ABSTRACT
Background and Aims
Psoriasis is an immune‐mediated disease affecting primarily the skin and joints, but which also has systemic implications. There are assumptions about a common genetic background between psoriasis and cardiovascular diseases. This narrative review aims to provide an overview of the existing data regarding the IL‐23 rs2066808, IL‐23R rs2201841, IL‐17RA rs4819554, and IL‐17A rs2275913 polymorphisms and their link to psoriasis and cardiovascular diseases.
Methods
PubMed, Web of Science, Scopus, and “SNPedia” searches were conducted to identify the relevant studies between 2007 and 2025 that investigated the association between psoriasis, cardiovascular diseases, and the IL‐23 rs2066808, IL‐23R rs2201841, IL‐17RA rs4819554, and IL17A rs2275913 polymorphisms.
Results
The existing data are conflicting. The polymorphisms could be implicated in psoriasis onset, disease susceptibility, and treatment response. Some of them are also linked to coronary artery disease. Contradictory data could result from the heterogeneity of the population included in the studies and the laboratory techniques used for polymorphism determination.
Conclusion
The interplay between IL‐17, IL‐23, and genetic polymorphisms illustrates a complex landscape in psoriasis and its associated comorbidities. More standardized case‐control studies are needed to confirm this hypothesis. Moreover, extensive studies in diverse populations would be important to clarify these relationships and the potential implications for treatment and management.
Keywords: coronary artery disease, IL17, IL23, polymorphisms, psoriasis
Abbreviations
- BC
base change
- BMI
body mass index
- BSA
body surface area
- C
codominant
- CAD
coronary artery disease
- CoD
codominant model
- CVD
cardiovascular disease
- CVM
cardiovascular mortality
- D
dominant model
- DC
dilated cardiomyopathy
- DCs
dendritic cells
- DLQI
dermatology life quality index
- GPSS
genital psoriasis symptoms scale
- HDL‐c
high‐density lipoprotein cholesterol
- HF
heart failure
- IL
interleukin
- LA
log‐additive
- LDL‐c
low‐density lipoprotein cholesterol
- LNPEP
leucyl and cystinyl aminopeptidase, an insulin‐responsive aminopeptidase
- NK
natural killer cells
- OR
odds ratio after adjustment
- OvD
overdominant
- p adj
p value adjusting for age, sex, BMI, hypertension, diabetes mellitus, smoking history
- PASI
psoriasis area and severity index
- p hwe
p value of the Hardy–Weinberg equilibrium tests
- p obs
observed p value
- R
recessive model
- RA
rheumatoid arthritis
- SNP(s)
single‐nucleotide polymorphism(s)
- TNF
tumor necrosis factor
- UVB
ultraviolet type B rays
1. Introduction
Psoriasis is an immune‐mediated disease with a global prevalence of 2%–3%, with significant inter‐regional variations [1]. Psoriasis affects primarily the skin and joints; however, it is now recognized as a systemic inflammatory disease [2]. The dermatological manifestations of psoriasis range from thick, red, scaly plaques (the plaque type) to multiple yellow sterile pustules on an erythematous skin, associated with systemic inflammation (generalized pustular psoriasis‐ von Zumbusch type) [3, 4, 5]. Other clinical presentations are inverse psoriasis, acrodermatitis continua of Hallopeau, palmoplantar pustulosis (localized pustular psoriasis), guttate psoriasis, and erythrodermic psoriasis [3, 4, 5]. The most common clinical presentation of the disease is psoriasis vulgaris (plaque‐type psoriasis) with elbows, knees, the umbilical area, the perianal region, and the scalp being most affected [6].
Psoriasis is a clinical diagnosis, based on the typical erythematous plaques with silvery scales affecting predilection areas, therefore a skin biopsy is rarely necessary. The severity of the disease is assessed using clinical scores such as psoriasis area and severity index (PASI), body surface area (BSA) [7], nail psoriasis severity index (NAPSI) [8], psoriasis scalp severity index (PSSI) [9], and the genital psoriasis symptoms scale (GPSS) [10]. A validated self‐reported questionnaire that evaluates the impact of the disease on the patient's quality of life is the dermatology life quality index (DLQI) [11]. Histopathological characteristics of the psoriasis plaque are epidermal hyperplasia (acanthosis) with hyperkeratosis and parakeratosis, hypogranulosis and dermal vessel dilation and elongation, and inflammatory infiltration consisting of dendritic cells (DCs), macrophages, T cells, and neutrophils [12, 13]. At the level of the parakeratotic epidermis, Munro micro‐abscesses containing polymorphonuclear leukocytes are present [13, 14].
From a pathophysiological perspective, psoriasis is characterized by disruptions in both innate and adaptative immune responses [15]. The disease is triggered or worsened by non‐specific danger signals consisting of infections, drugs, physical trauma (Koebner phenomenon), autoantigens [cathelicidin LL‐37, keratin 17, melanocytic ADAMTSL5, PLA2G4D, heterogeneous nuclear ribonucleoprotein A1 (hnRNP‐A1)] [15, 16, 17]. The natural killer cells (NK), NKT cells, neutrophils, mast cells, γδ T cells, and DCs are innate immune cells, increased in psoriatic lesions, and they are responsible for the release of cytokines (IL‐23, TNF‐alpha) [15, 18, 19]. In the initial stages of the disease, a major role belongs to DCs, antigen‐presenting cells which recognize antimicrobial peptides released by damaged keratinocytes [15, 18, 19]. Plasmacytoid DC activation is decisive in plaque formation. Consequently, IFN‐α ensure dermal DC maturation, which is responsible for Th1 and Th17 activation and differentiation [15, 18, 19]. After their activation, the myeloid DCs migrate into draining lymph nodes and secrete tumor necrosis factor (TNF)‐α, IL‐23, and IL‐12. The aforementioned cytokines induce Th17 and/or Th22 cell subsets, through activation of the IL‐23 and/or IL‐22 pathway [15, 18]. As a consequence, the abundance of psoriatic cytokines resulting from this process, namely TNF‐α, IFN‐γ, IL‐17, and IL‐22, disrupt the keratinocyte function and intensify the inflammation [18].
The TNFα‐IL‐23‐Th17 axis perpetuates inflammation in psoriasis [20]. The role played by IL‐17 in skin inflammation is well known and can be combated both directly and indirectly by blocking IL‐17 and its receptor (IL‐17R), respectively [20]. IL‐17 represents a family of proinflammatory cytokines consisting of 6 members, IL‐17A‐IL‐17F [15, 21]. In healthy skin, IL‐17 plays an important role in maintaining the integrity of the skin barrier and providing protection against fungal and bacterial infections [15, 21]. Dysfunctions of the IL‐17 signaling pathway drive the chronic inflammation characteristic of psoriasis [15, 21]. IL‐23 is produced both by keratinocytes and antigen‐presenting cells and induces Th17 activation and the secondary production of IL‐17, TNF‐alpha, and IL‐6 [22]. IL‐23 is a heterodimeric cytokine consisting of two subunits, p19 and p40 [23]. The IL‐23/IL‐17 axis is implicated in the pathogenesis of chronic inflammation that characterizes many autoimmune inflammatory diseases such as inflammatory bowel disease [24], rheumatoid arthritis (RA), Sjogren syndrome, and multiple sclerosis [25].
Of interest in recent years are the comorbidities associated with psoriasis, mainly cardiovascular diseases (CVDs) [26]. There is an independent association between psoriasis and myocardial infarction (MI), especially in young patients with severe psoriasis [27], as the prevalence of coronary atherosclerosis is twice higher in psoriasis patients compared to controls [28]. Cardiovascular mortality (CVM) was studied in a meta‐analysis by Armstrong et al. [29] among 4 cohorts of patients from the USA and Europe. The results showed a significant increase in CVM among patients with severe psoriasis. On the other hand, the risk of MI was increased in both mild and severe psoriasis [29]. Additionally, psoriasis is associated with an increased risk of stroke as shown in a meta‐analysis by Liu et al. [30].
The complexity of psoriasis pathogenesis and the characteristic chronic inflammatory state, objectified by serum biomarkers, justify the concept of a systemic disease [31]. The concept of multisystemic afflictions in psoriasis is generally accepted, with the development of guidelines that focus on managing these comorbidities, therefore every psoriasis patient should be explored beyond the skin [32]. While the association between psoriasis and CVDs is evident [33], accelerated atherosclerosis in patients with severe or longstanding psoriasis is responsible for the higher incidence of major cardiovascular events in these patients [34]. Garshick et al. [35] proposed a link between psoriasis and atherosclerosis, describing the overlapping inflammatory processes dominated by IFN‐gamma and TNF‐alpha that cause endothelial inflammation and the increased expression of the inflammatory chemokine CXCL10 and the adhesion protein VCAM‐1 [35]. Endothelial cells of psoriasis patients express a significant (2–8‐fold) upregulation of proinflammatory and chemotactic factors (VCAM‐1, IL‐1β, CXCL10, and COX‐2) compared to control subjects [35, 36]. The pattern of expression is similar to that of endothelial cells stimulated in vitro by specific combinations of cytokines like TNF‐α, IL‐17A, and IFNγ, thus supporting the pathogenetic similarities between psoriasis and atherosclerosis [35, 37]. The overlapping mechanisms of psoriasis and atherosclerotic disease are far more complex and extend beyond the focus of the present review [34, 38, 39].
Psoriasis is associated with diseases that represent risk factors for atherosclerosis, such as obesity, diabetes mellitus [40], hypertension, dyslipidemia, and metabolic syndrome [41]. Thus, the hypothesis of a common genetic background, in particular polymorphism in the IL‐23R and IL‐23 genes, of psoriasis and cardiovascular comorbidities has emerged [40]. The potential genetic link between psoriasis, hypertension, and diabetes mellitus, is supported by the presence of a missense mutation in the insulin‐responsive aminopeptidase LNPEP (leucyl and cystinyl aminopeptidase) gene, identified in a study conducted by Cheng et al. [42]. The results of a study conducted by Gupta et al. [43] contradict the aforementioned findings and support the genetic independence of psoriasis from the metabolic syndrome and coronary artery disease. The frequent association results from common environmental risk factors such as Western diet [43].
This review aims to provide an synopsis of the existing data regarding the IL‐23 rs2066808, IL‐23R rs2201841, IL‐17RA rs4819554, and IL17A rs2275913 polymorphisms that link psoriasis and its cardio‐metabolic comorbidities.
2. Materials and Methods
2.1. Literature Search
To identify all relevant studies that investigated the association between IL‐23, IL‐23R, and IL‐17 and IL‐17R polymorphisms (IL‐23 rs2066808, IL‐23R rs2201841, IL‐17RA rs4819554, and IL17A rs2275913) and psoriasis and cardiometabolic diseases, we conducted a PubMed, Web of Science and Scopus search using the terms “IL‐23R rs2201841 and psoriasis”, “IL‐23 rs2066808 and psoriasis“, “IL‐17RA rs4819554 and psoriasis”, “IL17A rs2275913 and psoriasis”, “IL‐23 rs2066808 and cardiovascular disease”, “IL‐23 rs2066808 and atherosclerosis”, “IL‐23 rs2066808 and heart failure”, “IL‐23 rs2066808 and coronary artery disease”, “IL‐23 rs2066808 and dilated cardiomyopathy”, “IL‐23 rs2066808 and metabolic syndrome”, “IL‐23R rs2201841 and cardiovascular disease”, “IL‐23R rs2201841 and atherosclerosis”, “IL‐23R rs2201841 and heart failure”, “IL‐23R rs2201841 and coronary artery disease”, “IL‐23R rs2201841 and dilated cardiomyopathy”, “IL‐23R rs2201841 and metabolic syndrome”, “IL‐17RA rs4819554 and cardiovascular disease”, “IL‐17RA rs4819554 and atherosclerosis”, “IL‐17RA rs4819554 and heart failure”, “IL‐17RA rs4819554 and coronary artery disease”, “IL‐17RA rs4819554 and dilated cardiomyopathy”, “IL‐17RA rs4819554 and metabolic syndrome”, “IL‐17A rs2275913 and cardiovascular disease”, “IL‐17A rs2275913 and atherosclerosis”, “IL‐17A rs2275913 and heart failure”, “IL‐17A rs2275913 and coronary artery disease”, “IL‐17A rs2275913 and dilated cardiomyopathy”, “IL‐17A rs2275913 and metabolic syndrome”.
We also search “SNPedia” for relevant studies. The reference lists of the included studies were checked for other relevant publications. We included in the review case‐control studies published between 2007 and 2025, written in English, which provided data regarding allele frequency or genotype distribution. We excluded duplicates, and studies that investigated family members. Of the 237 identified documents, after the full‐text review, only 22 articles were eligible for inclusion. The references of the studies were also screened for other studies suitable for inclusion in the present review. Figure 1 shows the literature search performed for the present study.
Figure 1.
Article selection protocol.
2.2. Data Collection
We extracted the following data from original studies: first author, year of publication, ethnicity, the number or ratio of cases and controls with minor allele, and the number or ratio of cases and controls with genotypes of the three variations, if available.
3. Results
3.1. IL‐23R rs2201841 and IL‐23 rs2066808 and Psoriasis
Indhumathi et al. [44] investigated the association between IL‐23R polymorphism and psoriatic risk in the Indian population. The case‐control study mentioned above included 360 psoriasis patients and 360 controls [44]. The results showed an increased risk of psoriasis associated with IL‐23R rs2201841 polymorphism in South Indian Tamils [44]. Additionally, the plasma level of IL‐23 was increased in the case of the minor variant genotype CC when compared with that of CT heterozygous and TT major variant genotypes of rs2201841 [44]. However, no significant association between IL‐23R rs2201841 polymorphism, IL‐23 levels and PASI score was found [44]. A more than twofold increase in the CC genotype in psoriasis patients compared to controls was also identified in a study conducted by Safrany et al. [45]. On the other hand, in a study also conducted by Safrany [46] in 2013, which investigated the differences between IL‐23 polymorphism rs220184 in ulcerative colitis compared to Crohn's disease and psoriasis, the only association found was with ulcerative colitis [46]. The association between psoriasis and IL‐23R rs2201841 polymorphism was demonstrated in a case‐control study by Nair et al. [47] in 2009. The results were further confirmed by another case‐control study by Eirís et al. [48]. The same case control‐study identified an association between the minor genotypic variant IL23R rs2201841‐GG and diabetes mellitus Type 2 (p = 0.027) [48]. Nikamo et al. [49] demonstrated that the rs2201841 is not only associated with severe forms (p = 0.004) of psoriasis but also with mild disease forms (p = 0.006). Contrarily, Popadic et al. [50] found an association between IL‐23R rs2201841 polymorphism and psoriatic arthritis, but not with psoriasis. No association between rs2201841 polymorphism and an increase in psoriasis risk [51, 52, 53, 54], psoriasis severity (PASI SCORE) [54], and no influence on treatment response (PASI value) was shown in another study either [53].
In a study conducted on the Spanish population, no association between rs2066808 (A > G) and psoriasis was found [48], but an association with psoriatic arthritis (AA homozygotes, p = 0.02) and nail psoriasis (p = 0.03) was present [48]. Contrarily, Nair et al. [47] in a case‐control study, found a strong association between the rs2066808 and psoriasis, further confirmed by an analysis of follow‐up samples (5048 cases, and 5051 controls) [47]. A higher risk of developing psoriasis in patients presenting the polymorphism above was demonstrated by Chen at el. [55] in a study conducted in the USA on 713 psoriasis cases and 2084 controls. IL‐23 rs2066808 is also associated with severe forms when analyzing severe versus control and severe versus mild cases [49]. The collected data regarding the implication of IL‐23R rs2201841 and IL‐23 rs2066808 polymorphisms in psoriasis development are presented in Table 1.
Table 1.
The association between the IL‐23R rs2201841 and IL‐23 rs2066808 polymorphisms and psoriasis.
| Gene and SNP | Year | Population | Genotype/allele | Psoriasis patients | Frequency of risk allele | Control group | Frequency of risk allele | p value | OR | 95% CI | Genotyping | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL‐23R rs2201841 | Capon (2007) | British | N = 318 | MAF 0.35 (218/624) | N = 288 | MAF = 0.32 (186/574) | N/S | 1.10 | Applied Biosystems‐ TaqMan assays or SnapShot assay protocol‐single‐base primer extension | [52] | ||
| Nair (2009) | USA | Risk allele: G | N = 1359 | N = 1400 | p = 2.9 × 10−7 | 1.347 | Applied Biosystems‐TaqMan assays or Sequenom single base extension assays or allele‐specific kinetic PCR. | [47] | ||||
| Non‐risk allele: A | 0.350 | 0.286 | ||||||||||
| G/G | 158 | 117 | ||||||||||
| G/A | 635 | 565 | ||||||||||
| A/A | 566 | 717 | ||||||||||
| USA | N = 1642 | N = 1101 | p = 0.0081 | 1.172 | [47] | |||||||
| Risk allele G | 0.325 | 0.291 | ||||||||||
| Non‐risk allele A | ||||||||||||
| G/G | 167 | 93 | ||||||||||
| G/A | 724 | 454 | ||||||||||
| A/A | 735 | 551 | ||||||||||
| Canada | Risk allele: G | N = 368 | N = 358 | p = 0.571 | 0.937 | [47] | ||||||
| Non‐risk allele: A | 0.314 | 0.328 | ||||||||||
| G/G | 33 | 44 | ||||||||||
| G/A | 154 | 138 | ||||||||||
| A/A | 163 | 162 | ||||||||||
| Canada | Risk allele: G | N = 691 | N = 217 | p = 0.730 | 1.041 | [47] | ||||||
| Non‐risk allele: A | 0.335 | 0.326 | ||||||||||
| G/G | 83 | 23 | ||||||||||
| G/A | 289 | 94 | ||||||||||
| A/A | 308 | 98 | ||||||||||
| 98 | ||||||||||||
| Germany | Risk allele: G | N = 718 | N = 1464 | p = 0.014 | 1.190 | [47] | ||||||
| Non‐risk allele: A | 0.312 | 0.276 | ||||||||||
| G/G | 71 | 122 | ||||||||||
| G/A | 301 | 559 | ||||||||||
| A/A | 337 | 772 | ||||||||||
| USA | Risk allele: G | N = 302 | N = 500 | p = 0.312 | 1.119 | [47] | ||||||
| Non‐risk allele: A | 0.332 | 0.307 | ||||||||||
| G/G | 31 | 49 | ||||||||||
| G/A | 137 | 205 | ||||||||||
| A/A | 132 | 239 | ||||||||||
| France | Risk allele: G | N = 346 | N = 486 | p = 0.521 | 1.214 | [47] | ||||||
| Non‐risk allele: A | 0.313 | 0.273 | ||||||||||
| G/G | 35 | 38 | ||||||||||
| G/A | 144 | 187 | ||||||||||
| A/A | 163 | 257 | ||||||||||
| USA | Risk allele: G | N = 498 | N = 498 | p = 0.468 | 1.073 | [47] | ||||||
| Non‐risk allele: A | 0.325 | 0.310 | ||||||||||
| G/G | 63 | 49 | ||||||||||
| G/A | 194 | 207 | ||||||||||
| A/A | 235 | 236 | ||||||||||
| USA | Risk allele: G | N = 483 | N = 427 | p = 0.276 | 1.115 | [47] | ||||||
| Non‐risk allele: A | 0.343 | 0.319 | ||||||||||
| G/G | 61 | 44 | ||||||||||
| G/A | 209 | 184 | ||||||||||
| A/A | 212 | 198 | ||||||||||
| Sarany (2011) | Hungarian | N = 214 | N = 189 | PCR‐RFLP | [45] | |||||||
| Genotype | ||||||||||||
| TT | 102 (47.7%) | 101 (53.4%) | ||||||||||
| TC | 87 (40.7%) | 79 (41.8%) | ||||||||||
| CC | 25 (11.7%) | 9 (4.76%) | p = 0.016 | 2.64 | 1.20–5.81 | |||||||
| TC + CC | 112 (52.3%) | 88 (46.6%) | ||||||||||
| MAF | 0.32 | 0.26 | ||||||||||
| Safrany (2013) | Hungarian | N = 263 | N = 253 | NS | PCR‐RFLP | [46] | ||||||
| Genotype | ||||||||||||
| TT | 33.8% | 48.6% | ||||||||||
| TC | 48.7% | 45.1% | ||||||||||
| CC | 17.5% | 6.32% | ||||||||||
| MAF | 0.42 | 0.29 | ||||||||||
| Popadic (2014) | Serbia | N = 103 | N = 291 | Applied Biosystems‐TaqMan assays | [50] | |||||||
| Allele: | ||||||||||||
| A | 136 | 0.660 | 403 | 0.692 | 0.393 | |||||||
| G | 70 | 0.340 | 179 | 0.308 | ||||||||
| Genotype | ||||||||||||
| AA | 44 | 0.427 | 145 | 0.498 | 0.215 | |||||||
| GA | 48 | 0.466 | 113 | 0.388 | 0.168 | |||||||
| GG | 11 | 0.107 | 33 | 0.113 | 0.862 | |||||||
| Eirís (2014) | Spanish | N = 405 | N = 426 | Applied Biosystems‐TaqMan assays | [48] | |||||||
| Allele: | ||||||||||||
| A | 0.70 | 0.65 | ||||||||||
| G | 0.30 | 0.35 | ||||||||||
| Genotype | ||||||||||||
| AA | 197 | 0.49 | 175 | 0.41 | ||||||||
| GG | 174 | 0.43 | 200 | 0.47 | ||||||||
| AG | 34 | 0.08 | 51 | 0.12 | ||||||||
| AA vs. (AG + GG) | p = 0.03 | 1.36 | 1.03–1.79 | |||||||||
| Nikamo (2015) | Caucasians/Sweden | N = 1411 | N = 1529 | Applied Biosystems‐TaqMan assays | [49] | |||||||
| Severe vs. Controls | Severe = 715 | p = 0.004 | 1.22 | (1.07–1.40) | ||||||||
| Mild vs. Controls | Mild = 696 | p = 0.006 | 0.99 | (0.86–1.14) | ||||||||
| Indhumathi (2016) | Indian | N = 360 | N = 360 | TaqMan assays | [44] | |||||||
| Genotype | ||||||||||||
| TT vs. CT + CC | 280 (77.8%) | 239 (66.4%) | p = 0.0009 | 1.77 | 1.27–2.47 | |||||||
| CC | 114 (31.7%) | 90 (25%) | p = 0.002 | 1.92 | 1.29–2.85 | |||||||
| CT | 166 (46.1%) | 149 (41.4%) | p = 0.006 | 1.69 | 1.18–2.41 | |||||||
| TT | 80 (22.2%) | 121 (33.6%) | ||||||||||
| Allele: | ||||||||||||
| C | 394 (54.7%) | 329 (45.7%) | p = 0.0007 | 1.44 | 1.17–1.77 | |||||||
| T | 326 (45.3%) | 391 (54.3%) | ||||||||||
| Bojko (2018) | Poland | N = 507 | N = 396 | p = 0.874 | 0.99 | 0.75–1.30 | Applied Biosystems‐TaqMan assays | [53] | ||||
| Risk allele | ||||||||||||
| G | MAF = 0.284 | MAF = 0.280 | ||||||||||
| Filiz (2019) | Turkish | N = 60 | N = 60 | p = 0.673 | Competitive allele‐specific PCR (KASP) assay | [51] | ||||||
| Allele/Genotype | ||||||||||||
| AA | 16 (26.7%) | 17 (28.4%) | ||||||||||
| GA | 29 (48.3%) | 32 (53.3%) | ||||||||||
| GG | 15 (25%) | 11 (18.3%) | ||||||||||
| Lin (2022) | Asian (China) | N = 43 | N = 24 | MALDI‐TOF mass spectrometry | [54] | |||||||
| Genotype | ||||||||||||
| GG | 29 | 15 | p = 0. 303 | |||||||||
| AG | 13 | 6 | ||||||||||
| AA | 1 | 3 | ||||||||||
| Allele | ||||||||||||
| A | 71 | 36 | p = 0.296 | 1.578 | 0.669–3.723 | |||||||
| G | 15 | 12 | 1.57 | |||||||||
| IL‐23 rs2066808 | Nair (2009) | USA | N = 1359 | N = 1400 | p = 1.7E‐05 | 1.676 | Applied Biosystems‐TaqMan assays or Sequenom single base extension assays or allele‐specific kinetic PCR. | [47] | ||||
| Allele: | ||||||||||||
| A‐risk | 0.958 | 0.931 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 1246 | 1215 | ||||||||||
| AG | 111 | 177 | ||||||||||
| GG | 2 | 8 | ||||||||||
| USA | N = 1642 | N = 1101 | p = 0.0014 | 1.447 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.951 | 0.930 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 1472 | 947 | ||||||||||
| AG | 151 | 139 | ||||||||||
| GG | 5 | 7 | ||||||||||
| Canada | N = 368 | N = 358 | p = 0.206 | 1.318 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.946 | 0.929 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 328 | 304 | ||||||||||
| AG | 38 | 50 | ||||||||||
| GG | 1 | 0 | ||||||||||
| Canada | N = 691 | N = 217 | p = 0.145 | 1.395 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.951 | 0.933 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 620 | 190 | ||||||||||
| AG | 65 | 25 | ||||||||||
| GG | 1 | 2 | ||||||||||
| Germany | N = 718 | N = 1464 | p = 0.014 | 1.405 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.948 | 0.928 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 635 | 1256 | ||||||||||
| AG | 72 | 185 | ||||||||||
| GG | 1 | 12 | ||||||||||
| USA | N = 302 | N = 500 | p = 0.0079 | 1.882 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.960 | 0.927 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 276 | 425 | ||||||||||
| AG | 24 | 68 | ||||||||||
| GG | 0 | 2 | ||||||||||
| France | N = 346 | N = 486 | p = 0.877 | 0.851 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.958 | 0.964 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 313 | 447 | ||||||||||
| AG | 29 | 35 | ||||||||||
| GG | 0 | 0 | ||||||||||
| USA | N = 498 | N = 498 | p = 0.953 | 0.990 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.924 | 0.925 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 420 | 420 | ||||||||||
| AG | 71 | 68 | ||||||||||
| GG | 2 | 3 | ||||||||||
| USA | N = 483 | N = 427 | p = 0.526 | 1.131 | [47] | |||||||
| Allele: | ||||||||||||
| A‐risk | 0.941 | 0.933 | ||||||||||
| G‐non‐risk | ||||||||||||
| Genotype: | ||||||||||||
| AA | 424 | 367 | ||||||||||
| AG | 57 | 52 | ||||||||||
| GG | 0 | 2 | ||||||||||
| Chen (2011) | USA | N = 731 | N = 2084 | p = 0.0001 | Illumina GoldenGate assay | [55] | ||||||
| Risk allele: T | ||||||||||||
| Nonrisk allele: C | 0.955 | 0.920 | 1.82 | 1.34–2.45 | ||||||||
| Eirís (2014) | Spanish | N = 405 | N = 426 | Applied Biosystems‐TaqMan assays | [48] | |||||||
| Allele: | ||||||||||||
| A | 0.93 | 0.92 | ||||||||||
| G | 0.07 | 0.08 | ||||||||||
| Genotype | ||||||||||||
| AA | 350 | 0.86 | 362 | 0.85 | ||||||||
| AG | 55 | 0.14 | 63 | 0.147 | ||||||||
| GG | 0 | 0 | 1 | 0.003 | ||||||||
| AA vs. (AG + GG) | p = 0.55 | 1.13 | 0.76–1.66 | |||||||||
| Nikamo (2015) | Caucasians/Sweden | N = 1411 | N = 1529 | Applied Biosystems‐TaqMan assays | [49] | |||||||
| Severe = 715 | ||||||||||||
| Mild = 696 | ||||||||||||
| Severe vs. Controls | p = 2 × 10−7 | 0.44 | (0.32–0.60) | |||||||||
| Mild vs. controls | p = 0.0005 | 0.78 | (0.61–1.01) |
Note: *p < 0.05, **p value for homozygosity of minor allele, N/S not significant, PCR‐RFLPolymerase chain reaction–restriction fragment length polymorphism
Abbreviation: MAF, minor allele frequency.
3.2. IL‐23R rs2201841 and IL‐23 rs2066808 and Cardiometabolic Diseases
Psoriasis patients have a higher prevalence of cardiovascular risk factors [56]. The risk of myocardial infarction and stroke is increased even in patients with mild psoriasis [26].
The IL‐23 receptor (IL‐23R) gene polymorphism rs2201841 has been implicated in CVD through its role in inflammatory pathways [57]. Research indicates that variations in IL‐23R can influence susceptibility to atherosclerosis and coronary artery disease (CAD) [57]. The Th17/IL‐23 axis is recognized for its role in inflammation, which is a critical factor in both CAD and other cardiometabolic disorders [57]. Eirís et al. [48] in their study published in 2014, showed an association between Type 2 diabetes mellitus in psoriasis patients and the minor genotypic variant of IL‐23R rs2201841‐GG. Additionally, factors such as age, dyslipidemia, hypertension or ischemic cardiomyopathy did not represent confounding factors after logistic regression [48]. There was no association found between IL‐23 rs2066808 and cardiometabolic diseases or cardiovascular risk factors [48, 58].
3.3. IL‐17RA rs4819554 and IL17A rs2275913 and Psoriasis
Batalla et al. [59] in a case‐control study, conducted in the Spanish population found an association between IL17RA rs4819554 SNP and the risk for psoriasis, namely the G carriers (AG + GG) were significantly more common (p = 0.017, OR = 1.33 (95% CI = 1.05–1.69). The presence of the aforementioned polymorphism had no impact on the psoriasis severity, presence of PsA, or demographics [59]. Additionally, the number of the IL17RA rs4819554 G‐carriers was higher in the Cw6‐positive subgroup, an association that lost statistical significance after adjusting for age of onset as a confounding factor [59]. Sabry et al. [60] found a significantly increased expression of IL‐17 in psoriasis patients vs. controls (p = 0.0006) and in patients with AA, GG and AG rs4819554 genotypes compared to the same genotypes controls (p = 0.0004). Also, the serum levels of IL‐17 and psoriasin were significantly higher in psoriasis patients compared to controls, especially in the AA and AG genotypes. No significant IL‐17 serum levels differences were found in GG genotypes in patients and controls [60].
A prospective, longitudinal, analytical cohort study by Puscaș et al. [61] published in 2023, including 81 patients with moderate to severe plaque psoriasis, found an association between IL‐17RA rs4819554 polymorphism, the GG genotype compared to those with GA/AA genotype and nail psoriasis and a higher BMI in the psoriasis patients.
A meta‐analysis by Villalpando‐Vargas et al. [62] showed that the presence of AG genotype of the rs4819554 polymorphism was associated with an almost significant reduction in the psoriasis risk when compared to the GG genotype (p = 0.050).
Lin et al. [54] included in a study conducted on Asian population 43 psoriasis patients and 24 controls. The results showed no association between 17RA rs4819554 polymorphism and psoriasis. In a case‐control study, conducted on the Polish population, Białecka et al. [63] found no association between IL17A rs2275913 polymorphisms and psoriasis, but they found that the IL‐17F polymorphisms could predict the treatment response to UVB‐nb phototherapy.
Kim et al. [64] in a case‐control study conducted on the Korean population, concluded that there is no association between the IL17A rs2275913 polymorphism and psoriasis. The same results were found in the Spanish population in a case‐control study by Prieto‐Pérez et al. [65] in 2015. The collected data regarding the implication of the IL‐17RA rs4819554 and IL17A rs2275913 polymorphisms in psoriasis development are presented in Table 2.
Table 2.
The association between the IL‐17RA rs4819554 and IL17A rs2275913 polymorphisms and psoriasis.
| Gene and SNP | Study | Population | Genotype/allele | Psoriasis patients | Control group | p value | OR | 95% CI | Genotyping | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4819554 | Batalla (2015) | Spanish | N = 580 | N = 567 | Applied Biosystems‐TaqMan assays | [59] | ||||
| Genotype: | ||||||||||
| AA | 57 | 63 | ||||||||
| AG | 38 | 33 | ||||||||
| GG | 5 | 4 | ||||||||
| Allele: | ||||||||||
| A | 0.76 | 0.80 | ||||||||
| G | 0.24 | 0.20 | ||||||||
| AG + GG vs. AA | p = 0.017 | 1.33 | 1.05–1.69 | |||||||
| Sabry (2020) | Egyptian | N = 100 | N = 100 | Applied Biosystems‐TaqMan assays | [60] | |||||
| Genotype: | ||||||||||
| AA | 20% | 48% | p = 0.007 | 2.283 | 1.321–3.946 | |||||
| GG | 22% | 6% | p = 0.029 | 0.344 | 0.132–0.895 | |||||
| AG | 58% | 46% | ||||||||
| Allele: | ||||||||||
| A | 49% | 71% | p = 0.003 | 2.283 | 1.321–3.946 | |||||
| Lin (2022) | Asian | N = 43 | N = 24 | MALDI‐TOF MS | [54] | |||||
| Genotype: | ||||||||||
| AG | 13 | 12 | p = 0.142 | |||||||
| AA | 21 | 6 | ||||||||
| GG | 9 | 6 | ||||||||
| Allele | ||||||||||
| A | 55 | 24 | p = 0.115 | 1.774 | 0.866–3.634 | |||||
| G | 31 | 24 | ||||||||
| rs2275913 | Prieto‐Pérez (2015) | Spanish | N = 194 | N = 197 | Applied Biosystems‐TaqMan assays | [65] | ||||
| Genotype: | ||||||||||
| AA | 16 (8%) | 24 (12%) | ||||||||
| GA | 93 (48%) | 77 (39%) | p = 0.135; C;GA | 1.40 | 0.92–2.13 | |||||
| GG | 83 (43%) | 96 (49%) | ||||||||
| Allele: | ||||||||||
| A | 125 (32.2%) | 125 (31.7%) | ||||||||
| Białecka (2016) | Polish | N = 407 | N = 205 | p = 0.70 | Applied Biosystems‐TaqMan assays | [63] | ||||
| Genotype: | ||||||||||
| AA | 53 (13.18%) | 28 (13.73%) | ||||||||
| GA | 173 (43.03%) | 94 (46.08%) | ||||||||
| GG | 176 (43.78%) | 82 (40.20%) | ||||||||
| AA + GA vs. GG | p = 0.43 | 0.86 | 0.61–1.22 | |||||||
| AA vs. GA + GG | p = 0.90 | 0.95 | 0.58–1.56 | |||||||
| AA vs. GG | p = 0.68 | 0.88 | 0.52–1.49 | |||||||
| Kim (2016) | Korean | N = 208 | N = 266 | PCR | [64] | |||||
| Alleles | ||||||||||
| G > A | 0.463 | 0.434 | p = 0.38 | 1.12 | 0.86–1.46 | |||||
| Ozkol (2020) | Turkish | N = 83 | N = 69 | Thermo Scientific, USA‐ TaqMan SNP Assay mix | [66] | |||||
| Genotype: | ||||||||||
| AA | 11 (57.9%) | 8 (42.1%) | ||||||||
| GA | 38 (55.9%) | 30 (44.1%) | ||||||||
| GG | 34 (53.1%) | 31 (46.9%) | p = 0.992 |
Abbreviations: C, codominant; MALDI‐TOF MS, matrix assisted laser desorption ionization‐time of flight mass spectrometry; PCR, polymerase chain reaction.
3.4. IL‐17RA rs4819554 and IL17A rs2275913 and Cardiometabolic Diseases
Psoriasis patients present a higher risk of CVDs and metabolic syndrome compared to the general population [67]. A common genetic background is speculated as presented in a review by Piaserico et al. [40]. One of the IL‐17RA polymorphisms, namely rs4819554, which is associated with psoriasis, is also linked to CVM, but not with heart failure [68]. Additionally, Coto et al. [69] found a higher IL‐17RA rs4819554 transcript in coronary artery disease patients, but no difference in genotype frequencies in patients and controls.
Regarding the IL‐17A rs2275913 and CVDs, no association was found with heart failure or cardiovascular death [68]. On the other hand, Geng et al. [70] found that IL‐17A rs2275913 may be implicated in the development of coronary artery disease (CAD) in codominant (p = 0.01) and recessive (p = 0.03) models. This association between the 17A rs2275913 and CAD was further found in another case‐control study [71] and seems to be linked to an early onset of CAD [71]. Zhang et al. [72] also studied the implications of the aforementioned polymorphism in CAD [72]. They included in their study 1031 patients with CAD and 935 controls. They defined the presence of CAD by 50% stenosis in major coronary arteries, or a prior history of angioplasty, major cardiac event, or bypass surgery. They did not find any association between the IL‐17A rs2275913 and coronary artery disease [72]. A statistically significant association between the IL‐17A rs2275913 and CAD was identified by Zhao et al. [73] but, after adjusting for confounding factors such as age, gender, smoking, drinking, hypertension, and Type 2 diabetes mellitus, the association was no longer valid. Su et al. [74] in a case‐control study conducted in the Chinese population in 2016, also found no association between the aforementioned polymorphism and CAD. Furthermore, there is no link between the aforementioned polymorphism and dilated cardiomyopathy [75]. The collected data regarding the implication of the IL‐17RA rs4819554 and IL17A rs2275913 polymorphisms in cardiometabolic diseases are presented in Table 3.
Table 3.
The association between the IL‐17RA rs4819554 and IL17A rs2275913 polymorphisms and cardiometabolic diseases.
| Gene and SNP | Study | Disease | Population | Genotype/allele | Patient group (%) | Control group (%) | Model | p value | OR | 95% CI | Genotyping | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL‐17RA rs4819554 | Sandip (2016) | HF | Chinese | N = 1713 | N = 1713 | p = 0.46 | 1.04 | 0.94–1.46 | Applied Biosystems‐TaqMan assays | [68] | ||
| Genotype: | 554 (32.3%) | 560 (32.7%) | ||||||||||
| GG | ||||||||||||
| GA | 769 (44.9%) | 780 (45.5%) | ||||||||||
| AA | 390 (22.8%) | 373 (21.8%) | ||||||||||
| MAF | 45.2% | 44.5% | ||||||||||
| IL17A rs2275913 | Zhang (2011) | CAD | Chinese | N = 1031 | N = 935 | PCR | [72] | |||||
| Genotype: | 290 (28.1%) | 266 (28.4%) | 0.92 | |||||||||
| GG | ||||||||||||
| GA | 535 (51.9%) | 489 (52.3%) | ||||||||||
| AA | 206 (20.0%) | 180 (19.3%) | ||||||||||
| AlleleG | 1115 (54.1%) | 1021 (54.6%) | 0.74 | 1.02 | 0.90‐1.15 | |||||||
| A | 947 (45.9%) | 849 (45.4%) | ||||||||||
| Peng (2013) | DC | Chinese | N = 288 | N = 421 | Applied Biosystems‐TaqMan assays | [75] | ||||||
| Genotype: | 96 (33.3%) | 155 (36.8%) | CoD | 0.64 | 1 | Ref. | ||||||
| GG | ||||||||||||
| AG | 142 (49.3%) | 193 (45.8%) | 0.85 | 0.61–1.19 | ||||||||
| AA | 50 (17.4%) | 73 (17.3%) | 0.91 | 0.58‐1.42 | ||||||||
| GG | 96 (33.3%) | 155 (36.8%) | D | 0.37 | 1 | Ref. | ||||||
| AG/AA | 192 (66.7%) | 266 (63.2%) | 0.87 | 0.63‐1.19 | ||||||||
| GG/AG | 238 (82.6%) | 348 (82.6%) | R | 1 | 1 | Ref. | ||||||
| AA | 50 (17.4%) | 73 (17.5%) | 1 | 0.67‐1.49 | ||||||||
| GG/AA | 146 (50.7%) | 228 (54.2%) | Ovd | 0.4 | 1 | Ref. | ||||||
| AG | 142 (49.3%) | 193 (45.8%) | 0.88 | 0.65‐1.19 | ||||||||
| — | — | — | LA | 0.54 | 0.94 | 0.75‐1.16 | ||||||
| Geng (2015) | CAD | Chinese | N = 306 | N = 306 | PCR‐RFLP | [70] | ||||||
| Genotype: GG | 123 (40.2%) | 146 (47.71%) | CoD | — | Ref. | |||||||
| GA | 140 (45.75%) | 134 (43.79%) | 0.21 | 1.24 | 0.87‐1.76 | |||||||
| AA | 43 (14.05%) | 26 (8.5%) | 0.01 | 1.96 | 1.10‐3.53 | |||||||
| GG | 123 (40.2%) | 146 (47.71%) | D | — | Ref. | |||||||
| GA+AA | 183 (59.8%) | 160 (52.29%) | 0.06 | 1.36 | 0.97‐1.89 | |||||||
| GG+GA | 263 (85.95%) | 280 (91.5%) | R | — | Ref. | |||||||
| AA | 43 (14.05%) | 26 (8.5%) | 0.03 | 1.76 | 1.02‐3.07 | |||||||
| Vargas‐Alarcón (2015) | CAD | Mexican | Genotypes | N = 900 | N = 667 | D | 0.55/0.62 | 0.91 | 0.70‐1.17 | Applied Biosystems‐TaqMan assays | [76] | |
| 616 (0.68) | 439 (0.66) | |||||||||||
| 259 (0.28) | 202 (0.30) | |||||||||||
| 23 (0.03) | 26 (0.04) | |||||||||||
| MAF | 0.16 | 0.19 | 0.44 | |||||||||
| Shuang (2015) | CAD | Asian/Chinese | N = 415 | N = 448 | PCR‐RFLP | [77] | ||||||
| Genotypes GG | 168 (40.48%) | 220 (49.11%) | — | 1.0 | Ref. | |||||||
| GA | 187 (45.06%) | 196 (43.75%) | 0.12 | 1.25 | 0.93‐1.68 | |||||||
| AA; BC: G>A | 60 (14.46%) | 36 (8.04%) | Phwe = 0.4 | 0.001 | 2.18 | 1.35‐3.56 | ||||||
| GA+AA | 247 (59.52%) | 232(51.79%) | 0.02 | 1.39 | 1.06‐1.84 | |||||||
| Sandip (2016) | HF | Chinese | N = 1713 | N = 1713 | P=0.30 | 0.95 | 0.85–1.05 | Applied Biosystems‐TaqMan assays | [68] | |||
| Genotype: GG | 494 (28.8%) | 461 (26.9%) | ||||||||||
| GA | 840 (49.0%) | 876 (51.1%) | ||||||||||
| AA | 379 (21.1%) | 376 (21.9%) | ||||||||||
| MAF | 46.6% | 47.5% | ||||||||||
| Zheng (2016) | CAD | Asian/Chinese | N = 372 | N = 372 | PCR‐RFLP | [78] | ||||||
| Grnotypes GG | 162 (43.55%) | 179 (48.12%) | 1.0 | Ref. | ||||||||
| GA | 170 (45.7%) | 163 (43.82%) | 1.15 | 0.84‐1.58 | ||||||||
| AA, BC: G>A | 40 (10.75%) | 30 (8.06%) | 0.4 | 1.47 | 0.85‐2.57 | |||||||
| GA+AA | 210 (56.45%) | 193 (51.88%) | 1.20 | 0.89‐1.62 | ||||||||
| Su (2016) | CAD | Chinese | N = 219 | N = 219 | SNaPshot SNP genotyping assay (Genesky, Shanghai, China) Td‐PCR | [74] | ||||||
| Genotypes/alleles: | 93 (42.5%) | 95 (43.4%) | 0.847 | 0.963 | 0.660‐1.407 | |||||||
| GG | ||||||||||||
| AG | 102 (46.6%) | 101 (46.1%) | 0.924 | 1.019 | 0.700‐1.483 | |||||||
| AA | 24 (11.0%) | 23 (10.5%) | 0.877 | 1.049 | 0.573‐1921 | |||||||
| G | 288 (65.8%) | 291 (66.4%) | 0.830 | 0.970 | 0.733‐1.283 | |||||||
| A | 150 (34.2%) | 147 (33.6%) | ||||||||||
| Zhao (2019) | CAD | Chinese | N = 191 | N = 131 | MALDI‐TOF MS | [73] | ||||||
| Genotype: | 30 (15.71%) | 33 (25.19%) | Ref. | Ref. | ||||||||
| GG | ||||||||||||
| AG | 112 (58.64%) | 73 (55.73%) | 1.45 | 0.78‐ 2.67 | ||||||||
| AA | 49 (25.64%) | 25 (19.08%) | 2.21 | 0.95‐5.19 | ||||||||
| AG/AA | 161 (84.29%) | 98 (74.81%) | 1.62 | 0.89‐2.93 | ||||||||
| Ghaznavi (2020) | CAD | Iran | N = 220 | N = 220 | PCR‐RFLP | [79] | ||||||
| Genotypes/alleles | 88 (40%) | 113 (51.36%) | ‐ | Ref. | ||||||||
| GG | ||||||||||||
| GA | 98 (44.54%) | 89 (40.45%) | 0.104 | 1.41 | 0.95‐2.10 | |||||||
| AA | 34 (15.46%) | 18 (8.19%) | 0.007 | 2.42 | 1.26‐4.54 | |||||||
| G | 274 (62.27) | 315 (71.60%) | ‐ | Ref. | ||||||||
| A | 166 (37.73%) | 125 (28.40%) | 0.041 | 1.53 | 1.16‐2.03 | |||||||
| Zhang (2023) | CAD | Chinese | N = 3042 | N = 3216 | PCR | [71] | ||||||
| Genotypes: Allele | 883/1950 | 907/179 | DOM | Pobs/padj = 0.059/0.255 | 1.078 | 0.947,1.226 | ||||||
| A | ||||||||||||
| 2269/564 | 2167/537 | REC | Pobs/padj = 0.964/0.247 | 0.914 | 0.786,1.064 | |||||||
| 883/1386/564 | 907/1260/537 | ADD | Pobs/padj = 0.137/0.920 | 1.004 | 0.923,1.093 | |||||||
| MAF | 0.44 | 0.43 | Phwe = 0.009 | 1.004 | 0.922‐1.095 | |||||||
| Pobs = 0.199 | ||||||||||||
| Padj = 0.919 |
Abbreviations: BC, base change; CAD, coronary artery disease; CoD, codominant model; D, dominant model; DC, dilated cardiomyopathy; HF, heart failure; LA, log‐additive; MALDI‐TOF MS, matrix assisted laser desorption ionization‐time of flight mass spectrometry; OR, odds ratio after adjustment; OvD, overdominant; p adj, p value adjusting for age, sex, BMI, hypertension, diabetes mellitus, smoking history, Tch, TG, HDL‐c, and LDL‐c; PCR, polymerase chain reaction; PCR‐RFLP, polymerase chain reaction combined with restriction fragment length polymorphism; p hwe, p value of the Hardy–Weinberg equilibrium tests; p obs, observed p value; R, recessive model; Td‐PCR‐touch‐down polymerase chain reaction.
4. Discussion
Interleukin‐17 plays a pivotal role in the pathogenesis of psoriasis. Its multifaceted involvement influences immune responses, keratinocyte behavior, and metabolic processes [80]. T helper cells produce IL‐17A, crucial in mediating psoriasis inflammatory responses [80]. The IL‐23/IL‐17 axis perpetuates the inflammation, leading to keratinocyte activation and hyperproliferation [81], together with IL‐17A‐induced metabolic reprogramming, which also contributes to their proliferation and the formation of psoriatic plaques [82]. IL‐23 triggers the production of IL‐17 [83]. Keratinocytes produce IL‐23, which further contributes to the inflammatory milieu in psoriasis [84]. Figure 2 presents the implication of IL‐17/IL‐23 axis in psoriasis and atherosclerosis.
Figure 2.
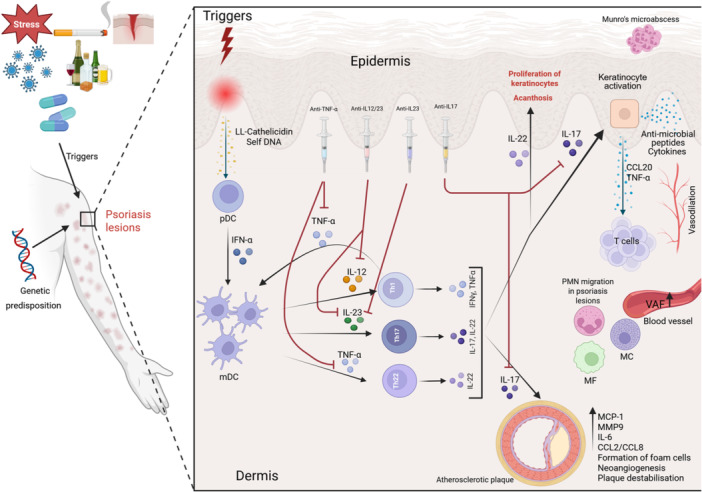
(Created in BioRender. Lazar, L. (2025) https://BioRender.com/ln3xljd). Note: The link between psoriasis and atherosclerosis: Psoriasis is triggered by non‐specific danger signals (Triggers) consisting of infections, drugs, physical trauma, alcohol, smoking, and autoantigens that act in an individual with a genetic predisposition for the disease. In the initial stages, a major role is played by DCs, antigen‐presenting cells, that recognize antimicrobial peptides (LL‐Cathelicidin) and self‐DNA released by damaged keratinocytes. The activation of plasmacytoid DC and IFN‐α release lead to dermal DC maturation and the release of TNF‐α, IL‐23, and IL‐12 responsible for Th1, Th17, and Th22 activation, differentiation, and the production of cytokines, namely TNF‐α, IFN‐γ, IL‐17, and IL‐22, that disrupt the keratinocyte function and intensify the inflammation. The TNFα‐IL‐23‐Th17 axis perpetuates inflammation in psoriasis. The keratinocyte activation induced by IL‐17 leads to their proliferation and the release of TNF‐α and CCL20, that chemoattracts T cells that produce more IL‐17. The expression of vascular adhesion factors is higher in psoriasis lesions, and it further enhances the adhesion of inflammatory cells to the endothelial cells, perpetuating the inflammation. The blood flow in psoriatic lesions is altered through the vasodilation of the blood vessels in the papillary dermis. The IL‐17 causes endothelial dysfunction, it mediates the expression of chemokines MCP‐1, MMP‐9, it upregulates the IL‐6 and also the chemokines CCL‐2 and CCL8, thus promoting atherosclerosis. The key cytokines implicated in these processes are the targets for the biological treatments, which could halt the detrimental process. pDC‐ Plasmacytoid DCs, IFN‐α‐Interferon alpha, mDC‐Myeloid DCs, TNF‐α‐Tumor necrosis factor alpha, IL‐12‐ Interleukin‐12, IL‐22‐ Interleukin‐22, IL‐23‐ Interleukin‐23, IL‐17‐ Interleukin‐17, IFN‐γ‐Interferon gamma, CCL‐20‐CC chemokine ligand 20, VAF‐ Vascular adhesion factors, PMN‐Polymorphonuclear neutrophils, MF‐Macrophage, MC‐Mast cells, MCP‐1‐Human monocyte chemoattractant protein‐1, MMP9‐Matrix metalloproteinase‐9, IL‐6‐ Interleukin‐6, CCL2‐C‐C motif ligand 2, CCL8‐C‐C motif ligand 8.
Biologics targeting IL‐23, such as risankizumab and guselkumab, have shown significant efficacy in treating psoriasis by inhibiting the aforementioned pathway [85]. Biologic therapies not only alleviate skin symptoms but may also impact the associated comorbidities [86]. As shown in the present review, the data regarding the association of those comorbidities and IL‐17 or IL‐23 polymorphisms are conflicting. While some studies showed the link between the presence of IL‐23 rs2066808 polymorphism [47, 53, 54], others deny the association [48]. On the other hand, the IL‐23R gene polymorphism rs2201841 has been studied in various autoimmune conditions, revealing mixed associations with disease susceptibility [87, 88, 89]. Conversely, another study found that rs2201841 was linked to higher inflammatory markers in RA patients [90]. Regarding the association with psoriasis, the rs2201841 polymorphism studies do not show a significant association with disease presence [51]. The IL‐17RA gene polymorphism rs4819554 has been implicated in the susceptibility and clinical manifestations of psoriasis [60]. Research indicates that this SNP is associated with increased psoriasis risk, particularly in specific populations such as Egyptian patients [60]. The study conducted by Sabry et al. [60] in 2020 found a strong association with psoriasis susceptibility. On the other hand, Villalpando‐Vargas et al. [62] contradict the aforementioned findings through the results of their meta‐analysis that showed a near‐significant reduction in psoriasis risk associated with the AG genotypes in comparison to the GG genotype (OR = 0.604, p = 0.050).
IL‐17RA is crucial for the signaling of IL‐17 cytokines, which are central to the inflammatory processes in psoriasis [91]. These receptors’ polymorphisms may alter their function, impacting disease severity and treatment response [91]. No significant differences in genotype frequencies of IL17A rs2275913 were found between psoriatic patients and controls, suggesting it may not be a direct risk factor for psoriasis [66]. However, the AA genotype was associated with shorter disease duration and better treatment responses, indicating a potential role in disease management rather than susceptibility [91].
Polymorphisms of the genes encoding the interleukins implicated in the psoriasis pathophysiology may represent the missing key to the puzzle in the case of non‐responders. For instance, there is an association between the interleukin 17 family polymorphisms, the severity of the psoriasis and response to both classical and biological treatment [92]. In addition, these polymorphisms are also implicated in the risk of developing other conditions such as autoimmune diseases, including inflammatory bowel disease [93], multiple sclerosis [94], rheumatoid arthritis [95], and hematologic neoplasias [96] or carcinomas [97].
The influence of polymorphisms on treatment response has been studied. Thus, Prieto‐Pérez, in a review published in 2013 [98], identified that the G allele of the SNP rs610604 and the T allele of the SNP rs2230926 are associated with a good response to TNFi therapy [98, 99]. A good response to etanercept, but not to infliximab and adalimumab, is associated with the polymorphisms TNF‐a –857 (rs1799724) and TNFRSF1B (rs1061622) [100]. In 2015, Prieto‐Pérez et al. [65] in a study conducted on 194 psoriasis patients and 197 controls, found an association between IL‐17F SNP rs763780 and the treatment response to ustekinumab and infliximab and a link between rs763780 and the response to adalimumab. Batalla et al. [101] conducted a study on the Caucasian population and investigated the IL‐17RA rs4819554 and rs879577 polymorphisms [101]. The IL‐17RA rs4819554 (allele A) polymorphism was associated with the anti‐TNF‐α treatment response at week 12 (p = 0.01) and 24 (p = 0.04) [101]. In a cohort study conducted on 250 Caucasian Greek patients with psoriasis, Masouri et al. [102] showed that the presence of Rs10484554 polymorphism was associated with a good response to anti‐TNF‐α agents but not to the anti‐IL12/IL23 agent, Ustekinumab. On the other hand, the rs151823 and rs26653 are linked to a good response to anti‐IL‐12/23 therapy [102]. However, the aforementioned study has several limitations, namely the small sample size and the fact that the follow‐up was performed only at 6 months, without further evaluations of the included subjects [102]. A good response to Ustekinumab treatment was also associated with the presence of HLA‐Cw6 [103, 104]. The success of phototherapy treatment could also be predicted by the presence of the IL17A and IL17F polymorphisms in psoriasis patients [63]. The patients presenting the IL17F rs2397084 (allele C) polymorphism were more resistant to phototherapy and required a higher number of treatment sessions when compared to the TT homozygotes [63]. Ozkol et al. [66] did not find any association between IL‐17A rs2275913 and IL‐17F rs763780 and psoriasis in the Turkish population. However, the rs2275913 polymorphism was linked to a shorter disease duration and the treatment response (AA genotype) [66].
Contrarily, van Vugt et al. [105] in a cohort study conducted in the European population, concluded that the genetic variations of the IL‐17A gene did not explain the differences in the treatment response to anti‐IL17 biologics.
Given that IL17A's proinflammatory role is common in the pathogenesis of psoriasis, metabolic diseases, and CVDs, anti‐IL17 therapy has a positive effect not only on the cutaneous manifestations of psoriasis, but also on the cardiovascular system and concomitant metabolic disorders [106]. The IL‐17A effect on the cardiovascular system is promoted by IL‐6 and TNF‐α [107]. Psoriasis patients present subclinical endothelial alterations detectable by flow‐mediated dilation (FMD) [108]. Compared to healthy controls, the psoriasis patients, without known CVDs, present a lower FMD [108]. Interestingly, after 52 weeks’ treatment with secukinumab 300 mg the FMD was significantly improved compared to the baseline measurements when compared to placebo, suggesting that secukinumab improves endothelial function in such patients [108]. Additionally, the biological treatments has a beneficial effect on the reduction of the coronary non‐calcified atherosclerotic plaques [109]. The most effective biological treatment for reducing the coronary atherosclerotic burden are the therapies targeting IL‐17 when compared to anti‐IL12/23 and anti‐TNF treatments [109]. Furthermore, the biological treatment was associated with a reduction in coronary inflammation [110]. Other studies also evidenced the superiority of anti‐IL17 and anti‐IL‐23 targeted therapies for skin disease and the associated cardiovascular comorbidities or metabolic syndrome [37, 106, 111].
Jain et al. [112] published a meta‐analysis regarding the presence of coronary artery microvascular dysfunction assessed by Echocardiographic Coronary Flow Reserve Parameters. In psoriasis patients, the coronary flow velocity reserve was reduced, which pointed to a coronary microvascular dysfunction [112]. Additionally, the psoriasis patients present reduced GLS and GCS, which suggests a subclinical cardiac dysfunction that may predict the future cardiovascular events [113]. Moreover, the atrial fibrillation risk seems to be higher in psoriatic patients [114]. However, there are still conflicting data regarding the link between the anti‐IL12/23 therapies and cardiovascular risk [115]. A global population‐based retrospective cohort study showed no differences regarding the all‐cause mortality or major cardiac events risk among biologic therapies, which reduce the risk of death and CVD when compared to classical anti‐psoriatic drugs and apremilast [116]. The aforementioned findings are crucial taking into consideration that psoriasis patients, including young patients with severe forms of the disease, have a higher risk of myocardial infarction [27]. Importantly, the association of severe forms of psoriasis with cardiac mortality is independent of the traditional risk factors for CVDs [117]. Psoriasis was also associated with a higher risk of stroke [117]. A case control study, conducted on the Chinese population, showed an association between the IL‐17A rs2275913 polymorphism and the risk of stroke [118]. Additionally, the IL‐17A levels were higher in patients carrying the rs2275913 GA or GG genotype [118].
As IL‐17 and IL‐23 are implicated in both psoriasis and atherosclerotic disease, more studies should focus on investigating the presence of their polymorphisms in patients with both psoriasis and coronary artery disease vs. psoriasis patients without coronary artery disease.
Some of the polymorphisms implicated in the psoriasis development are also linked to CVD. One of them is the IL‐23 rs2066808 polymorphism that is linked to premature coronary artery disease in specific populations, such as Mexicans [58]. In a Middle Eastern Chinese cohort, polymorphisms in the IL‐23 receptor were associated with increased CAD risk, suggesting a broader role of IL‐23 in cardiovascular pathology [71, 73]. Research indicates that the IL‐23/Th17 pathway plays a significant role in the pathogenesis of psoriasis, which is associated with increased cardiovascular and metabolic disease risk [57]. Elevated IL‐23 levels have been found in atherosclerotic plaques and correlate with disease severity [57]. Additionally, the rs2201841 variant has shown associations with ulcerative colitis and RA, indicating its role in inflammatory processes that may contribute to cardiometabolic diseases [119, 120]. However, the evidence supports a connection between IL‐23R variants and inflammatory diseases, the direct impact of rs2201841 on cardiometabolic outcomes remains to be fully elucidated, necessitating more targeted studies.
In the present review we found that the IL‐17 receptor A (IL‐17RA) polymorphism rs4819554 is associated with increased CVM in heart failure patients, highlighting its potential as a genetic risk factor in this population [68]. The IL17A rs2275913 polymorphism has been linked to increased susceptibility to CAD, particularly in early‐onset cases [71].
The polymorphisms analysis could be a step further for a personalized management of psoriasis patients, especially non‐responders to conventional treatment.
5. Major Open Questions
The data regarding the implication of the above mentioned polymorphisms in the development of psoriasis and cardiometabolic diseases are conflicting. One of the factors that could contribute to contradictory results is the subjects' ethnicity; most of the studies were conducted on Asian populations. It is known that the prevalence of psoriasis varies among populations, and it is highest in Caucasians (3.6%), followed by African Americans 1.9%, and Hispanics 1.6%, and 1.2% in other ethnic groups [121]. One explanation for the discrepancies regarding the psoriasis prevalence in different ethnic groups with a higher frequency in the Caucasioan population, could be explained by the genetic drift [122]. The psoriatic plaques could represent a “chemical shield” for infection control, which favors energy conservation, wound healing, low‐grade inflammation [122]. Another interesting fact is that psoriasis is more frequent in high‐income countries [123]. Additionally, the exacerbating factors differ among different ethnic groups, with stress being a more important exacerbating factor for Asians compared to Hispanics/Latinos and Caucasians [124]. On the other hand, medication was more frequently employed as an exacerbating factor for Caucasians compared to Asians [124].
Furthermore, the techniques used for the polymorphism determination vary between studies, and there is no consensus regarding the type of psoriasis that the subjects in these studies have. Moreover, there are few studies conducted in the European population.
Other important limitations are the small sample sizes of some of the included studies, the sample size differences, the methodological inconsistencies, and the lack of studies that demonstrate the causality relationship.
6. Conclusion and Perspectives
In conclusion, psoriasis and cardiometabolic diseases could have a common genetic background that explains the higher frequency of cardiovascular comorbidities in psoriasis patients. The IL17A rs2275913 polymorphism could be implicated in psoriasis, influencing both disease susceptibility and treatment response. Some of the polymorphisms implicated in the psoriasis development are also linked to CVD. Research indicates that variations in the IL‐23 gene may influence susceptibility to coronary artery disease and other cardiovascular conditions. Furthermore, a larger sample size across diverse populations would provide a more comprehensive understanding of the hypothesis in question. Ultimately, the establishment of robust evidence through these standardized studies will contribute significantly to the field and help in drawing more definitive conclusions regarding the validity of the hypothesis. To validate the hypothesis presented, it is essential to conduct more standardized case‐control studies. These studies should be designed with rigorous methodologies and uniform protocols to ensure consistency and reliability in the results. Understanding the genetic basis of psoriasis and its cardiovascular comorbidities could enhance treatment strategies, particularly in selecting biologic therapies that target IL‐17 and IL‐23 pathways.
Author Contributions
Andrada‐Luciana Lazar: conceptualization, investigation, writing – original draft, methodology, writing – review and editing, formal analysis. Cleopatra Romana Vulturar: supervision, data curation, visualization, validation, writing – review and editing. Adela‐Viviana Sitar‐Tăut: writing – review and editing, formal analysis, supervision, data curation. Ramona Suharoschi: writing – original draft, formal analysis, supervision. Olga Hilda Orășan: conceptualization, methodology, formal analysis. Adina Ancuța Chiș: conceptualization, study design, revising article – intellectual content. Adriana Fodor: validation, formal analysis, writing – review and editing. Brîndușa Tiperciuc: writing – review and editing, formal analysis, visualization. Cezar Login: visualization, writing – review & editing, formal analysis. Angela Cozma: supervision, formal analysis, visualization, writing – review and editing.
Conflicts of Interest
The authors declare no conflicts of interest.
Transparency Statement
The lead author Cleopatra Romana Vulturar affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Acknowledgments
Open access publishing facilitated by Anelis Plus (the official name of “Asociatia Universitatilor, a Institutelor de Cercetare – Dezvoltare si a Bibliotecilor Centrale Universitare din Romania”), as part of the Wiley ‐ Anelis Plus agreement.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Damiani G., Bragazzi N. L., Karimkhani Aksut C., et al., “The Global, Regional, and National Burden of Psoriasis: Results and Insights From the Global Burden of Disease 2019 Study,” Frontiers in Medicine 8 (2021): 743180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boehncke W. H., “Systemic Inflammation and Cardiovascular Comorbidity in Psoriasis Patients: Causes and Consequences,” Frontiers in Immunology 9 (2018): 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.P. A. Nair and T. Badri, “Psoriasis.” Apr 3, 2023. StatPearls [Internet] (Treasure Island (FL): StatPearls Publishing, 2023 Jan).
- 4. Sarac G., Koca T. T., and Baglan T., “A Brief Summary of Clinical Types of Psoriasis,” Northern Clinics of Istanbul 3, no. 1 (2016): 79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dhabale A. and Nagpure S., “Types of Psoriasis and Their Effects on the Immune System,” Cureus 14, no. 9 (2022): e29536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luba K. M. and Stulberg D. L., “Chronic Plaque Psoriasis,” American Family Physician 73, no. 4 (2006): 636–644. [PubMed] [Google Scholar]
- 7. Salgado‐Boquete L., Carrascosa J. M., Llamas‐Velasco M., Ruiz‐Villaverde R., de la Cueva P., and Belinchón I., “A New Classification of the Severity of Psoriasis: What's Moderate Psoriasis?,” Life 11, no. 7 (2021): 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rich P. and Scher R. K., “Nail Psoriasis Severity Index: A Useful Tool for Evaluation of Nail Psoriasis,” Journal of the American Academy of Dermatology 49, no. 2 (2003): 206–212. [DOI] [PubMed] [Google Scholar]
- 9. Wozel G., Klein E., Mrowietz U., Reich K., Sebastian M., and Streit V., “Scalp Psoriasis,” JDDG: Journal der Deutschen Dermatologischen Gesellschaft 9, no. 1 (2011): 70–74. [DOI] [PubMed] [Google Scholar]
- 10. Gottlieb A. B., Kirby B., Ryan C., et al., “The Development of a Patient‐Reported Outcome Measure for Assessment of Genital Psoriasis Symptoms: The Genital Psoriasis Symptoms Scale (GPSS),” Dermatology and Therapy 8, no. 1 (2018): 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liluashvili S. and Kituashvili T., “Dermatology Life Quality Index and Disease Coping Strategies in Psoriasis Patients,” Advances in Dermatology and Allergology 36, no. 4 (2019): 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mihu C., Neag M. A., Bocşan I. C., et al., “Novel Concepts in Psoriasis: Histopathology and Markers Related to Modern Treatment Approaches,” Romanian Journal of Morphology and Embryology 62, no. 4 (2022): 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King D. T., “Munro Microabscess: Let's Spell It Right,” Archives of Dermatology 115, no. 7 (1979): 816–817.378139 [Google Scholar]
- 14. Xu X., Zhang Y., Pan Z., et al., “Genome‐Wide DNA Methylation of Munro's Microabscess Reveals the Epigenetic Regulation in the Pathogenesis of Psoriasis,” Frontiers in Immunology 13 (2022): 1057839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rendon A. and Schäkel K., “Psoriasis Pathogenesis and Treatment,” International Journal of Molecular Sciences 20, no. 6 (2019): 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu P., Wang M., Gao H., et al., “The Role of Helper T Cells in Psoriasis,” Frontiers in Immunology 12 (2021): 788940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ten Bergen L. L., Petrovic A., Aarebrot A. K., and Appel S., “Current Knowledge on Autoantigens and Autoantibodies in Psoriasis,” Scandinavian Journal of Immunology 92, no. 4 (2020): e12945. [DOI] [PubMed] [Google Scholar]
- 18. Hu P., Wang M., Gao H., et al., “The Role of Helper T Cells in Psoriasis,” Frontiers in Immunology 12 (2021): 788940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schön M. P., “Adaptive and Innate Immunity in Psoriasis and Other Inflammatory Disorders,” Frontiers in Immunology 10 (2019): 1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ten Bergen L. L., Petrovic A., Krogh Aarebrot A., and Appel S., “The TNF/IL‐23/IL‐17 Axis‐Head‐to‐Head Trials Comparing Different Biologics in Psoriasis Treatment,” Scandinavian Journal of Immunology 92, no. 4 (2020): e12946. [DOI] [PubMed] [Google Scholar]
- 21. Fletcher J. M., Moran B., Petrasca A., and Smith C. M., “IL‐17 in Inflammatory Skin Diseases Psoriasis and Hidradenitis Suppurativa,” Clinical and Experimental Immunology 201, no. 2 (2020): 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu T., Li S., Ying S., et al., “The IL‐23/IL‐17 Pathway in Inflammatory Skin Diseases: From Bench to Bedside,” Frontiers in Immunology 11 (2020): 594735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lupardus P. J. and Garcia K. C., “The Structure of Interleukin‐23 Reveals the Molecular Basis of p40 Subunit Sharing With Interleukin‐12,” Journal of Molecular Biology 382, no. 4 (2008): 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iwakura Y., “The IL‐23/IL‐17 Axis in Inflammation,” Journal of Clinical Investigation 116, no. 5 (2006): 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schinocca C., Rizzo C., Fasano S., et al., “Role of the IL‐23/IL‐17 Pathway in Rheumatic Diseases: An Overview,” Frontiers in Immunology 12 (2021): 637829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Armstrong E. J., Harskamp C. T., and Armstrong A. W., “Psoriasis and Major Adverse Cardiovascular Events: A Systematic Review and Meta‐Analysis of Observational Studies,” Journal of the American Heart Association 2, no. 2 (2013): e000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelfand J. M., Neimann A. L., Shin D. B., Wang X., Margolis D. J., and Troxel A. B., “Risk of Myocardial Infarction in Patients With Psoriasis,” Journal of the American Medical Association 296, no. 14 (2006): 1735–1741. [DOI] [PubMed] [Google Scholar]
- 28. Ludwig R. J., Herzog C., Rostock A., et al., “Psoriasis: A Possible Risk Factor for Development of Coronary Artery Calcification,” British Journal of Dermatology 156 (2007): 271–276. [CrossRef] [PubMed]. [DOI] [PubMed] [Google Scholar]
- 29. Armstrong E. J., Harskamp C. T., and Armstrong A. W., “Psoriasis and Major Adverse Cardiovascular Events: A Systematic Review and Meta‐Analysis of Observational Studies,” Journal of the American Heart Association 2, no. 2 (2013): e000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L., Cui S., Liu M., Huo X., Zhang G., and Wang N., “Psoriasis Increased the Risk of Adverse Cardiovascular Outcomes: A New Systematic Review and Meta‐Analysis of Cohort Study,” Frontiers in Cardiovascular Medicine 9 (2022): 829709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan B. X., Chen X. Y., Ye L. R., Chen J. Q., Zheng M., and Man X. Y., “Cutaneous and Systemic Psoriasis: Classifications and Classification for the Distinction,” Frontiers in Medicine 8 (2021): 649408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elmets C. A., Leonardi C. L., Davis D. M. R., et al., “Joint AAD‐NPF Guidelines of Care for the Management and Treatment of Psoriasis With Awareness and Attention to Comorbidities,” Journal of the American Academy of Dermatology 80, no. 4 (2019): 1073–1113. [DOI] [PubMed] [Google Scholar]
- 33. Masson W., Lobo M., and Molinero G., “Psoriasis and Cardiovascular Risk: A Comprehensive Review,” Advances in Therapy 37, no. 5 (2020): 2017–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sajja A. P., Joshi A. A., Teague H. L., Dey A. K., and Mehta N. N., “Potential Immunological Links Between Psoriasis and Cardiovascular Disease,” Frontiers in Immunology 9 (2018): 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garshick M. S., Barrett T. J., Wechter T., et al., “Inflammasome Signaling and Impaired Vascular Health in Psoriasis,” Arteriosclerosis, Thrombosis, and Vascular Biology 39 (2019): 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Machoń N. J., Zdanowska N., Klimek‐Trojan P., and Owczarczyk‐Saczonek A., “Vascular Cell Adhesion Molecule 1 and E‐Selectin as Potential Cardiovascular Risk Biomarkers in Psoriasis,” International Journal of Molecular Sciences 26, no. 2 (2025. Jan 18): 792, 10.3390/ijms26020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garshick M. S., Ward N. L., Krueger J. G., and Berger J. S., “Cardiovascular Risk in Patients With Psoriasis,” Journal of the American College of Cardiology 77, no. 13 (2021): 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C., Chen H., Liu Y., et al., “Immunity: Psoriasis Comorbid With Atherosclerosis,” Frontiers in Immunology 13 (2022): 1070750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Allam G., Abdel‐Moneim A., and Gaber A. M., “The Pleiotropic Role of Interleukin‐17 in Atherosclerosis,” Biomedicine & Pharmacotherapy 106 (2018): 1412–1418, 10.1016/j.biopha.2018.07.110. [DOI] [PubMed] [Google Scholar]
- 40. Piaserico S., Orlando G., and Messina F., “Psoriasis and Cardiometabolic Diseases: Shared Genetic and Molecular Pathways,” International Journal of Molecular Sciences 23, no. 16 (2022): 9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takeshita J., Grewal S., Langan S. M., et al., “Psoriasis and Comorbid Diseases,” Journal of the American Academy of Dermatology 76, no. 3 (2017): 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng H., Li Y., Zuo X. B., et al., “Identification of a Missense Variant in LNPEP That Confers Psoriasis Risk,” Journal of Investigative Dermatology 134, no. 2 (2014): 359–365. [DOI] [PubMed] [Google Scholar]
- 43. Gupta Y., Möller S., Zillikens D., Boehncke W. H., Ibrahim S. M., and Ludwig R. J., “Genetic Control of Psoriasis Is Relatively Distinct From That of Metabolic Syndrome and Coronary Artery Disease,” Experimental Dermatology 22, no. 8 (2013): 552–553. [DOI] [PubMed] [Google Scholar]
- 44. Indhumathi S., Rajappa M., Chandrashekar L., Ananthanarayanan P. H., Thappa D. M., and Negi V. S., “Investigation of Association of the IL‐12B and IL‐23R Genetic Variations With Psoriatic Risk in a South Indian Tamil Cohort,” Human Immunology 77, no. 1 (2016): 54–62. [DOI] [PubMed] [Google Scholar]
- 45. Safrany E., Szell M., Csongei V., et al., “Polymorphisms of the IL23R Gene Are Associated With Psoriasis but Not With Immunoglobulin A Nephropathy in a Hungarian Population,” Inflammation 34, no. 6 (2011): 603–608. [DOI] [PubMed] [Google Scholar]
- 46. Safrany E., Szabo M., Szell M., et al., “Difference of Interleukin‐23 Receptor Gene Haplotype Variants in Ulcerative Colitis Compared to Crohn's Disease and Psoriasis,” Inflammation Research 62, no. 2 (2013): 195–200. [DOI] [PubMed] [Google Scholar]
- 47. Nair R. P., Duffin K. C., Helms C., et al., “Genome‐Wide Scan Reveals Association of Psoriasis With IL‐23 and NF‐κB Pathways,” Nature Genetics 41, no. 2 (2009): 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eirís N., González‐Lara L., Santos‐Juanes J., Queiro R., Coto E., and Coto‐Segura P., “Genetic Variation at IL12B, IL23R and IL23A Is Associated With Psoriasis Severity, Psoriatic Arthritis and Type 2 Diabetes Mellitus,” Journal of Dermatological Science 75, no. 3 (2014): 167–172. [DOI] [PubMed] [Google Scholar]
- 49. Nikamo P., Lysell J., and Ståhle M., “Association With Genetic Variants in the IL‐23 and NF‐κB Pathways Discriminates Between Mild and Severe Psoriasis Skin Disease,” Journal of Investigative Dermatology 135, no. 8 (2015): 1969–1976. [DOI] [PubMed] [Google Scholar]
- 50. Popadic S., Ramic Z., Medenica L., Pravica V., and Popadic D., “IL‐23R Gene Polymorphism rs2201841 Is Associated With Psoriatic Arthritis,” International Journal of Immunogenetics 41, no. 4 (2014): 335–337. [DOI] [PubMed] [Google Scholar]
- 51. Filiz B., Yildirim M., Hekimler Öztürk K., et al., “Evaluation of Interleukin‐23 Receptor (IL‐23R) Gene Polymorphisms and Serum IL‐23 Levels in Patients With Psoriasis,” Turkish Journal of Medical Sciences 49, no. 5 (2019): 1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Capon F., Di Meglio P., Szaub J., et al., “Sequence Variants in the Genes for the Interleukin‐23 Receptor (IL23R) and Its Ligand (IL12B) Confer Protection Against Psoriasis,” Human Genetics 122, no. 2 (2007): 201–206. [DOI] [PubMed] [Google Scholar]
- 53. Bojko A., Ostasz R., Białecka M., et al., “IL12B, IL23A, IL23R and HLA‐C*06 Genetic Variants in Psoriasis Susceptibility and Response to Treatment,” Human Immunology 79, no. 4 (2018): 213–217. [DOI] [PubMed] [Google Scholar]
- 54. Lin L., Wang Y., Lu X., et al., “The Inflammatory Factor SNP May Serve as a Promising Biomarker for Acitretin to Alleviate Secondary Failure of Response to TNF‐a Monoclonal Antibodies in Psoriasis,” Frontiers in Pharmacology 13 (2022): 937490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen H., Poon A., Yeung C., et al., “A Genetic Risk Score Combining Ten Psoriasis Risk Loci Improves Disease Prediction,” PLoS One 6, no. 4 (2011): e19454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hu S. and Lan C. C. E., “Psoriasis and Cardiovascular Comorbidities: Focusing on Severe Vascular Events, Cardiovascular Risk Factors and Implications for Treatment,” International Journal of Molecular Sciences 18, no. 10 (2017): 2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Egeberg A., Gisondi P., Carrascosa J. M., Warren R. B., and Mrowietz U., “The Role of the Interleukin‐23/Th17 Pathway in Cardiometabolic Comorbidity Associated With Psoriasis,” Journal of the European Academy of Dermatology and Venereology 34, no. 8 (2020): 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vázquez‐Vázquez C., Posadas‐Sánchez R., Pérez‐Hernández N., et al., “The rs2066808 Polymorphism Located Near the IL‐23A Gene Is Associated With Premature Coronary Artery Disease in Mexican Population (GEA Study),” DNA and Cell Biology 38, no. 8 (2019): 880–886. [DOI] [PubMed] [Google Scholar]
- 59. Batalla A., Coto E., González‐Lara L., et al., “Association Between Single Nucleotide Polymorphisms IL17RA rs4819554 and IL17E rs79877597 and Psoriasis in a Spanish Cohort,” Journal of Dermatological Science 80, no. 2 (2015): 111–115. [DOI] [PubMed] [Google Scholar]
- 60. Sabry D., Aboraia N., and Samir M., “A Potential Association Between Psoriasin to rs4819554 of IL‐17RA Gene Polymorphism in Psoriasis Egyptian Patients,” Archives of Dermatological Research 312, no. 4 (2020): 273–281. [DOI] [PubMed] [Google Scholar]
- 61. Pușcaș A. D., Morar I. I., Vesa C., et al., “Association Between IL‐17F, IL‐17RA Gene Polymorphisms and Response to Biological Drugs in Psoriasis and Beyond,” Genes 14, no. 5 (2023): 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Villalpando‐Vargas F. V., Rivera‐Valdés J. J., Alvarado‐Navarro A., et al., “Association Between IL‐17A, IL‐17F and IL‐17RA Gene Polymorphisms and Susceptibility to Psoriasis and Psoriatic Arthritis: A Meta‐Analysis,” Inflammation Research 70, no. 10–12 (2021): 1201–1210. [DOI] [PubMed] [Google Scholar]
- 63. Białecka M., Ostasz R., Kurzawski M., et al., “IL17A and IL17F Gene Polymorphism Association With Psoriasis Risk and Response to Treatment in a Polish Population,” Dermatology 232, no. 5 (2016): 592–596. [DOI] [PubMed] [Google Scholar]
- 64. Kim S. Y., Hur M. S., Choi B. G., et al., “A Preliminary Study of New Single Polymorphisms in the T Helper Type 17 Pathway for Psoriasis in the Korean Population,” Clinical and Experimental Immunology 187, no. 2 (2017): 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prieto‐Pérez R., Solano‐López G., Cabaleiro T., et al., “The Polymorphism rs763780 in the IL‐17F Gene Is Associated With Response to Biological Drugs in Patients With Psoriasis,” Pharmacogenomics 16, no. 15 (2015): 1723–1731. [DOI] [PubMed] [Google Scholar]
- 66. Ozkol H., Gorgisen G., Ates C., et al., “Evaluation of the Relationship of IL17A and IL17F Gene Polymorphisms With the Response to Treatment in Psoriatic Patients Using Biological Drugs: Case‐Control Study in Patients in Eastern Turkey,” Advances in Dermatology and Allergology 38, no. 5 (2021): 780–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Furue M., Tsuji G., Chiba T., and Kadono T., “Cardiovascular and Metabolic Diseases Comorbid With Psoriasis: Beyond the Skin,” Internal Medicine 56, no. 13 (2017): 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sandip C., Tan L., Huang J., et al., “Common Variants in IL‐17A/IL‐17RA Axis Contribute to Predisposition to and Progression of Congestive Heart Failure,” Medicine 95, no. 27 (2016): e4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Coto E., Pascual I., Avanzas P., et al., “IL17RA in Early‐Onset Coronary Artery Disease: Total Leukocyte Transcript Analysis and Promoter Polymorphism (rs4819554) Association,” Cytokine 136 (2020): 155285. [DOI] [PubMed] [Google Scholar]
- 70. Geng G. Y., Liu H. L., Zhao Y. J., Wu L., Mao L., and Ba N., “Correlation Between Polymorphisms in the IL‐17A and IL‐17F Genes and Development of Coronary Artery Disease,” Genetics and Molecular Research 14, no. 3 (2015): 11488–11494. [DOI] [PubMed] [Google Scholar]
- 71. Zhang H., Nie S., Chen Q., et al., “Gene Polymorphism in IL17A and Gene‐Gene Interaction in the IL23R/IL17A Axis Are Associated With Susceptibility to Coronary Artery Disease,” Cytokine 164 (2023): 156142. [DOI] [PubMed] [Google Scholar]
- 72. Zhang X., Pei F., Zhang M., et al., “Interleukin‐17A Gene Variants and Risk of Coronary Artery Disease: A Large Angiography‐Based Study,” Clinica Chimica Acta 412, no. 3–4 (2011): 327–331. [DOI] [PubMed] [Google Scholar]
- 73. Zhao Q., Jiang H., Ma T., Qiu H., Guo M., and Zhang X., “The Association Between IL‐17A and IL‐23R Polymorphisms and Coronary Artery Disease Risk in a Middle Eastern Chinese Population,” Journal of Clinical Laboratory Analysis 33, no. 6 (2019): e22893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Su G. B., Guo X. L., Liu X. C., Cui Q. T., and Zhou C. Y., “Association Between Interleukin‐17A Polymorphism and Coronary Artery Disease Susceptibility in the Chinese Han Population,” Genetics and Molecular Research 15, no. 3 (2016), 10.4238/gmr.15038235. [DOI] [PubMed] [Google Scholar]
- 75. Peng Y., Zhou B., Wang Y. Y., et al., “Analysis of IL‐17 Gene Polymorphisms in Chinese Patients With Dilated Cardiomyopathy,” Human Immunology 74, no. 5 (2013): 635–639. [DOI] [PubMed] [Google Scholar]
- 76. Vargas‐Alarcón G., Angeles‐Martínez J., Villarreal‐Molina T., et al., “Interleukin‐17A Gene Haplotypes Are Associated With Risk of Premature Coronary Artery Disease in Mexican Patients From the Genetics of Atherosclerotic Disease (GEA) Study,” PLoS One 10, no. 1 (2015): e0114943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shuang L., Li Z., Chen F., et al., “Association Between Interleukin‐17 Gene Polymorphisms and Risk of Coronary Artery Disease,” International Journal of Clinical and Experimental Pathology 8, no. 9 (2015): 11653–11658. [PMC free article] [PubMed] [Google Scholar]
- 78. Zheng X. S., Wang S., and Ni M., “Association Between Interleukin 17A Gene Polymorphisms and Risk of Coronary Artery Disease,” Genetics and Molecular Research 15, no. 1 (2016). [DOI] [PubMed] [Google Scholar]
- 79. Ghaznavi H. and Soltanpour M. S., “Association Study Between rs2275913 Genetic Polymorphism and Serum Levels of IL‐17A With Risk of Coronary Artery Disease,” Molecular Biology Research Communications 9, no. 1 (2020): 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brembilla N. C. and Boehncke W. H., “Revisiting the Interleukin 17 Family of Cytokines in Psoriasis: Pathogenesis and Potential Targets for Innovative Therapies,” Frontiers in Immunology 14 (2023): 1186455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dascălu R. C., Bărbulescu A. L., Stoica L. E., et al., “Review: A Contemporary, Multifaced Insight Into Psoriasis Pathogenesis,” Journal of Personalized Medicine 14, no. 5 (2024): 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dhamija B., Marathe S., Sawant V., et al., “IL‐17A Orchestrates Reactive Oxygen Species/HIF1α‐Mediated Metabolic Reprogramming in Psoriasis,” The Journal of Immunology 212, no. 2 (2024): 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fragoulis G. E. and Siebert S., “The Role of IL‐23 and the Use of IL‐23 Inhibitors in Psoriatic Arthritis,” Musculoskeletal care 20, no. Suppl 1 (2022): S12–S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Park Y. J., Kim Y. H., Lee E. S., and Kim Y. C., “Comparative Analysis of Single‐Cell Transcriptome Data Reveals a Novel Role of Keratinocyte‐Derived IL‐23 in Psoriasis,” Frontiers in Immunology 13 (2022): 905239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Daniele S. G., Eldirany S. A., Damiani G., Ho M., and Bunick C. G., “Structural Basis for p19 Targeting by Anti‐IL‐23 Biologics: Correlations With Short‐ and Long‐Term Efficacy in Psoriasis. Biorxiv [Preprint] 2023;531913,” JID Innovations: Skin Science From Molecules to Population Health 4, no. 2 (2024): 100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Riaz S., Emam S., Wang T., and Gniadecki R., “Negative Impact of Comorbidities on All‐Cause Mortality of Patients With Psoriasis Is Partially Alleviated by Biologic Treatment: A Real‐World Case‐Control Study,” Journal of the American Academy of Dermatology 91, no. 1 (2024): 43–50. [DOI] [PubMed] [Google Scholar]
- 87. Zou Q., Zhao Y., Wang Y., Fang Y., and Liu Y., “Associations Between IL‐23R Gene Polymorphisms and the Susceptibility of Rheumatoid Arthritis: A Meta‐Analysis,” Artificial Cells, Nanomedicine, and Biotechnology 47, no. 1 (2019): 951–956. [DOI] [PubMed] [Google Scholar]
- 88. Duerr R. H., Taylor K. D., Brant S. R., et al., “A Genome‐Wide Association Study Identifies IL23R as An Inflammatory Bowel Disease Gene,” Science 314, no. 5804 (2006): 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Huber A. K., Jacobson E. M., Jazdzewski K., Concepcion E. S., and Tomer Y., “Interleukin (IL)‐23 Receptor Is a Major Susceptibility Gene for Graves' Ophthalmopathy: The IL‐23/T‐Helper 17 Axis Extends to Thyroid Autoimmunity,” The Journal of Clinical Endocrinology & Metabolism 93, no. 3 (2008): 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Soysal E., Ulutaş F., Tepeli E., Kaymaz S., and Çobankara V., “IL‐23R Gene Polymorphisms in Rheumatoid Arthritis,” Rheumatology International 42, no. 3 (2022): 555–562. [DOI] [PubMed] [Google Scholar]
- 91. Vidal S., Puig L., Carrascosa‐Carrillo J. M., González‐Cantero Á., Ruiz‐Carrascosa J. C., and Velasco‐Pastor A. M., “From Messengers to Receptors in Psoriasis: The Role of IL‐17RA in Disease and Treatment,” International Journal of Molecular Sciences 22, no. 13 (2021): 6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pușcaș A. D., Cătană A., Pușcaș C., et al., “Psoriasis: Association of Interleukin‐17 Gene Polymorphisms With Severity and Response to Treatment,” Experimental and Therapeutic Medicine 18, no. 2 (2019): 875–880, 10.3892/etm.2019.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Arisawa T., Tahara T., Shibata T., et al., “The Influence of Polymorphisms of Interleukin‐17A and interleukin‐17F Genes on the Susceptibility to Ulcerative Colitis,” Journal of Clinical Immunology 28, no. 1 (2008): 44–49, 10.1007/s10875-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 94. Salah S., Sadeq Y. I., Mosaad Y. M., Elmenshawi I. E. H., and Tawhid Z. M. E., “Association of Interleukin‐17F (rs763780) Single Nucleotide Polymorphism With Multiple Sclerosis and Optic Neuritis,” Scientific Reports 14, no. 1 (2024): 13643, 10.1038/s41598-024-62736-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen P., Li Y., Li L., et al., “Association Between the Interleukin (IL)‐17A rs2275913 Polymorphism and Rheumatoid Arthritis Susceptibility: A Meta‐Analysis and Trial Sequential Analysis,” Journal of International Medical Research 49, no. 10 (2021): 3000605211053233, 10.1177/03000605211053233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wróbel T., Gębura K., Wysoczańska B., et al., “IL‐17F Gene Polymorphism Is Associated With Susceptibility to Acute Myeloid Leukemia,” Journal of Cancer Research and Clinical Oncology 140, no. 9 (2014): 1551–1555, 10.1007/s00432-014-1674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lu Y., Gu J., Lu H., et al., “Association Between IL‐17A +197 G/A Polymorphism and Cancer Risk: A Meta‐Analysis,” Genetic Testing and Molecular Biomarkers 20, no. 1 (2016): 24–30, 10.1089/gtmb.2015.0143. [DOI] [PubMed] [Google Scholar]
- 98. Prieto‐Pérez R., Cabaleiro T., Daudén E., and Abad‐Santos F., “Gene Polymorphisms That Can Predict Response to Anti‐TNF Therapy in Patients With Psoriasis and Related Autoimmune Diseases,” The Pharmacogenomics Journal 13, no. 4 (2013): 297–305, 10.1038/tpj.2012.53. [DOI] [PubMed] [Google Scholar]
- 99. Tejasvi T., Stuart P. E., Chandran V., et al., “TNFAIP3 Gene Polymorphisms Are Associated With Response to TNF Blockade in Psoriasis,” Journal of Investigative Dermatology 132, no. 3 Pt 1 (2012): 593–600, 10.1038/jid.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vasilopoulos Y., Manolika M., Zafiriou E., et al., “Pharmacogenetic Analysis of TNF, TNFRSF1A, and TNFRSF1B Gene Polymorphisms and Prediction of Response to Anti‐TNF Therapy in Psoriasis Patients in the Greek Population,” Molecular Diagnosis & Therapy 16, no. 1 (2012): 29–34, 10.1007/BF03256427. [DOI] [PubMed] [Google Scholar]
- 101. Batalla A., Coto E., Gómez J., et al., “IL17RA Gene Variants and Anti‐TNF Response Among Psoriasis Patients,” The Pharmacogenomics Journal 18 (2018): 76–80, 10.1038/tpj.2016.70. [DOI] [PubMed] [Google Scholar]
- 102. Masouri S., Stefanaki I., Ntritsos G., et al., “A Pharmacogenetic Study of Psoriasis Risk Variants in a Greek Population and Prediction of Responses to Anti‐TNF‐α and Anti‐IL‐12/23 Agents,” Molecular Diagnosis & Therapy 20, no. 3 (2016): 221–225, 10.1007/s40291-016-0198-z. [DOI] [PubMed] [Google Scholar]
- 103. Talamonti M., Botti E., Galluzzo M., et al., “Pharmacogenetics of Psoriasis: HLA‐Cw6 But not LCE3B/3C Deletion nor TNFAIP3 Polymorphism Predisposes to Clinical Response to Interleukin 12/23 Blocker Ustekinumab.” Br J Dermatol (2013). 169, 458–463. 2, 10.1111/bjd.12331. [DOI] [PubMed] [Google Scholar]
- 104.Erratum in: Br J Dermatol. 2016. Jul;175(1):228‐9, 10.1111/bjd.14747. [DOI] [Google Scholar]
- 105. van Vugt L. J., van den Reek J. M. P. A., Meulewaeter E., et al., “Response to IL‐17A Inhibitors Secukinumab and Ixekizumab Cannot Be Explained by Genetic Variation in the Protein‐Coding and Untranslated Regions of the IL‐17A Gene: Results From a Multicentre Study of Four European Psoriasis Cohorts.” J Eur Acad Dermatol Venereol (2020). 34, 112–118. 1, 10.1111/jdv.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. von Stebut E., Boehncke W. H., Ghoreschi K., et al., “IL‐17A in Psoriasis and Beyond: Cardiovascular and Metabolic Implications,” Frontiers in Immunology 10 (2020): 3096, 10.3389/fimmu.2019.03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Karbach S., Croxford A. L., Oelze M., et al., “Interleukin 17 Drives Vascular Inflammation, Endothelial Dysfunction, and Arterial Hypertension in Psoriasis‐Like Skin Disease,” Arteriosclerosis, Thrombosis, and Vascular Biology 34 (2014): 2658–2668, 10.1161/ATVBAHA.114.304108. [DOI] [PubMed] [Google Scholar]
- 108. von Stebut E., Reich K., Thaçi D., et al., “Impact of Secukinumab on Endothelial Dysfunction and Other Cardiovascular Disease Parameters in Psoriasis Patients Over 52 Weeks,” Journal of Investigative Dermatology 139, no. 5 (2019): 1054–1062, 10.1016/j.jid.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 109. Elnabawi Y. A., Dey A. K., Goyal A., et al., “Coronary Artery Plaque Characteristics and Treatment With Biologic Therapy in Severe Psoriasis: Results From a Prospective Observational Study,” Cardiovascular Research 115, no. 4 (2019): 721–728, 10.1093/cvr/cvz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Elnabawi Y. A., Oikonomou E. K., Dey A. K., et al., “Association of Biologic Therapy With Coronary Inflammation in Patients With Psoriasis as Assessed by Perivascular Fat Attenuation Index,” JAMA Cardiology 4, no. 9 (2019): 885–891, 10.1001/jamacardio.2019.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Trovato E., Rubegni P., and Prignano F., “Place in Therapy of Anti‐IL‐17 and 23 in Psoriasis According to the Severity of Comorbidities: A Focus on Cardiovascular Disease and Metabolic Syndrome,” Expert Opinion on Biological Therapy 22, no. 12 (2022): 1443–1448, 10.1080/14712598.2022.2093106. [DOI] [PubMed] [Google Scholar]
- 112. Jain H., Tariq M. D., Khan A. M., et al., “Assessment of Subclinical Atherosclerosis in Patients With Psoriasis Using Echocardiographic Coronary Flow Reserve Parameters: A Systematic Review and Meta‐Analysis,” British Journal of Hospital Medicine 86, no. 2 (2025): 1–16, 10.12968/hmed.2024.0618. [DOI] [PubMed] [Google Scholar]
- 113. Jain H., Jain J., Dey D., et al., “Subclinical Myocardial Dysfunction Assessment Using Speckle Tracking Echocardiography in Patients With Psoriasis: A Pilot Meta‐Analysis,” Clinical Cardiology 47, no. 12 (2024): e70047, 10.1002/clc.70047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jain H., Odat R. M., Goyal A., et al., “Association Between Psoriasis and Atrial Fibrillation: A Systematic Review and Meta‐Analysis,” Current Problems in Cardiology 49, no. 6 (2024): 102538, 10.1016/j.cpcardiol.2024.102538. [DOI] [PubMed] [Google Scholar]
- 115. Mehta H., Narang T., Dogra S., Handa S., Hatwal J., and Batta A., “Cardiovascular Considerations and Implications for Treatment in Psoriasis: An Updated Review,” Vascular health and Risk Management 20 (2024): 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kridin K., Bieber K., Vorobyev A., et al., “Biological, as Opposed to Classic Antipsoriatic Drug or Apremilast, Treatment Mitigates the Risk of Death and Cardiovascular Disease in Psoriasis,” EBioMedicine 111 (2025): 105485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mehta N. N., Azfar R. S., Shin D. B., Neimann A. L., Troxel A. B., and Gelfand J. M., “Patients With Severe Psoriasis Are at Increased Risk of Cardiovascular Mortality: Cohort Study Using the General Practice Research Database,” European Heart Journal 31, no. 8 (2010): 1000–1006, 10.1093/eurheartj/ehp567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Huang H. T., Lu Y. L., Wang R., et al., “The Association of IL‐17A Polymorphisms With IL‐17A Serum Levels and Risk of Ischemic Stroke,” Oncotarget 8, no. 61 (2017): 103499–103508, 10.18632/oncotarget.21498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Peng L. L., Wang Y., Zhu F. L., Xu W. D., Ji X. L., and Ni J., “IL‐23R Mutation Is Associated With Ulcerative Colitis: A Systemic Review and Meta‐Analysis,” Oncotarget 8, no. 3 (2017): 4849–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Faragó B., Magyari L., Sáfrány E., et al., “Functional Variants of Interleukin‐23 Receptor Gene Confer Risk for Rheumatoid Arthritis but Not for Systemic Sclerosis,” Annals of the Rheumatic Diseases 67, no. 2 (2008): 248–250. [DOI] [PubMed] [Google Scholar]
- 121. Abrouk M., Lee K., Brodsky M., et al., “Ethnicity Affects the Presenting Severity of Psoriasis,” Journal of the American Academy of Dermatology 77, no. 1 (2017): 180–182, 10.1016/j.jaad.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 122. Romaní de Gabriel J., “Medicina Darwiniana Y Psoriasis,” Actas Dermo‐Sifiliográficas 106, no. 3 (2015): 189–194, 10.1016/j.ad.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 123. Parisi R., Iskandar I. Y. K., Kontopantelis E., Augustin M., Griffiths C. E. M., and Ashcroft D. M., “National, Regional, and Worldwide Epidemiology of Psoriasis: Systematic Analysis and Modelling Study,” BMJ 369 (2020): m1590, 10.1136/bmj.m1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yan D., Afifi L., Jeon C., Cordoro K. M., and Liao W., “A Cross‐Sectional Study of Psoriasis Triggers Among Different Ethno‐Racial Groups,” Journal of the American Academy of Dermatology 77, no. 4 (2017): 756–758.e1, 10.1016/j.jaad.2017.04.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


