypical teratoid/rhabdoid tumors. Neurooncol Adv 2:vdaa051. 10.1093/noajnl/vdaa051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wanior M, Krämer A, Knapp S, Joerger AC (2021) Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. Oncogene 40:3637–3654. 10.1038/s41388-021-01781-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Paassen I, Williams J, Ríos Arceo C et al (2023) Atypical teratoid/rhabdoid tumoroids reveal subgroup-specific drug vulnerabilities. Oncogene 42:1661–1671. 10.1038/s41388-023-02681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Pauck D, Picard D, Maue M et al (2025) An in vitro pharmacogenomic approach reveals subtype-specific therapeutic vulnerabilities in atypical teratoid/rhabdoid tumors (AT/RT). Pharmacol Res 213:107660. 10.1016/j.phrs.2025.107660 [DOI] [PubMed] [Google Scholar]
- 333.Romagosa C, Simonetti S, López-Vicente L, Mazo A, Lleonart ME, Castellvi J, Ramon y Cajal S (2011) p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30:2087–2097. 10.1038/onc.2010.614 [DOI] [PubMed] [Google Scholar]
- 334.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR (1997) Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A 94:7245–7250. 10.1073/pnas.94.14.7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP (2008) SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 28:3457–3464. 10.1128/MCB.02019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Venneti S, Le P, Martinez D, Eaton KW, Shyam N, Jordan-Sciutto KL, Pawel B, Biegel JA, Judkins AR (2011) p16INK4A and p14ARF tumor suppressor pathways are deregulated in malignant rhabdoid tumors. J Neuropathol Exp Neurol 70:596–609. 10.1097/NEN.0b013e31822146ca [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kim KH, Roberts CW (2014) Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet 207:365–372. 10.1016/j.cancergen.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Bracken AP, Brien GL, Verrijzer CP (2019) Dangerous liaisons: interplay between SWI/SNF, nurd, and polycomb in chromatin regulation and cancer. Genes Dev 33:936–959. 10.1101/gad.326066.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Bergwell M, Park J, Kirkland JG (2024) Differential modulation of polycomb-associated histone marks by cBAF, pBAF, and gBAF complexes. Life Sci Alliance 7:e202402715. 10.26508/lsa.202402715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Pollock RM, Kuntz KW, Keilhack H (2013) Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 110:7922–7927. 10.1073/pnas.1303800110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Feng S, Marhon SA, Sokolowski DJ et al (2024) Inhibiting EZH2 targets atypical teratoid rhabdoid tumor by triggering viral mimicry via both RNA and DNA sensing pathways. Nat Commun 15:9321. 10.1038/s41467-024-53515-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Sasaki M, Kato D, Murakami K, Yoshida H, Takase S, Otsubo T, Ogiwara H (2024) Targeting dependency on a paralog pair of CBP/p300 against de-repression of KREMEN2 in SMARCB1-deficient cancers. Nat Commun 15:4770. 10.1038/s41467-024-49063-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B (2003) Genomic targets of the human c-Myc protein. Genes Dev 17:1115–1129. 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Jakobsen ST, Siersbæk R (2025) Transcriptional regulation by MYC: an emerging new model. Oncogene 44:1–7. 10.1038/s41388-024-03174-2 [DOI] [PubMed] [Google Scholar]
- 345.Jones CA, Tansey WP, Weissmiller AM (2022) Emerging themes in mechanisms of tumorigenesis by SWI/SNF subunit mutation. Epigenet Insights 15:25168657221115656. 10.1177/25168657221115656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Liu NQ, Paassen I, Custers L, Zeller P, Teunissen H, Ayyildiz D, He J, Buhl JL, Hoving EW, van Oudenaarden A, de Wit E, Drost J (2023) SMARCB1 loss activates patient-specific distal oncogenic enhancers in malignant rhabdoid tumors. Nat Commun 14:7762. 10.1038/s41467-023-43498-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Stojanova A, Tu WB, Ponzielli R, Kotlyar M, Chan PK, Boutros PC, Khosravi F, Jurisica I, Raught B, Penn LZ (2016) MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation. Cell Cycle 15:1693–1705. 10.1080/15384101.2016.1146836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Weissmiller AM, Wang J, Lorey SL, Howard GC, Martinez E, Liu Q, Tansey WP (2019) Inhibition of MYC by the SMARCB1 tumor suppressor. Nat Commun 10:2014. 10.1038/s41467-019-10022-5 [DOI] [PMC free article] [PubMed]
- 349.Mora-Blanco EL, Mishina Y, Tillman EJ, Cho YJ, Thom CS, Pomeroy SL, Shao W, Roberts CW (2014) Activation of β-catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene 33:933–938. 10.1038/onc.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Chakravadhanula M, Hampton CN, Chodavadia P, Ozols V, Zhou L, Catchpoole D, Xu J, Erdreich-Epstein A, Bhardwaj RD (2015) Wnt pathway in atypical teratoid rhabdoid tumors. Neuro Oncol 17:526–535. 10.1093/neuonc/nou229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Jagani Z, Mora-Blanco EL, Sansam CG et al (2010) Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med 16:1429–1433. 10.1038/nm.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Hooper JE, Scott MP (2005) Communicating with hedgehogs. Nat Rev Mol Cell Biol 6:306–317. 10.1038/nrm1622 [DOI] [PubMed] [Google Scholar]
- 353.Jiang J, Hui CC (2008) Hedgehog signaling in development and cancer. Dev Cell 15:801–812. 10.1016/j.devcel.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Ge T, Gu X, Jia R, Ge S, Chai P, Zhuang A, Fan X (2022) Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities. Cancer Commun (Lond) 42:1049–1082. 10.1002/cac2.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Hertwig F, Meyer K, Braun S, Ek S, Spang R, Pfenninger CV, Artner I, Prost G, Chen X, Biegel JA, Judkins AR, Englund E, Nuber UA (2012) Definition of genetic events directing the development of distinct types of brain tumors from postnatal neural stem/progenitor cells. Cancer Res 72:3381–3392. 10.1158/0008-5472.CAN-11-3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Carugo A, Minelli R, Sapio L et al (2019) p53 is a master regulator of proteostasis in SMARCB1-deficient malignant rhabdoid tumors. Cancer Cell 35:204–220e9. 10.1016/j.ccell.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH (2000) Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A 97:13796–13800. 10.1073/pnas.250492697 [DOI] [PMC free article] [PubMed]
- 358.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, Muchardt C, Hue L, Pontoglio M, Yaniv M, Klochendler-Yeivin A (2005) The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J 24:3313–3324. 10.1038/sj.emboj.7600802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Radko-Juettner S, Yue H, Myers JA et al (2024) Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF. Nature 628:442–449. 10.1038/s41586-024-07250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Kohashi K, Oda Y, Yamamoto H, Tamiya S, Izumi T, Ohta S, Taguchi T, Suita S, Tsuneyoshi M (2007) Highly aggressive behavior of malignant rhabdoid tumor: a special reference to SMARCB1/INI1 gene alterations using molecular genetic analysis including quantitative real-time PCR. J Cancer Res Clin Oncol 133:817–824. 10.1007/s00432-007-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Forrest SJ, Al-Ibraheemi A, Doan D, Ward A, Clinton CM, Putra J, Pinches RS, Kadoch C, Chi SN, DuBois SG, Leavey PJ, LeBoeuf NR, Mullen E, Collins N, Church AJ, Janeway KA (2020) Genomic and immunologic characterization of INI1-deficient pediatric cancers. Clin Cancer Res 26:2882–2890. 10.1158/1078-0432.CCR-19-3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Asai K, Tani S, Mineharu Y, Tsurusaki Y, Imai Y, Agawa Y, Iwaki K, Matsumoto N, Sakai N (2015) Familial schwannomatosis with a germline mutation of SMARCB1 in Japan. Brain Tumor Pathol 32:216–220. 10.1007/s10014-015-0213-9 [DOI] [PubMed] [Google Scholar]
- 363.Smith MJ, Walker JA, Shen Y, Stemmer-Rachamimov A, Gusella JF, Plotkin SR (2012) Expression of SMARCB1 (INI1) mutations in familial schwannomatosis. Hum Mol Genet 21:5239–5245. 10.1093/hmg/dds370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Hulsebos TJ, Kenter S, Verhagen WI, Baas F, Flucke U, Wesseling P (2014) Premature termination of SMARCB1 translation may be followed by reinitiation in schwannomatosis-associated schwannomas, but results in absence of SMARCB1 expression in rhabdoid tumors. Acta Neuropathol 128:439–448. 10.1007/s00401-014-1281-3 [DOI] [PubMed] [Google Scholar]
- 365.Lindeboom RG, Supek F, Lehner B (2016) The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48:1112–1118. 10.1038/ng.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Patil S, Perry A, Maccollin M, Dong S, Betensky RA, Yeh TH, Gutmann DH, Stemmer-Rachamimov AO (2008) Immunohistochemical analysis supports a role for INI1/SMARCB1 in hereditary forms of schwannomas, but not in solitary, sporadic schwannomas. Brain Pathol 18:517–519. 10.1111/j.1750-3639.2008.00155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Caltabiano R, Magro G, Polizzi A et al (2017) A mosaic pattern of INI1/SMARCB1 protein expression distinguishes schwannomatosis and NF2-associated peripheral schwannomas from solitary peripheral schwannomas and NF2-associated vestibular schwannomas. Childs Nerv Syst 33:933–940. 10.1007/s00381-017-3340-2 [DOI] [PubMed] [Google Scholar]
- 368.Kehrer-Sawatzki H, Farschtschi S, Mautner VF, Cooper DN (2017) The molecular pathogenesis of schwannomatosis, a paradigm for the co-involvement of multiple tumour suppressor genes in tumorigenesis. Hum Genet 136:129–148. 10.1007/s00439-016-1753-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Patel M, Binsuwaidan R, Surti M, Alshammari N, Ibrahim AMM, Adnan M (2025) Predicting high-risk clinical missense variants of SMARCB1 in rare neurogenetic disorder schwannomatosis (nerve tumor) through sequence, structure, and molecular dynamics analyses. Neurogenetics 26:31. 10.1007/s10048-025-00812-z [DOI] [PubMed] [Google Scholar]
- 370.Smith MJ, Bowers NL, Banks C, Coates-Brown R, Morris KA, Ewans L, Wilson M, Pinner J, Bhaskar SS, Cammarata-Scalisi F, Wallace AJ, Evans DGR (2020) A deep intronic SMARCB1 variant associated with schwannomatosis. Clin Gene 97:376–377. 10.1111/cge.13637 [DOI] [PubMed] [Google Scholar]
- 371.Tauziède-Espariat A, Masliah-Planchon J, Brugières L, Puget S, Dufour C, Schneider P, Laquerrière A, Frebourg T, Bodet D, Lechapt-Zalcman E, Pierron G, Delattre O, Varlet P, Bourdeaut F (2017) Deep intronic hotspot variant explaining rhabdoid tumor predisposition syndrome in two patients with atypical teratoid and rhabdoid tumor. Eur J Hum Genet 25:1170–1172. 10.1038/ejhg.2017.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Perez-Becerril C, Evans DG, Smith MJ (2021) Pathogenic noncoding variants in the neurofibromatosis and schwannomatosis predisposition genes. Hum Mutat 42:1187–1207. 10.1002/humu.24261 [DOI] [PubMed] [Google Scholar]
- 373.Thomson G, Filser M, Guerrini-Rousseau L et al (2024) Postzygotic mosaicism of SMARCB1 variants in patients with rhabdoid tumors: A not-so-rare condition exposing to successive tumors. Neuro Oncol 26:2102–2112. 10.1093/neuonc/noae122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Fleischmann LS, Nemes K, Glaser S, Kouroukli AG, Boros M, Bens S, Dahlum S, Kretzmer H, Oyen F, Gerss J, Hasselblatt M, Frühwald MC, Siebert R (2025) Constitutional mosaicism of pathogenic variants in SMARCB1 in a subset of patients with sporadic rhabdoid tumors. Neuro Oncol 27:533–544. 10.1093/neuonc/noae188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Gigante L, Paganini I, Frontali M, Ciabattoni S, Sangiuolo FC, Papi L (2016) Rhabdoid tumor predisposition syndrome caused by SMARCB1 constitutional deletion: prenatal detection of new case of recurrence in siblings due to gonadal mosaicism. Fam Cancer 15:123–126. 10.1007/s10689-015-9836-6 [DOI] [PubMed] [Google Scholar]
- 376.Hulsebos TJ, Kenter SB, Jakobs ME, Baas F, Chong B, Delatycki MB (2010) SMARCB1/INI1 maternal germ line mosaicism in schwannomatosis. Clin Genet 77:86–91. 10.1111/j.1399-0004.2009.01249.x [DOI] [PubMed] [Google Scholar]
- 377.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M (2000) The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep 1:500–506. 10.1093/embo-reports/kvd129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN (2001) Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol 21:3598–3603. 10.1128/MCB.21.10.3598-3603.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Stemmer-Rachamimov AO, Ino Y, Lim ZY, Jacoby LB, MacCollin M, Gusella JF, Ramesh V, Louis DN (1998) Loss of the NF2 gene and merlin occur by the tumorlet stage of schwannoma development in neurofibromatosis 2. J Neuropathol Exp Neurol 57:1164–1167. 10.1097/00005072-199812000-00008 [DOI] [PubMed] [Google Scholar]
- 380.Hadfield KD, Smith MJ, Urquhart JE, Wallace AJ, Bowers NL, King AT, Rutherford SA, Trump D, Newman WG, Evans DG (2010) Rates of loss of heterozygosity and mitotic recombination in NF2 schwannomas, sporadic vestibular schwannomas and schwannomatosis schwannomas. Oncogene 29:6216–6221. 10.1038/onc.2010.363 [DOI] [PubMed] [Google Scholar]
- 381.Kaufman DL, Heinrich BS, Willett C, Perry A, Finseth F, Sobel RA, MacCollin M (2003) Somatic instability of the NF2 gene in schwannomatosis. Arch Neurol 60:1317–1320. 10.1001/archneur.60.9.1317 [DOI] [PubMed] [Google Scholar]
- 382.Lekanne Deprez RH, Bianchi AB, Groen NA et al (1994) Frequent NF2 gene transcript mutations in sporadic meningiomas and vestibular schwannomas. Am J Hum Genet 54:1022–1029 [PMC free article] [PubMed] [Google Scholar]
- 383.Håvik AL, Bruland O, Myrseth E, Miletic H, Aarhus M, Knappskog PM, Lund-Johansen M (2018) Genetic landscape of sporadic vestibular schwannoma. J Neurosurg 128:911–922. 10.3171/2016.10.JNS161384 [DOI] [PubMed] [Google Scholar]
- 384.Tsuchiya T, Miyawaki S, Teranishi Y, Ohara K, Hirano Y, Ogawa S, Torazawa S, Sakai Y, Hongo H, Ono H, Saito N (2025) Current molecular Understanding of central nervous system schwannomas. Acta Neuropathol Commun 13:24. 10.1186/s40478-025-01937-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Yidian C, Chen L, Hongxia D, Yanguo L, Zhisen S (2022) Single-cell sequencing reveals the cell map and transcriptional network of sporadic vestibular Schwannoma. Front Mol Neurosci 15:984529. 10.3389/fnmol.2022.984529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Barrett TF, Patel B, Khan SM, Mullins RDZ, Yim AKY, Pugazenthi S, Mahlokozera T, Zipfel GJ, Herzog JA, Chicoine MR, Wick CC, Durakovic N, Osbun JW, Shew M, Sweeney AD, Patel AJ, Buchman CA, Petti AA, Puram SV, Kim AH (2024) Single-cell multi-omic analysis of the vestibular schwannoma ecosystem uncovers a nerve injury-like state. Nat Commun 15:478. 10.1038/s41467-023-42762-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Huo Z, Wang Z, Luo H, Maimaitiming D, Yang T, Liu H, Li H, Wu H, Zhang Z (2024) Single-cell transcriptomes reveal the heterogeneity and microenvironment of vestibular schwannoma. Neuro Oncol 26:444–457. 10.1093/neuonc/noad201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Gonzalez Castro LN, Gavish A, Bussema L et al (2025) A single-cell atlas of schwannoma across genetic backgrounds and anatomic locations. Genome Med 17:37. 10.1186/s13073-025-01462-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Gavish A, Tyler M, Greenwald AC et al (2023) Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours. Nature 618:598–606. 10.1038/s41586-023-06130-4 [DOI] [PubMed] [Google Scholar]
- 390.Agnihotri S, Jalali S, Wilson MR et al (2016) The genomic landscape of schwannoma. Nat Genet 48:1339–1348. 10.1038/ng.3688 [DOI] [PubMed] [Google Scholar]
- 391.Mossink B, Negwer M, Schubert D, Nadif Kasri N (2021) The emerging role of chromatin remodelers in neurodevelopmental disorders: a developmental perspective. Cell Mol Life Sci 78:2517–2563. 10.1007/s00018-020-03714-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Gallagher JE, Saeed-Vafa D, Bui MM, Makanji R (2024) Coffin-Siris syndrome and SMARCB1 mutation presenting with schwannomatosis: a case report and literature review. Cureus 16:e67333. 10.7759/cureus.67333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Ha TT, Burgess R, Newman M et al (2023) Aicardi syndrome is a genetically heterogeneous disorder. Genes (Basel) 14:1565. 10.3390/genes14081565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.van der Sluijs PJ, Joosten M, Alby C et al (2022) Discovering a new part of the phenotypic spectrum of Coffin-Siris syndrome in a fetal cohort. Genet Med 24:1753–1760. 10.1016/j.gim.2022.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Brugmans AK, Walter C, Moreno N et al (2023) A carboxy-terminal smarcb1 point mutation induces hydrocephalus formation and affects AP-1 and neuronal signalling pathways in mice. Cell Mol Neurobiol 43:3511–3526. 10.1007/s10571-023-01361-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Ka M, Chopra DA, Dravid SM, Kim WY (2016) Essential roles for ARID1B in dendritic arborization and spine morphology of developing pyramidal neurons. J Neurosci 36:2723–2742. 10.1523/JNEUROSCI.2321-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Narayanan R, Tuoc TC (2014) Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming. Cell Tissue Res 356:575–584. 10.1007/s00441-013-1791-7 [DOI] [PubMed] [Google Scholar]
- 398.Evans DG, Messiaen LM, Foulkes WD, Irving REA, Murray AJ, Perez-Becerril C, Rivera B, McDonald-McGinn DM, Stevenson DA, Smith MJ (2021) Typical 22q11.2 deletion syndrome appears to confer a reduced risk of schwannoma. Genet Med 23:1779–1782. 10.1038/s41436-021-01175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Smith MJ, Urquhart JE, Harkness EF, Miles EK, Bowers NL, Byers HJ, Bulman M, Gokhale C, Wallace AJ, Newman WG, Evans DG (2016) The contribution of whole gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum Mutat 37:250–256. 10.1002/humu.22938 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.