Abstract
DNA guanine (G)-quadruplexes (G4s) are unique secondary structures formed by two or more stacked G-tetrads in G-rich DNA sequences. These structures have been found to play a crucial role in highly transcribed genes, especially in cancer-related oncogenes, making them attractive targets for cancer therapeutics. Significantly, targeting oncogene promoter G4 structures has emerged as a promising strategy to address the challenge of undruggable and drug-resistant proteins, such as MYC, BCL2, KRAS, and EGFR. Natural products have long been an important source of drug discovery, particularly in the fields of cancer and infectious diseases. Noteworthy progress has recently been made in the discovery of naturally occurring DNA G4-targeting drugs. Numerous DNA G4s, such as MYC-G4, BCL2-G4, KRAS-G4, PDGFR-β-G4, VEGF-G4, and telomeric-G4, have been identified as potential targets of natural products, including berberine, telomestatin, quindoline, sanguinarine, isaindigotone, and many others. Herein, we summarize and evaluate recent advancements in natural and nature-derived DNA G4 binders, focusing on understanding the structural recognition of DNA G4s by small molecules derived from nature. We also discuss the challenges and opportunities associated with developing drugs that target DNA G4s.
Keywords: G-quadruplex, Natural products, Alkaloids, Cancer, Promoter
1. Introduction
The concept that DNA guanine (G)-quadruplexes (G4s) are a novel class of biomolecular targets for cancer drug discovery originated in the late 1980s [1,2], although the ability of guanine derivatives to self-assemble into four-stranded structures has been recognized since 1962 [3]. Telomeric-G4, formed in the single-strand telomeric guanine-rich DNA sequences in eukaryotic chromosomes under near-physiological conditions, was the first example of G4 formation in organisms [1,4,5]. In 1997, human telomerase was found to be inhibited by telomeric-G4-interactive small molecules [6]. Subsequently, the formation of G4 in the promoter region of the human oncogene MYC (MYC-G4) was reported in 2002, which provided initial evidence for the biological significance of DNA G4 in oncogene expression and demonstrated that their stabilization by G4-interactive small molecules can cause the downregulation of oncogenes [7]. This study sparked interest in non-telomeric -G4s and established the targeting of oncogene promoter G4s as an alternative strategy for cancer therapy, with MYC-G4 as a model system. Subsequently, a large number of oncogene promoter G4s have been discovered, including KRAS-, PDGFR-β-, BCL-2-, c-KIT-, VEGF-, and EGFR-G4s [8–15]. The 2009 Nobel Prize in Physiology or Medicine was awarded to Elizabeth Blackburn, Carol Greider, and Jack Szostak for the discovery of telomeres and telomerase. Since then, G4s have garnered significant attention as promising anticancer drug targets. In addition to their association with cancers, G4s are linked to various other diseases, including neurodegenerative diseases [16,17] and viral and parasitic infections [18,19]. The expansion of massive GGGGCC repeats has been implicated in frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) [17]. Helicase defects can lead to the accumulation of G4s in both the transcriptome and genome, resulting in many severe congenital diseases, such as Fanconi anemia (FANCJ-deficient), primordial dwarfism (BLM-deficient), Werner syndrome (WRN-deficient), and so forth [17,20]. The strong correlation between G4 disruption/formation and disease onset—including sporadic Alzheimer’s disease, severe familiar coagulopathy, atopic dermatitis, myocardial infarction, and deafness—has also been reported recently [21]. Therefore, it is of great significance to comprehend G4 structures, unravel the biological functions of G4s, and explore novel G4-targeting compounds for disease treatment.
DNA G4s are secondary structures of nucleic acid consisting of two or more stacked G-tetrads, which are formed by four guanines in G-rich sequences arranged in a circular manner and connected via Hoogsteen hydrogen bonds. The stable stacking of G-tetrads also requires the coordination of monovalent cations, such as K+ or Na+, with the guanine O6 (Fig. 1(a)) [22,23]. Although G4s can form intermolecularly or intramolecularly, most biologically relevant G4s are intramolecular and consist of a core comprising three tetrads [24]. Interestingly, G4s are characterized by high structural diversity, in contrast to the rather uniform duplex structures (Fig. 1(b)) [24], as they can vary in terms of the directionality of G-tracts, loop type, loop length, or loop sequences. Moreover, G4s exhibit diversity in the capping structures that cover the external tetrads. For example, MYC-G4 and VEGF-G4 are canonical right-handed parallel-strand G4s, adhering to the well-established rules for G4 formation with a consensus sequence of G≥3(N1–7G≥3)≥3 (Fig. 1(c)) [24]. Telomeric (Tel) hybrid-1 and hybrid-2 G4s are hybrid structures consisting of three parallel-oriented G-tracts and one antiparallel-oriented G-tract, arranged in different order (Fig. 1(c)) [24]. In contrast, KRAS-G4, PDGFR-β-G4, and EGFR-G4 represent bulged G4, fill-in vacancy G4, and snap-back G4 topologies, respectively, which deviate from the canonical G4 structures (Fig. 1(c)) [15,24]. Moreover, hTERT-G4 is a distinctive tertiary DNA G4 structure consisting of an end-to-end stacked pair of G4s with a 26-base long loop, making it the first reported instance of such a configuration (Fig. 1(c)) [25]. These distinct G4 structures are proposed to interact with various proteins to perform specific biological functions [26]. The unique diversity of G4s among DNA secondary structures presents opportunities for the development of specific drugs that target individual G4s.
Fig. 1.

The secondary DNA G4 structures. (a) G-rich sequences form G-tetrads that make up the core of DNA G4s. The four guanine bases in the G-tetrad are connected by Hoogsteen hydrogen bonds. The stacked tetrads are stabilized by the coordination of K+ or Na+ (black ball). Xm: different numbers (m) of loop residues (X); R: (deoxy)ribose-phosphate backbones; M: metal cations. (b) Schematic representation of parallel, antiparallel, and hybrid unimolecular DNA G4 structures. The different types of loops are marked. (c) Schematic representation of several human genome G4s with distinct topologies. Tel: telomeric; dGMP: deoxyguanosine-5′-monophosphate.
By using G4-specific antibodies and chemical probes, DNA G4s have been mapped in the human genome and visualized in living human cells [27–31]. Under G4-inducing conditions, the human genome has the potential to form over 70 000 G4s [32]. However, only around 10 000 G4 structures have been identified in human chromatin and living cells [30,31]. These findings suggest that DNA G4 formation is a dynamic process that is highly associated with specific chromosome structures, genomic features, and cell status. Furthermore, G4-forming sequences are highly enriched in key human gene regulatory regions, such as 5′-untranslated regions, oncogene promoters, and telomeres [30,31]. The biological function of DNA G4s, which can be seen as epigenetic modulators, involves the regulation of different cellular processes, including gene transcription, translation, and replication; genome stability; and telomere maintenance [26,27,33]. The presence of DNA G4s is substantially associated with highly transcribed genes involved in cancer and neurodegenerative disorders [27,34]. The formation of DNA G4 structures in human cancer cells is prominent in specific cell cycles and occurs in a dynamic equilibrium between folded and unfolded states. This equilibrium can be shifted toward a folded state by using G4-stabilizing compounds, which then suppress oncogene expression by interfering with the interaction between G4s and transcriptional factors or functional proteins, ultimately leading to cancer cell death [31]. Moreover, the formation of DNA G4 structures is associated with increased heterogeneity in breast cancers, thereby facilitating stratification and enabling the discovery of personalized cancer treatment strategies [35]. Taken together, DNA G4s have been recognized as distinct targets for cancer treatment, particularly in targeting “undruggable” and drug-resistant proteins such as MYC, EGFR, KRAS, and PDGFR-β [36,37].
Natural products and their derivatives have historically played an important role in the discovery of drugs, particularly for cancer and infectious diseases [38,39]. In fact, more than 60% of approved anticancer drugs are derived from natural sources, including the famous antitumor drugs paclitaxel, teniposide, camptothecin, and vincristine [39]. Natural products have intrinsic advantages in structural complexity and functional diversity [38]. Notably, many natural and nature-derived compounds, including telomestatin, quindoline i, berberine, coptisine, epiberberine, and sanguinarine, have been found to bind DNA G4 structures with potent affinity and anti-cancer activity (Fig. 2) [37,40–44], while the compounds listed above are just one tip of the iceberg for natural products to be excavated of their DNA G4-targeting activities.
Fig. 2.

Chemical structures of reported important natural and nature-derived small molecules that target DNA G4s.
In this review, we summarize and evaluate the recent progress related to natural and nature-derived DNA G4 binders, with an emphasis on structural studies of DNA G4s in complex with natural small molecules. We then discuss the challenges and opportunities met in the development of DNA G4-targeting drugs based on natural products.
2. Telomestatin and its derivatives are potent telomeric-G-quadruplex stabilizers
Human telomeres are typically made up of 5–8 kilobases (kb) d(TTAGGG)n G-rich DNA repeats with a single-stranded 3′-end chromosomal overhang of 150–200 nucleotides [45–47]. In general, 50–200 bases of telomeres are lost per round of replication, and the cell ultimately undergoes senescence and apoptosis when the telomeric DNA reaches a critical length [46,48]. However, the telomerase enzyme, which is selectively activated in most tumor cells, maintains the telomere length through its reverse transcriptase activity and thus has a critical role in cellular transformation and immortalization [48,49]. G-rich human telomeres can form two distinct types of DNA G4s—namely, hybrid-1 and hybrid-2—in physiologically relevant solution conditions. The stabilization of telomeric G4s by small molecules could hinder telomere extension mediated by telomerase or alternative lengthening of telomeres (ALT), leading to the shortening of the telomeric DNA and finally causing cancer cell death [24,50,51]. Therefore, developing telomeric-G4 stabilizers has emerged as an effective anti-cancer strategy and has been supported by extensive studies [24,52–58].
Telomestatin, a natural macrocyclic compound isolated from Streptomyces annulus 3533-SV4, consists of seven oxazole rings and a thiazoline ring. It exhibits potent inhibitory activity against telomerase (Fig. 2) [59]. This compound has been found to specifically inhibit telomerase activity by stabilizing telomeric-G4s with an a half maximal inhibitory concentration value with telomerase repeat amplification protocol (IC50-TRAP) of 5 nmol·L−1—much more potent than any other reported G4-interactive molecules [42,59]. In addition, telomestatin exhibits antitumor activity against a number of cancer cells, while displaying negligible toxicity to normal cells. Therefore, telomestatin has been considered as a promising anticancer drug candidate and has attracted great research attention [57,60]. However, its low water solubility, together with the difficulty of obtaining large amounts of telomestatin, limits further clinical evaluation [61,62].
Notably, a number of telomestatin derivatives with improved physicochemical and biological properties have been reported [57,63–69]. Among them, one compound called L2H2–6M(2)OTD (L2H) (Fig. 2) containing six oxazole rings and two alkyl amine side chains showed comparable bioactivity to telomestatin (IC50-TRAP = 20 nmol·L−1) [57]. The alkyl amine side chains are positively charged under physiological conditions, which facilitates electrostatic interaction with the negatively charged phosphate backbone of telomeric-G4 and improves water solubility. Importantly, the nuclear magnetic resonance (NMR) solution structure of Tel-hybrid-1 G4 in complex with L2H has been resolved by Chung et al. [60] (Fig. 3). The determined complex structure showed that L2H disrupts the original T–A base pair in the 5′-end capping structure of the Tel-hybrid-1 G4. Instead, L2H is stacked on the 5′-end outer G-tetrad for extensive π-stacking interactions. The proximity of the two alkyl amine side chains to the phosphate groups of nucleotides facilitates electrostatic interactions, which play a critical role in establishing strong binding affinity. Collectively, this solution structure is the only available complex structure of a human telomeric-G4 bound to a telomestatin derivative, which provides valuable insight into the design of telomeric-G4-targeting drugs. Hence, many L2H derivatives with various functional groups have been synthesized [63,66–68,70].
Fig. 3.

(a) NMR-based solution structure of a telomestatin derivative (L2H, yellow) in complex with the Tel-hybrid 1 G4. L2H covers the entire 5′-end outer G-tetrad core for extensive π-stacking interactions. (b) NMR-based solution structure of quindoline i (yellow) in complex with the human MYC oncogene promoter G4. Quindoline i recruits the flanking residues A6 and T23 to form a ligand–base co-plane that stacks on the outer G-tetrads for adaptive binding pocket formation. PDB: protein data bank.
3. Quindoline and its derivatives as MYC G-quadruplex stabilizers
Aside from telomeric DNA G4s, oncogene promoter G4s have attracted great attention, especially the MYC oncogene promoter G4 [22]. Stabilizing oncogene promoter G4 structures to modulate downstream gene expression has emerged as an alternative therapeutic strategy for cancer treatment [8,22]. Pioneering work from the Hurley group reported that a DNA G4 formed in the MYC proximal-promoter region (MYC-G4) [7]; it works as a gene transcription repressor and can be stabilized by G4-targeting small molecules for MYC transcription inhibition. This work established a paradigm for subsequent promoter G4 studies for genes such as KRAS, PDGFR-β, BCL-2, c-KIT, VEGF, and many others [8–14].
MYC, a validated “driver” oncogene, is highly deregulated in human cells and exhibits overexpression in more than 80% of human cancers [71–73]. The MYC oncoprotein plays pivotal roles in tumor cell proliferation, differentiation, metastases, and drug resistance and is considered an attractive cancer therapeutic target [71–73]. However, it is well-known to be “undruggable” due to its flat surface, which lacks a compound binding pocket [72–74]. Significantly, the binding and stabilization of small molecules to MYC-G4 can effectively reduce MYC expression, ultimately leading to the death of cancer cells [7]. Therefore, MYC-G4 represents an alternative target for MYC-signaling downregulation and has been attracting great attention from the cancer community.
The major free MYC-G4 structure was determined by our lab in 2005 [75]. However, finding a small molecule that binds to MYC-G4 with sufficient affinity and specificity for structure determination has been challenging [24]. Although we have been extensively working on finding a specific MYC-G4 stabilizer, it took six more years before we determined the first high-resolution NMR solution structure of the MYC-G4 in complex with two molecules of quindoline i (a quindoline derivative). This is also the first complex structure of a biologically relevant promoter G4 recognized by small molecules (Fig. 3(b)) [76]. Quindoline i is a derivative of the naturally occurring indoloquinoline alkaloid cryptolepine [77]. Cryptolepine was isolated from Cryptolepis triangularis N.E.Br. It has a variety of bioactivities, including antimicrobial, antibacterial, anti-inflammatory, and anticancer activities [77,78]. Moreover, cryptolepine has been used as an antimalarial drug in Central and Western African nations [41,78]. In the year 2000, cryptolepine and its derivatives were shown to have antitumor activity, which may be connected with their DNA G4 interactions [79]. Since then, a number of cryptolepine derivatives have been synthesized, some of which have shown a potent MYC-G4 stabilization effect, including quindoline i [41,80,81].
The NMR titration data of quindoline i to MYC-G4 exhibited a high spectral quality that was suitable for structure determination [76]. Therefore, the 2:1 quindoline i–MYC-G4 complex structure was determined and showed several unexpected features, such as the rearrangement of the flanking residues induced by quindoline i to form a binding pocket (Fig. 3(b)) [76]. At each end, quindoline i recruits the adjacent flanking residue A6 or T23 to form a “quasi-triad plane” that stacks on the outer G-tetrads of the MYC-G4. The binding of quindoline i involves both electrostatic and π-stacking interactions. Importantly, the diethylamino functional groups may have electrostatic interactions with the DNA phosphate backbone. It is notable that this is the first structural study showing the simultaneous binding of a DNA G4 by two small molecules. It should be pointed out that the crescent-shaped quindoline only covers two guanines of a G-tetrad for π-stacking interactions, which is different from previously described and more extensive symmetric cyclic ring-fused compounds, including telomestatin and TMPyP4, that overlap evenly with all four guanines in the external G-tetrad for maximum stacking interactions.
However, compounds with symmetric cyclic fused rings appear to bind G4s with less specificity, whereas asymmetric compounds bearing a smaller stacking skeleton—such as quindoline and quarfloxin—are more likely to bind to a specific G4 in a defined manner. Therefore, it is of great interest to develop more crescent-shaped compounds for specific G4-targeting drug discovery. Indeed, inspired by quindoline, a growing number of compounds that are specific for a particular G4 have been discovered from both natural and synthetic compounds in subsequent studies [36,82–90].
4. Berberine and its derivatives as telomeric and promoter G-quadruplex stabilizers
In general, the selectivity of G4-targeting compounds is unsatisfactory, as it is often difficult for such compounds to effectively differentiate among multiple DNA G4s. A well-known example is berberine, a natural isoquinoline alkaloid found in many medicinal plants, including Hydrastis canadensis, Coptis chinensis, and Berberis aristate [91,92]. Berberine has a broad array of pharmacological functions implicated in its protein and nucleic acid interactions, such as anti-cancer, anti-inflammatory, antimicrobial, and antidiabetic activities, and has been used in traditional Chinese prescriptions for hundreds of years [91,92]. Recently, berberine and its derivatives were found to be effective G4 stabilizers, uncovering a new functional mechanism of berberine scaffolds for drug development [37,93]. Notably, berberine is non-selective for DNA G4s and can bind to both the telomeric-G4 and a number of gene promoter G4s.
4.1. Berberine binds to human parallel telomeric-G4 with a 6:2 stoichiometry
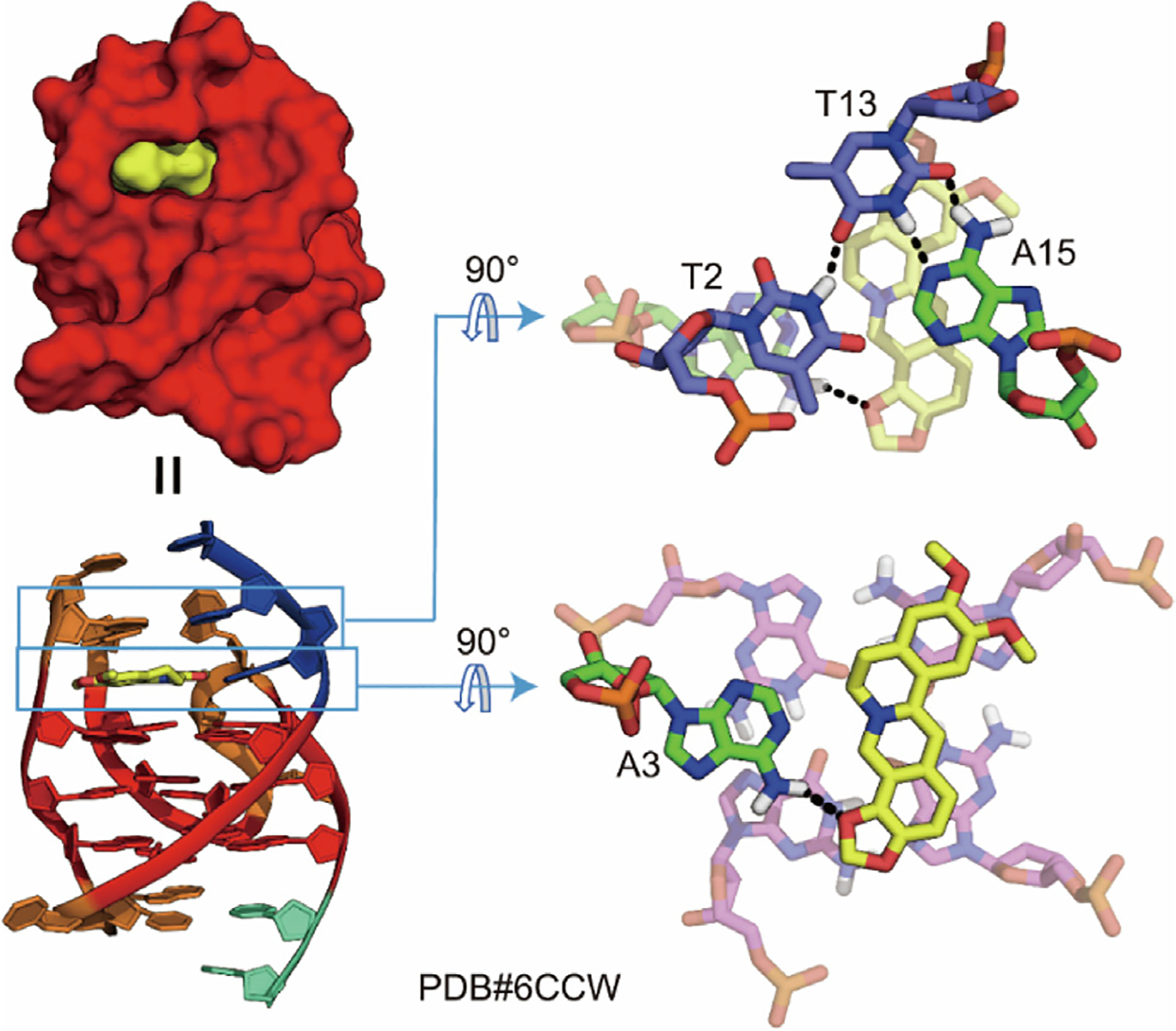
The first DNA G4 in complex with berberine was determined by X-ray crystal diffraction in 2013, when six berberine molecules were found to bind to a parallel G4 dimer formed by the human telomere sequence [94]. In the determined crystal structure, two molecules of berberine match each other and form a co-plane with their concave sites in the center, which then stack onto the outer G-tetrad of the parallel telomeric-G4 (Fig. 4). Interestingly, the A2 and T13 residues from two different DNA monomers are connected by Watson–Crick hydrogen bonds, creating a binding pocket at the two 5′-end sites where the paired berberine is located [94]. In this way, a drug-stabilized 5′-end-to-5′-end DNA G4 dimer is formed with a total of 6:2 berberine–G4 binding stoichiometry [94]. Notably, the biological significance of this determined crystal structure is limited due to its potential association with crystal packing rather than being a unique type of 6:2 berberine–G4 complex in solution. Nevertheless, the dimeric binding mode derived from the crystal structure has been used in many studies for developing G4-targeting berberine derivatives [95–97].
Fig. 4.

X-ray structure of berberine (yellow) in complex with a parallel human telomeric-G4. Two berberine stack onto one outer G-tetrad without any flanking residue interactions.
4.2. Berberine and coptisine bind to MYC and KRAS promoter G-quadruplexes
We investigated the binding of berberine with various DNA G4s using 1H NMR titration experiments. The results showed that berberine preferably binds to parallel DNA G4s, including the MYC and KRAS promoter G4s. Unlike the crystal complex structure, which shows a dimeric binding mode, we found that berberine binds to MYC-G4 (dissociation constant Kd ≈ 9.9 μmol·L−1) in a monomeric form with a 2:1 berberine–MYC-G4 binding stoichiometry [97]. The mass spectra used in this study clearly show that there are two major high-affinity binding sites of berberine to MYC-G4, which differs from the previously reported 6:2 binding mode in the crystal solid state [94,97]. The determined NMR solution structure of the 2:1 berberine–MYC-G4 complex shows that berberine recruits the flanking residue to form a ligand–base co-plane that stacks on the 5′- or 3′-external G-tetrad, and the coexistence of two different berberine orientations can be observed at each binding site (Figs. 5(a) and (b)). Interestingly, two distinct conformations of berberine to MYC-G4 at each binding site are clearly defined and related via rotational symmetry of about 180° (Figs. 5(a) and (b)). The wide binding pocket formed by the outer G-tetrad and the flanking residues in parallel G4s makes the bound ligand more flexible and able to adopt different conformations.
Fig. 5.

(a, b) NMR-based solution structures of berberine (yellow) in complex with the human MYC oncogene promoter G4. Berberine recruits the immediate flanking adenine at both the 5′- and 3′-end to form intercalated ligand–base planes, with two distinct berberine orientations being observed at each binding site.
KRAS-G4 is another actively studied promoter G4, which is a transcriptional regulator and amenable for small-molecule targeting to downregulate KRAS expression [9]. The KRAS oncogene is one of the highly mutated genes in the human genome and contributes to a number of human cancers [37,98]. Although KRAS-G4 was discovered over ten years ago, and a number of KRAS-G4-binding compounds were reported, no high-resolution KRAS-G4–ligand complex structure had as yet been solved, which severely impeded the further development of KRAS-G4-targeting drugs [37]. In 2022, we determined two NMR solution structures of KRAS-G4 in complex with the natural products berberine (Kd ≈ 0.55 μmol·L−1) and coptisine (Kd ≈ 0.50 μmol·L−1) [37]. In these two complex structures, the 2:1 binding stoichiometry and base-recruiting mechanism can again be observed, and appear to be the key features of berberine derivatives bound to parallel G4s in the solution state (Fig. 6). Notably, the KRAS-G4 contains a unique thymine bulge, which is base-paired with the flanking residue adenine by Watson–Crick hydrogen bonds in the ligand-free form. However, with berberine and coptisine binding, the original A–T base pair is disrupted to form an adenine–ligand co-plane that stacks on the 3′-end G-tetrad. The bulge base thymine partially covers the bound ligand and participates in the formation of a 3′end binding pocket (Fig. 6). This unique thymine bulge could serve as a binding moiety to enhance the ligand selectivity for KRAS-G4. Moreover, the KRAS-G4-coptisine complex structure is stabilized by an extra H-bond that forms between the methylenedioxy five-member ring of coptisine and adenine H61. Nevertheless, the 4 nucleotide (nt) loop residues are not involved in the binding pocket formation, which may be worth exploring further. In addition, novel berberine derivatives can be designed by introducing different side chains at the C1, C12, and C13 positions to achieve higher affinity and selectivity for KRAS-G4 based on the determined complex structures.
Fig. 6.

NMR-based solution structure of (a) berberine (yellow) and (b) coptisine (yellow) in complex with the human KRAS oncogene promoter G4. Berberine and coptisine recruit the immediate flanking adenine at both the 5′- and 3′-end to form intercalated ligand–base planes. The bulge base T10 partially covers the bound ligand and participates in the formation of the 3′-end binding pocket. A potential hydrogen bond is shown as a dashed line in the coptisine-A3 plane.
4.3. Berberine binds to a dGMP-fill-in vacancy G-quadruplex formed in the PDGFR-β gene promoter
The vacancy G4 (vG4) is a unique type of DNA G4 that is formed by three G3 tracts and one G2 tract and thus has a G-vacancy site in an incomplete G-tetrad [99,100]. vG4s are less stable than the canonical G4s due to the presence of the G-vacancy site [99]. However, the G-vacancy site can be specifically filled in by guanine derivatives, such as cyclic guanosine monophosphate (cGMP) and deoxyguanosine-5′-monophosphate (dGMP), which provides an opportunity for developing selective vG4-targeting drugs by designing small-molecule-guanine conjugates that utilize the G-vacancy site as an anchor point [101,102]. Moreover, the formation of G-fill-in vG4s indicates potential gene-regulatory mechanisms associated with the guanine metabolite concentration in cells and implies new opportunities for novel drug development [101,103–105]. In 2020, we determined the NMR solution structure of the first dGMP fill-in vG4 from the PDGFR-β gene promoter [101]. We also found the natural alkaloid berberine as a suitable small molecule that could specifically bind and stabilize this distinct type of dGMP fill-in vG4 (Kd ≈ 1.6 μmol·L−1) [93]. The NMR structure of the ternary berberine–dGMP-vG4 complex in potassium solution was determined (Fig. 7) [93]. This is the first complex structure of a small molecule bound to a fill-in vG4. Like the berberine–MYC-G4 complex structure, each berberine recruits the adenine residue from the two flanking sequences to form a “quasi-triad plane” that stacks on the two outer G-tetrads of the fill-in vG4. The binding involves π-stacking and electrostatic interactions. The coexistence of a minor ligand conformation in the two binding sites is also observed in the berberine–dGMP–PDGFR-β-vG4 complex, just like the berberine–MYC-G4 binary complex (Figs. 5 and 7) [93,97]. This study reveals the interactions of berberine with a biologically relevant vG4 and contributes structural insight for the design of vG4-targeting berberine derivatives.
Fig. 7.

NMR-based solution structure of the ternary complex of dGMP–PDGFR-β-vG4 with berberine (yellow). Berberine recruits the immediate flanking adenine at both the 5’- and 3′-end to form intercalated ligand–base planes, which significantly stabilizes the 5′-end dGMP-fill-in vG4 complex.
4.4. Epiberberine specifically binds to human Tel-hybrid-2 G-quadruplex
Epiberberine is a berberine derivative that differs in the positions of the methoxy and methylenedioxy groups (Fig. 2) [106]. Our lab determined the structure of the biologically relevant human Tel-hybrid-2 G4 (Tel2G4) back in 2007 [51], and it took almost ten years to find a suitable small molecule that can specifically bind to this unique structure [106]. In 2018, we solved the solution structure of Tel2G4 in complex with epiberberine (Kd ≈ 0.016 μmol·L−1) using NMR (Fig. 8) [53,106]. Unlike berberine, which binds to the promoter G4 with a 2:1 binding stoichiometry, epiberberine specifically binds to the 5′-end G-tetrad of the Tel2G4. This specific binding induces a significant rearrangement of the flanking residues and the TTA loop at the 5′-end site, forming a well-fitted binding pocket. Epiberberine recruits the flanking adenine to form an H-bonded ligand–base co-plane that stacks on top of the 5′-end outer G-tetrad. Simultaneously, this region is covered by a T:T:A triad layer and another T:T base pair through π-stacking interactions. Such an extensive four-layer binding pocket has never been described in G4–ligand complexes before.
Fig. 8.
NMR-based solution structure of epiberberine (yellow) in complex with the human Tel2G4. Epiberberine recruits the A3 residue at the 5′-end to form a ligand–base co-plane. Potential H-bonds are indicated by dashed lines.
It is notable that the structurally similar alkaloids berberine (Kd ≈ 1.99 μmol·L−1), coptisine (Kd ≈ 0.33 μmol·L−1), and palmatine (Kd ≈ 0.74 μmol·L−1) cannot bind to Tel2G4 well [53], suggesting that the positions of the methoxy and methylenedioxy groups have a crucial impact on the specific recognition. Significantly, epiberberine exhibits such high specificity toward Tel2G4 that it can convert other telomeric-G4 structures into hybrid-2 under physiological conditions, making it the first reported example of this kind. Overall, this study provides structural insight into ligand interaction with the human telomeric-G4 and contributes a model system for developing specific Tel2G4-targeting drugs.
5. Many other natural products as DNA G-quadruplex binders
Given the structural diversity and polymorphism of the human DNA G4s, it is reasonable to look for potent and selective G4 binders among natural products. Indeed, a growing list of naturally occurring DNA G4 binders has been reported beyond the above-discussed G4-binding natural alkaloids, including distamycin A [107], Fe(III)-protoporphyrin IX (hemin) [108], colchicine [109], pegaharmine D [84], sanguinarine [40], chelerythrine [40], piperine [110], magnoflorine [111], triptolide [112], jatrorrhizine [113], fangchinoline [114], evodiamine [114], isaindigotone [58], quinazoline [115], and schizocommunin derivatives [116].
Distamycin A, a canonical DNA minor groove binder, was found to bind to the two opposite grooves of an intermolecular parallel [d(TGGGGT)]4 G4 in 4:1 binding stoichiometry with a binding constant (Kb) value of (4.0 ± 3.0) ×106 L·mol−1 [107]. This finding suggests that the design of novel G4-binding compounds can be achieved by combining G4 ligands with DNA duplex binders, thereby enhancing both G4 specificity and affinity. Furthermore, a flexible DNA duplex binder can be used to link two G4-binding compounds, forming a clamp-like ligand for stable anchoring inspired by the artificial G4 probe (G4P) protein [31]. Hemin, a rigid natural macrocycle compound chelated with a metal ion, can be captured by G4 structures formed in the expanded hexanucleotide repeat RNA (Kd ≈ 3 μmol·L−1) and DNA of the C9orf72 gene. This interaction enhances peroxidase activity and is associated with the development of neurodegenerative diseases such as ALS and FTD [117]. The positive ion center and aromatic macrocycle skeleton are natural characteristics of G4 binding compounds that have attracted intense attention from researchers in structural modification. In general, the cation center and the side chains are the main modified objects, represented by the pentacationic manganese(III)–porphyrin complex (association constant (Ka) = 108 L·mol−1 human telomeric-G4) and the porphyrin-bridged tetranuclear platinum complexes with significant G4-binding specificity [118–123]. Among the discovered G4-binding natural products, many have crescent-shaped skeletons with significant G4-stabilizing activity, such as sanguinarine (Ka ≈ 1.16 × 106 L·mol−1-KRAS-G4) [124], jatrorrhizine (Ka ≈ 0.90 × 106 L·mol−1-KRAS-G4) [124], and chelerythrine (Kd ≈ 0.25 μmol·L−1-VEGFA-G4) [125]. The drug potential of sanguinarine and chelerythrine is restrained due to their toxicity against normal cells [126,127]. However, structural modifications may help to attenuate such side effects. It is interesting to note that schizocommunin is not a rigid crescent-shaped compound and has a relatively flexible carbon skeleton, whereas the intramolecular hydrogen bond contributes greatly to the compound’s planarity and thus its G4-binding affinity [116,128]. Therefore, it is worth considering the introduction of H-bond-forming fragments to amplify the planarity of other possible compounds while ensuring a certain degree of flexibility. The tertiary nitrogen centers could also be quaternized to introduce positive charges, thereby enhancing the interactions with electron-rich G4s [129,130]. Many studies have investigated the structural modifications of crescent-shaped natural products for higher G4-binding specificity and better pharmacological activity [131–133]. Given the lack of determined structures for many G4–ligand complexes, it is crucial to comprehend the molecular recognition of such products for specific G4s and utilize this knowledge to develop novel nature-derived drugs.
6. Challenges and opportunities for DNA G4-targeting drug design based on natural products
Structural studies on natural small-molecule-G4 complexes have provided valuable structural basis and insight into targeting human promoter G4s and telomeric-G4s. Macrocyclic molecules, such as telomestatin derivatives, are similar in size to a G-tetrad and cover the entire four guanines of the outer G-tetrad in the Tel-hybrid-1 G4 for extensive π-stacking and electrostatic interactions. These large macrocyclic molecules generally have high affinity and low selectivity for the different topologies of DNA G4s, which is challenging to bind a specific G4. In contrast, small crescent-shaped pharmacophores with suitable functional groups, such as berberine, epiberberine, and quindolines, are more likely to bind to biologically relevant intramolecular G4s in a specific manner. Since crescent-shaped small molecules only cover two guanines, they often recruit one flanking residue to form a “quasi-triad plane” that stacks over the outer G-tetrad for specific G4 binding. Distinct from DNA minor-groove binders, which emphasize skeleton length and flexibility for sequence-specific targeting and to adapt to the helical topology of duplex DNA [134–137], crescent-shaped G4-binding compounds are characterized by extended aromatic ring systems, positively charged centers, and modifiable side chains. The central positively charged nitrogen in a crescent-shaped pharmacophore would normally be positioned above the negatively polarized carbonyl groups of the G-tetrad, resulting in strong electrostatic interactions. Moreover, the possible hydrogen bonding interaction between the ligands and the recruited flanking residues is an important factor that reinforces the specific interaction, as in the cases of quindoline i and berberine to MYC-G4, epiberberine to telomeric-G4, and coptisine to KRAS-G4. Meanwhile, the modifiable cationic side chains would interact with the grooves or phosphate backbones of the G4s for steric and electrostatic interactions, contributing additional forces for the specific recognition of distinct DNA G4s. Collectively, the optimal small molecules utilize a combination of interactions—including steric effects, π-stacking, H-bonds, and electrostatic interactions, to recognize individual G4s specifically—which can only be identified in NMR solution structure studies.
The main challenge of current G4-targeting drug discovery is G4 selectivity. Structural studies of the human genomic G4s have shown that many G4s share the same general features—that is, a stacked G-tetrad core with several short loops. The reported G4-targeting compounds stack on the outer G-tetrad for extensive π-stacking interactions; therefore, it is difficult for these compounds to distinguish among different G4s, especially with similar G4 topologies. Remarkably, the recently discovered unique G4s, such as vG4s [101,104,105], bulge-containing G4s [37], and stem-loop-containing G4s [138–140], may provide opportunities to develop particular G4-targeting drugs, because these G4s have features distinct from the canonical G4s that can be utilized for more selective ligand design. For example, inspired by a dual-specific targeting approach [141,142], natural G4-binding ligands could be conjugated with specific DNA duplex binders to target specific stem-loop G4s or G4s with suitable grooves. Similarly, natural ligands could be modified with guanine analogs for vG4-specific binding by partly contributing to the integrity of vG4s [93,102,104,143]. For G4s with bulges or loops, natural compounds could be conjugated with complementary base analogs linked by flexible carbon chains for possible complementary pairing and hydrogen bonding. Apart from G4s with special structural topologies or sequence compositions, the unwound single DNA strands adjacent to G4s could be targeted by ligand–”oligonucleotide” complexes, such as ligand–peptide nucleic acid (PNA) conjugates, for individual G4 targeting [144,145]. It was reported that the conjugate of the G4 ligand naphthalene diimide (NDI) with PNA, which can hybridize with G4 flanking sequences, bound specifically to the G4 within the human immunodeficiency virus (HIV)-1 long terminal repeat (LTR) region, implying its potential in acquired immune deficiency syndrome (AIDS) treatment [144]. Notably, the cell membrane permeability should be considered when designing ligand–PNA conjugates. Platinum halide modification could also be utilized to realize covalent binding with flanking or loop bases and to reduce off-target effects for potent cancer therapies [146,147]. In addition, carbohydrates could be added to the ligand for higher selectivity toward cancer cell G4s [87,148,149]. Since studies of ligand-conjugates have rarely involved natural products, further experiments should be carried out to verify the feasibility of the above strategies. On the other hand, the enantioselectivity of natural and nature-derived compounds could be considered for higher specificity toward targeted G4s. Many synthetic metal complexes have exhibited enantioselectivity toward specific G4s [150–155]; however, the chirality of natural and nature-derived compounds associated with G4-binding activity is less understood. For ligand skeletons with chiral carbons, the chiral effects should be investigated further. For example, the derivative (S)-telomestatin was reported to have much higher telomeric-G4 binding and telomerase-inhibitory activity than natural (R)-telomestatin [52]. Pegaharmine D, a pair of racemates, could bind parallel G4s specifically and showed remarkable inhibitory activity against cancer cells, while the effects of its chirality on G4 binding and bioactivity have not been investigated yet [84]. Since many alkaloids are chiral with distinct pharmacological activity, it would be of great significance to dissect the relationships between ligand enantioselectivity and G4 affinity–selectivity through specific structural analysis. Altogether, the above strategies could be combined for much more specific G4 targeting as well as higher therapeutic effects.
Another challenge confronted by G4-targeting drug discovery is the timely circulation and sharing of newly found natural alkaloids all over the world. Hundreds to thousands of novel alkaloids with various skeletons have been isolated from terrestrial and marine organisms, including plants, fungi, and bacteria [156–163]. Many of them have aromatic planes and positively charged centers. In particular, many marine alkaloids have natural halogen groups, which have been found to stabilize π-stacking by withdrawing electrons [164–166]. Halogen groups could also help improve the lipo-solubility and bioavailability of ligands [116]. Tajuddeen et al. [167] reported a versatile new class of axially chiral N,C-coupled naphthylisoquinoline alkaloids isolated from plants, which might have the potential to bind G4s. However, the G4-binding activity screening of these newly discovered natural alkaloids is hampered by the limitations of their sources, synthesis, and commercialization. Another reason may be that G4s are newly recognized targets for small molecules and are not as well-known as targets such as duplex DNA and proteins. In fact, telomestatin is the only known G4-binding natural ligand isolated from microorganisms since its discovery in 2001, and it has been derivatized substantially for better bioactivity [57,59]. Therefore, numerous novel natural products remain to be tested and derivatized for their G4-binding potential, which will advance the field of natural or nature-derived drug discovery based on G4 interactions.
At present, a momentous argument is whether it is necessary to target a specific G4 for disease treatment. Cancer and other disease cells have been found to have more G4s, along with more obvious genomic instability, than normal cells. Many reported G4-binding compounds have been found to bind different G4s and exhibit prominent cancer-inhibitory activities with little or no toxicity toward normal cells [20,168–170]. Berberine, which was discussed earlier in this review, has been found to bind the G4s formed in telomeres and the promoters of MYC, KRAS, and PDGFR-β, and inhibit the proliferation of non-small cell lung cancer (NSCLC) cells. Thus, it is worth considering the pursuit of multiple G4 targeting for complex disease treatment. However, for diseases caused by one or several canonical aberrant genes (e.g., EGFR and KRAS mutations, and C9orf72 hexanucleotide repeats) or those related to synthetic lethality [8,27,117,171], individual G4 targeting would be an effective therapy with higher safety and thus could still be an important direction for drug design. The “oncogene addiction” hypothesis suggests that some cancers depend on a driver gene or genes for growth and viability, and define appropriate cancer targets [172,173]. Therefore, targeting aberrantly expressed genes by acting on promoter G4s could be a fruitful way for novel drug development in cancer therapy, especially as an alternative strategy for “undruggable” and drug-resistant proteins such as MYC, KRAS, and EGFR. Collectively, more experimental verifications on the cellular and animal levels are needed to clarify the advantages and disadvantages of individual-G4 targeting and multiple-G4 targeting strategies.
7. Conclusions
Natural products have clear importance and advantages in drug design, as they represent a huge inspiring chemical library to be tested and derivatized. Numerous studies have revealed that G4s participate in various aberrant biological processes as epigenetic and regulatory elements in replication, transcription, and translation. The formation of G4s can inhibit DNA methylation and influence the nucleosome assembly, which renders G4s as special epigenetic markers. In addition, G4 formation can cause site mutagenesis, gene deletion–junction, transposition, rearrangement, and copy number alterations, which are important sources of genome instability and disease genesis [174,175]. G4s in the promoter regions can induce the binding of transcription factors and may even change the chromatin architecture and regulate gene expression by promoting long-range interactions including promoter–enhancer interactions and chromatin looping, mediated by chromatin remodeler proteins and long-loop G4 formation [176–180]. The formation and stabilization of R-loops are also closely related to G4 formation [176,181]. The long-range interactions and R-loops mediated by G4s, together with regulatory proteins, may promote liquid–liquid phase separation (LLPS) to ensure efficient biological processes [26], which are prominent in cancer cells for rampant proliferation and nutrient depredation. However, the specific mechanisms still need experimental confirmation. Furthermore, the aberrant formation of G4s in coding regions would induce replication fork stalling, transcription and translation termination, like a simple “roadblock,” thereby affecting cellular homeostasis and causing pathological lesions. To maintain orderly vital movements, helicases, and other nucleic acid binding proteins are frequently needed to unwind G4s in normal cells, which also involves DNA damage repair pathways [174,182–184]. Once the balance of G4-formation–unwinding is disrupted and DNA repair errors occur due to congenital or environmental factors, diseases such as cancers can happen. Therefore, aside from protein targeting, it is judicious to develop G4-targeting drugs by taking advantage of quadruplex-structural features distinct from double-stranded DNA (dsDNA). Natural drugs that target G4s can compete with G4-binding proteins and impede gene expression, acting as a “brake,” thus offering a promising strategy for disease treatment.
Structural analyses of G4–ligand complexes can provide information about ligand binding mechanisms and novel drug design. Based on the determined complex structures, it is evident that natural small molecules specifically recognize G4s through a combination of interactions, including π-stacking, H-bonds, electrostatic interactions, and steric effects. These specific binding modes of ligand–G4 complexes in pseudo-physiological conditions can be best studied via NMR solution structure analysis, because crystallization commonly produces artificial ligand–G4 complexes due to crystal packing, as in the case of the 6:2 berberine–G4 crystal structure observed under crystalline conditions. Unlike macrocyclic molecules, which directly cover the entire G-tetrad for both π-stacking and electrostatic interactions, small crescent-shaped pharmacophores can recruit flanking residues to form a “quasi-triad plane” that stacks over the outer G-tetrad, facilitating extensive π-stacking and electrostatic interactions. Specific H-bond interactions are observed in the “quasi-triad plane,” which promote higher binding affinity. Specific structural modifications based on natural products are greatly needed to improve the selectivity and bioactivity of these compounds toward G4s.
The development of various small molecule modification strategies to enhance G4-binding specificity and affinity has made it more feasible to target individual G4s, while targeting multiple G4s is emerging as an important strategy for treating complex diseases. Pidnarulex (CX-5461), a first-in-class G4-targeting compound, is in clinical evaluation for treating BRCA1/2 deficient breast and ovarian cancer patients [169]. Due to the significant correlation between G4 formation and tumor progression, G4-binding drug development will open up a new window for disease treatments. More novel natural alkaloids—especially those from marine organisms and endophytic fungi—should be tested for their G4-binding ability and pharmacological activity; meanwhile, ligand–G4 complex structure analysis is imperative for instructive drug derivatization. Since natural products have historically been an important source of therapeutic drug leads, we believe more naturally occurring G4-targeting drugs will be discovered in the future, with exceptional G4 selectivity and high efficacy in disease treatment.
Acknowledgment
This research was supported by the National Institutes of Health (R01CA177585, U01CA240346, and R01CA153821) (DY), the Purdue Center for Cancer Research (P30CA023168), the National Natural Science Foundation of China (82173707 and 82322065), the Program for Jiangsu Province Innovative Research Scholar (JSSCRC2021512), and the “Double First-Class” University Project (CPUQNJC22_08).
Footnotes
Compliance with ethics guidelines
Kai-Bo Wang, Yingying Wang, Jonathan Dickerhoff, and Danzhou Yang declare that they have no conflict of interest or financial conflicts to disclose.
References
- [1].Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988;334(6180):364–6. [DOI] [PubMed] [Google Scholar]
- [2].Hurley LH. DNA and associated targets for drug design. J Med Chem 1989;32(9):2027–33. [DOI] [PubMed] [Google Scholar]
- [3].Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA 1962;48(12):2013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989;342(6251):825–9. [DOI] [PubMed] [Google Scholar]
- [5].Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature 1991;350(6320):718–20. [DOI] [PubMed] [Google Scholar]
- [6].Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem 1997;40(14):2113–6. [DOI] [PubMed] [Google Scholar]
- [7].Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA 2002;99(18):11593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Neidle S Quadruplex nucleic acids as novel therapeutic targets. J Med Chem 2016;59(13):5987–6011. [DOI] [PubMed] [Google Scholar]
- [9].Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res 2008;36(11):3765–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor β (PDGFR-β) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-β expression. Biochemistry 2010;49(19):4208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen Y, Agrawal P, Brown RV, Hatzakis E, Hurley L, Yang D. The major G-quadruplex formed in the human platelet-derived growth factor receptor β promoter adopts a novel broken-strand structure in K+ solution. J Am Chem Soc 2012;134(32):13220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, et al. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc 2006;128(4):1096–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, et al. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc 2005;127(30):10584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res 2013;41(22):10584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Y, Li J, Zhang Y, Wang Y, Chen J, Bian Y, et al. Structure of the major G-quadruplex in the human EGFR oncogene promoter adopts a unique folding topology with a distinctive snap-back loop. J Am Chem Soc 2023;145(29):16228–37. [DOI] [PubMed] [Google Scholar]
- [16].Mirkin SM. Expandable DNA repeats and human disease. Nature 2007;447 (7147):932–40. [DOI] [PubMed] [Google Scholar]
- [17].Maizels N G4-associated human diseases. EMBO Rep 2015;16(8):910–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Amrane S, Kerkour A, Bedrat A, Vialet B, Andreola ML, Mergny JL. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: a potential target for anti-HIV drug development. J Am Chem Soc 2014;136(14):5249–52. [DOI] [PubMed] [Google Scholar]
- [19].Belmonte-Reche E, Martínez-García M, Guédin A, Zuffo M, Arévalo-Ruiz M, Doria F, et al. G-quadruplex identification in the genome of protozoan parasites points to naphthalene diimide ligands as new antiparasitic agents. J Med Chem 2018;61(3):1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Muller S, Rodriguez R. G-quadruplex interacting small molecules and drugs: from bench toward bedside. Expert Rev Clin Pharmacol 2014;7(5):663–79. [DOI] [PubMed] [Google Scholar]
- [21].Lorenzatti A, Piga EJ, Gismondi M, Binolfi A, Margarit E, Calcaterra NB, et al. Genetic variations in G-quadruplex forming sequences affect the transcription of human disease-related genes. Nucleic Acids Res 2023;51(22):12124–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 2011;10(4):261–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Spiegel J, Adhikari S, Balasubramanian S. The structure and function of DNA G-quadruplexes. Trends Chem 2020;2(2):123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen L, Dickerhoff J, Sakai S, Yang D. DNA G-quadruplex in human telomeres and oncogene promoters: structures, functions, and small molecule targeting. Acc Chem Res 2022;55(18):2628–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc 2009;131(31):10878–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Robinson J, Raguseo F, Nuccio SP, Liano D, Di Antonio M. DNA G-quadruplex structures: more than simple roadblocks to transcription? Nucleic Acids Res 2021;49(15):8419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol 2017;18(5):279–84. [DOI] [PubMed] [Google Scholar]
- [28].Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 2013;5(3):182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Di Antonio M, Ponjavic A, Radzevičius A, Ranasinghe RT, Catalano M, Zhang X, et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat Chem 2020;12(9):832–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet 2016;48(10):1267–72. [DOI] [PubMed] [Google Scholar]
- [31].Zheng KW, Zhang J, He Y, Gong J, Wen C, Chen J, et al. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res 2020;48(20):11706–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 2015;33(8):877–81. [DOI] [PubMed] [Google Scholar]
- [33].Fleming AM, Burrows CJ. Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J Am Chem Soc 2020;142(3):1115–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang E, Thombre R, Shah Y, Latanich R, Wang J. G-quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res 2021;49(9):4816–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hänsel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet 2020;52(9):878–83. [DOI] [PubMed] [Google Scholar]
- [36].Wang KB, Elsayed MSA, Wu G, Deng N, Cushman M, Yang D. Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J Am Chem Soc 2019;141(28):11059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang KB, Liu Y, Li J, Xiao C, Wang Y, Gu W, et al. Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine. Nat Commun 2022;13(1):6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Atanasov AG, Zotchev SB, Dirsch VM, Supuran CT. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov 2021;20(3):200–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 2020;83(3):770–803. [DOI] [PubMed] [Google Scholar]
- [40].Gao Y, Peng H, Li L, Wang F, Meng J, Huang H, et al. Screening of high-efficiency and low-toxicity antitumor active components in Macleaya cordata seeds based on the competitive effect of drugs on double targets by a new laminar flow chip. Analyst 2021;146(15):4934–44. [DOI] [PubMed] [Google Scholar]
- [41].Zhou JL, Lu YJ, Ou TM, Zhou JM, Huang ZS, Zhu XF, et al. Synthesis and evaluation of quindoline derivatives as G-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J Med Chem 2005;48(23):7315–21. [DOI] [PubMed] [Google Scholar]
- [42].Kim MY, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc 2002;124(10):2098–9. [DOI] [PubMed] [Google Scholar]
- [43].Zhang L, Liu H, Shao Y, Lin C, Jia H, Chen G, et al. Selective lighting up of epiberberine alkaloid fluorescence by fluorophore-switching aptamer and stoichiometric targeting of human telomeric DNA G-quadruplex multimer. Anal Chem 2015;87(1):730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu L, Li J, He Y. Multifunctional epiberberine mediates multi-therapeutic effects. Fitoterapia 2020;147:104771. [DOI] [PubMed] [Google Scholar]
- [45].Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 1997;88(5):657–66. [DOI] [PubMed] [Google Scholar]
- [46].Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 2013;14(2):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Blackburn EH. Telomere states and cell fates. Nature 2000;408(6808):53–6. [DOI] [PubMed] [Google Scholar]
- [48].Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell 2001;106(3):275–86. [DOI] [PubMed] [Google Scholar]
- [49].Blackburn EH. Telomeres and telomerase: the means to the end (Nobel lecture). Angew Chem Int Ed Engl 2010;49(41):7405–21. [DOI] [PubMed] [Google Scholar]
- [50].Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res 2006;34(9):2723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res 2007;35(7):2440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Doi T, Shibata K, Yoshida M, Takagi M, Tera M, Nagasawa K, et al. (S)-stereoisomer of telomestatin as a potent G-quadruplex binder and telomerase inhibitor. Org Biomol Chem 2011;9(2):387–93. [DOI] [PubMed] [Google Scholar]
- [53].Deng N, Xia J, Wickstrom L, Lin C, Wang K, He P, et al. Ligand selectivity in the recognition of protoberberine alkaloids by hybrid-2 human telomeric G-quadruplex: binding free energy calculation, fluorescence binding, and NMR experiments. Molecules 2019;24(8):1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ji X, Sun H, Zhou H, Xiang J, Tang Y, Zhao C. The interaction of telomeric DNA and C-MYC22 G-quadruplex with 11 natural alkaloids. Nucleic Acid Ther 2012;22(2):127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadré J, Koebel P, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res 2006;66(14):6908–12. [DOI] [PubMed] [Google Scholar]
- [56].Binz N, Shalaby T, Rivera P, Shin-ya K, Grotzer MA. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur J Cancer 2005;41(18):2873–81. [DOI] [PubMed] [Google Scholar]
- [57].Tera M, Ishizuka H, Takagi M, Suganuma M, Shin-ya K, Nagasawa K. Macrocyclic hexaoxazoles as sequence- and mode-selective G-quadruplex binders. Angew Chem Int Ed Engl 2008;47(30):5557–60. [DOI] [PubMed] [Google Scholar]
- [58].Tan JH, Ou TM, Hou JQ, Lu YJ, Huang SL, Luo HB, et al. Isaindigotone derivatives: a new class of highly selective ligands for telomeric G-quadruplex DNA. J Med Chem 2009;52(9):2825–35. [DOI] [PubMed] [Google Scholar]
- [59].Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, et al. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc 2001;123(6):1262–3. [DOI] [PubMed] [Google Scholar]
- [60].Chung WJ, Heddi B, Tera M, Iida K, Nagasawa K, Phan AT. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J Am Chem Soc 2013;135(36):13495–501. [DOI] [PubMed] [Google Scholar]
- [61].Linder J, Garner TP, Williams HE, Searle MS, Moody CJ. Telomestatin: formal total synthesis and cation-mediated interaction of its seco-derivatives with G-quadruplexes. J Am Chem Soc 2011;133(4):1044–51. [DOI] [PubMed] [Google Scholar]
- [62].Doi T, Yoshida M, Shin-ya K, Takahashi T. Total synthesis of (R)-telomestatin. Org Lett 2006;8(18):4165–7. [DOI] [PubMed] [Google Scholar]
- [63].Iida K, Nagasawa K. Macrocyclic polyoxazoles as G-quadruplex ligands. Chem Rec 2013;13(6):539–48. [DOI] [PubMed] [Google Scholar]
- [64].Tera M, Iida K, Ishizuka H, Takagi M, Suganuma M, Doi T, et al. Synthesis of a potent G-quadruplex-binding macrocyclic heptaoxazole. ChemBioChem 2009;10(3):431–5. [DOI] [PubMed] [Google Scholar]
- [65].Iida K, Nakamura T, Yoshida W, Tera M, Nakabayashi K, Hata K, et al. Fluorescent-ligand-mediated screening of G-quadruplex structures using a DNA microarray. Angew Chem Int Ed Engl 2013;52(46):12052–5. [DOI] [PubMed] [Google Scholar]
- [66].Ma Y, Iida K, Sasaki S, Hirokawa T, Heddi B, Phan A, et al. Synthesis and telomeric G-quadruplex-stabilizing ability of macrocyclic hexaoxazoles bearing three side chains. Molecules 2019;24(2):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Abraham Punnoose J, Ma Y, Li Y, Sakuma M, Mandal S, Nagasawa K, et al. Adaptive and specific recognition of telomeric G-quadruplexes via polyvalency induced unstacking of binding units. J Am Chem Soc 2017;139(22):7476–84. [DOI] [PubMed] [Google Scholar]
- [68].Iida K, Majima S, Nakamura T, Seimiya H, Nagasawa K. Evaluation of the interaction between long telomeric DNA and macrocyclic hexaoxazole (6OTD) dimer of a G-quadruplex ligand. Molecules 2013;18(4):4328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rzuczek SG, Pilch DS, Liu A, Liu L, LaVoie EJ, Rice JE. Macrocyclic pyridyl polyoxazoles: selective RNA and DNA G-quadruplex ligands as antitumor agents. J Med Chem 2010;53(9):3632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sakuma M, Ma Y, Tsushima Y, Iida K, Hirokawa T, Nagasawa K. Design and synthesis of unsymmetric macrocyclic hexaoxazole compounds with an ability to induce distinct G-quadruplex topologies in telomeric DNA. Org Biomol Chem 2016;14(22):5109–16. [DOI] [PubMed] [Google Scholar]
- [71].Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol 2011;18(3):219–28. [DOI] [PubMed] [Google Scholar]
- [72].Dang CV. MYC on the path to cancer. Cell 2012;149(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer 2009;9(12):849–61. [DOI] [PubMed] [Google Scholar]
- [74].Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 2017;17(8):502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005;44(6):2048–58. [DOI] [PubMed] [Google Scholar]
- [76].Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline–c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J Am Chem Soc 2011;133(44):17673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Tackie AN, Boye GL, Sharaf MHM, Schiff PL Jr, Crouch RC, Spitzer TD, et al. Cryptospirolepine, a unique spiro-nonacyclic alkaloid isolated from Cryptolepis sanguinolenta. J Nat Prod 1993;56(5):653–70. [Google Scholar]
- [78].Bierer DE, Fort DM, Mendez CD, Luo J, Imbach PA, Dubenko LG, et al. Ethnobotanical-directed discovery of the antihyperglycemic properties of cryptolepine: its isolation from Cryptolepis sanguinolenta, synthesis, and in vitro and in vivo activities. J Med Chem 1998;41(6):894–901. [DOI] [PubMed] [Google Scholar]
- [79].Caprio V, Guyen B, Opoku-Boahen Y, Mann J, Gowan SM, Kelland LM, et al. A novel inhibitor of human telomerase derived from 10H-indolo[3,2-b]-quinoline. Bioorg Med Chem Lett 2000;10(18):2063–6. [DOI] [PubMed] [Google Scholar]
- [80].Guyen B, Schultes CM, Hazel P, Mann J, Neidle S. Synthesis and evaluation of analogues of 10H-indolo[3,2-b]quinoline as G-quadruplex stabilising ligands and potential inhibitors of the enzyme telomerase. Org Biomol Chem 2004;2(7):981–8. [DOI] [PubMed] [Google Scholar]
- [81].Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J Med Chem 2007;50(7):1465–74. [DOI] [PubMed] [Google Scholar]
- [82].Funke A, Dickerhoff J, Weisz K. Towards the development of structure-selective G-quadruplex-binding indolo[3,2-b]quinolines. Chemistry 2016;22(9):3170–81. [DOI] [PubMed] [Google Scholar]
- [83].Zhai Q, Gao C, Ding J, Zhang Y, Islam B, Lan W, et al. Selective recognition of c-MYC Pu22 G-quadruplex by a fluorescent probe. Nucleic Acids Res 2019;47(5):2190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang KB, Li DH, Hu P, Wang WJ, Lin C, Wang J, et al. A series of β-carboline alkaloids from the seeds of Peganum harmala show G-quadruplex interactions. Org Lett 2016;18(14):3398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liu HY, Chen AC, Yin QK, Li Z, Huang SM, Du G, et al. New disubstituted quindoline derivatives inhibiting burkitt’s lymphoma cell proliferation by impeding c-MYC transcription. J Med Chem 2017;60(13):5438–54. [DOI] [PubMed] [Google Scholar]
- [86].Amato J, Morigi R, Pagano B, Pagano A, Ohnmacht S, De Magis A, et al. Toward the development of specific G-quadruplex binders: synthesis, biophysical, and biological studies of new hydrazone derivatives. J Med Chem 2016;59(12):5706–20. [DOI] [PubMed] [Google Scholar]
- [87].Li ML, Yuan JM, Yuan H, Wu BH, Huang SL, Li QJ, et al. Design, synthesis, and evaluation of new sugar-substituted imidazole derivatives as selective c-MYC transcription repressors targeting the promoter G-quadruplex. J Med Chem 2022;65(19):12675–700. [DOI] [PubMed] [Google Scholar]
- [88].Hu MH, Lin JH. New dibenzoquinoxalines inhibit triple-negative breast cancer growth by dual targeting of topoisomerase 1 and the c-MYC G-quadruplex. J Med Chem 2021;64(10):6720–9. [DOI] [PubMed] [Google Scholar]
- [89].Ray S, Tillo D, Boer RE, Assad N, Barshai M, Wu G, et al. Custom DNA microarrays reveal diverse binding preferences of proteins and small molecules to thousands of G-quadruplexes. ACS Chem Biol 2020;15(4):925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Pelliccia S, Amato J, Capasso D, Di Gaetano S, Massarotti A, Piccolo M, et al. Bio-inspired dual-selective BCL-2/c-MYC G-quadruplex binders: design, synthesis, and anticancer activity of drug-like imidazo[2,1–i]purine derivatives. J Med Chem 2020;63(5):2035–50. [DOI] [PubMed] [Google Scholar]
- [91].Zhang C, Sheng J, Li G, Zhao L, Wang Y, Yang W, et al. Effects of berberine and its derivatives on cancer: a systems pharmacology review. Front Pharmacol 2019;10:1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tillhon M, Guamán Ortiz LM, Lombardi P, Scovassi AI. Berberine: new perspectives for old remedies. Biochem Pharmacol 2012;84(10):1260–7. [DOI] [PubMed] [Google Scholar]
- [93].Wang KB, Dickerhoff J, Yang D. Solution structure of ternary complex of berberine bound to a dGMP-fill-in vacancy G-quadruplex formed in the PDGFR-β promoter. J Am Chem Soc 2021;143(40):16549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res 2013;41(1):632–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhou CQ, Yang JW, Dong C, Wang YM, Sun B, Chen JX, et al. Highly selective, sensitive and fluorescent sensing of dimeric G-quadruplexes by a dimeric berberine. Org Biomol Chem 2016;14(1):191–7. [DOI] [PubMed] [Google Scholar]
- [96].Ferraroni M, Bazzicalupi C, Papi F, Fiorillo G, Guamán-Ortiz LM, Nocentini A, et al. Solution and solid-state analysis of binding of 13-substituted berberine analogues to human telomeric G-quadruplexes. Chem Asian J 2016;11(7):1107–15. [DOI] [PubMed] [Google Scholar]
- [97].Dickerhoff J, Brundridge N, McLuckey SA, Yang D. Berberine molecular recognition of the parallel MYC G-quadruplex in solution. J Med Chem 2021;64(21):16205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19(8):533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Li XM, Zheng K, Zhang J, Liu H, He Y, Yuan B, et al. Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc Natl Acad Sci USA 2015;112(47):14581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Heddi B, Martín-Pintado N, Serimbetov Z, Kari TM, Phan AT. G-quadruplexes with (4n–1) guanines in the G-tetrad core: formation of a G-triad water complex and implication for small-molecule binding. Nucleic Acids Res 2016;44(2):910–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang KB, Dickerhoff J, Wu G, Yang D. PDGFR-β promoter forms a vacancy G-quadruplex that can be filled in by dGMP: solution structure and molecular recognition of guanine metabolites and drugs. J Am Chem Soc 2020;142(11):5204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang KB, Liu Y, Li Y, Dickerhoff J, Li J, Yang MH, et al. Oxidative damage induces a vacancy G-quadruplex that binds guanine metabolites: solution structure of a cGMP fill-in vacancy G-quadruplex in the oxidized BLM gene promoter. J Am Chem Soc 2022;144(14):6361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Winnerdy FR, Das P, Heddi B, Phan AT. Solution structures of a G-quadruplex bound to linear- and cyclic-dinucleotides. J Am Chem Soc 2019;141(45):18038–47. [DOI] [PubMed] [Google Scholar]
- [104].He YD, Zheng K, Wen C, Li X, Gong J, Hao Y, et al. Selective targeting of guanine-vacancy-bearing G-quadruplexes by G-quartet complementation and stabilization with a guanine-peptide conjugate. J Am Chem Soc 2020;142(26):11394–403. [DOI] [PubMed] [Google Scholar]
- [105].Chen JN, He Y, Liang H, Cai T, Chen Q, Zheng K. Regulation of PDGFR-β gene expression by targeting the G-vacancy bearing G-quadruplex in promoter. Nucleic Acids Res 2021;49(22):12634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lin C, Wu G, Wang K, Onel B, Sakai S, Shao Y, et al. Molecular recognition of the hybrid-2 human telomeric G-quadruplex by epiberberine: insights into conversion of telomeric G-quadruplex structures. Angew Chem Int Ed Engl 2018;57(34):10888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Martino L, Virno A, Pagano B, Virgilio A, Di Micco S, Galeone A, et al. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J Am Chem Soc 2007;129(51):16048–56. [DOI] [PubMed] [Google Scholar]
- [108].Li W, Li Y, Liu Z, Lin B, Yi H, Xu F, et al. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res 2016;44(15):7373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wang F, Wang C, Liu Y, Lan W, Han H, Wang R, et al. Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR. Chem Commun 2020;56(14):2099–102. [DOI] [PubMed] [Google Scholar]
- [110].Tawani A, Amanullah A, Mishra A, Kumar A. Evidences for Piperine inhibiting cancer by targeting human G-quadruplex DNA sequences. Sci Rep 2016;6(1):39239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Han T, Cao X, Xu J, Pei H, Zhang H, Tang Y. Separation of the potential G-quadruplex ligands from the butanol extract of Zanthoxylum ailanthoides Sieb. & Zucc. by countercurrent chromatography and preparative high performance liquid chromatography. J Chromatogr A 2017;1507:104–14. [DOI] [PubMed] [Google Scholar]
- [112].Qian C, Fu H, Kovalchik KA, Li H, Chen DDY. Specific binding constant and stoichiometry determination in free solution by mass spectrometry and capillary electrophoresis frontal analysis. Anal Chem 2017;89(17):9483–90. [DOI] [PubMed] [Google Scholar]
- [113].Yang P, Wang X, Gu Z, Li H, Chen DDY, Yang X. Evaluation of the binding of natural products with thrombin binding aptamer G-quadruplex using electrospray ionization mass spectrometry and spectroscopic methods. Talanta 2019;200:424–31. [DOI] [PubMed] [Google Scholar]
- [114].Cui X, Lin S, Yuan G. Spectroscopic probing of recognition of the G-quadruplex in c-kit promoter by small-molecule natural products. Int J Biol Macromol 2012;50(4):996–1001. [DOI] [PubMed] [Google Scholar]
- [115].Wang SK, Wu Y, Wang XQ, Kuang GT, Zhang Q, Lin SL, et al. Discovery of small molecules for repressing cap-independent translation of human vascular endothelial growth factor (hVEGF) as novel antitumor agents. J Med Chem 2017;60(13):5306–19. [DOI] [PubMed] [Google Scholar]
- [116].Che T, Chen SB, Tu JL, Wang B, Wang YQ, Zhang Y, et al. Discovery of novel schizocommunin derivatives as telomeric G-quadruplex ligands that trigger telomere dysfunction and the deoxyribonucleic acid (DNA) damage response. J Med Chem 2018;61(8):3436–53. [DOI] [PubMed] [Google Scholar]
- [117].Grigg JC, Shumayrikh N, Sen D. G-quadruplex structures formed by expanded hexanucleotide repeat RNA and DNA from the neurodegenerative disease-linked C9orf72 gene efficiently sequester and activate heme. PLoS One 2014;9(9):e106449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Yaku H, Murashima T, Miyoshi D, Sugimoto N. Specific binding of anionic porphyrin and phthalocyanine to the G-quadruplex with a variety of in vitro and in vivo applications. Molecules 2012;17(9):10586–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Georgiades SN, Abd Karim NH, Suntharalingam K, Vilar R. Interaction of metal complexes with G-quadruplex DNA. Angew Chem Int Ed 2010;49(24):4020–34. [DOI] [PubMed] [Google Scholar]
- [120].Dixon IM, Lopez F, Tejera AM, Estève JP, Blasco MA, Pratviel G, et al. A G-quadruplex ligand with 10 000-fold selectivity over duplex DNA. J Am Chem Soc 2007;129(6):1502–3. [DOI] [PubMed] [Google Scholar]
- [121].Alzeer J, Vummidi BR, Roth PJ, Luedtke NW. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew Chem Int Ed Engl 2009;48(49):9362–5. [DOI] [PubMed] [Google Scholar]
- [122].Cao Q, Li Y, Freisinger E, Qin PZ, Sigel RKO, Mao ZW. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs. Inorg Chem Front 2017;4(1):10–32. [Google Scholar]
- [123].Zheng XH, Cao Q, Ding YL, Zhong YF, Mu G, Qin PZ, et al. Platinum(II) clovers targeting G-quadruplexes and their anticancer activities. Dalton Trans 2015;44(1):50–3. [DOI] [PubMed] [Google Scholar]
- [124].Wen LN, Xie MX. Spectroscopic investigation of the interaction between G-quadruplex of KRAS promoter sequence and three isoquinoline alkaloids. Spectrochim Acta A Mol Biomol Spectrosc 2017;171:287–96. [DOI] [PubMed] [Google Scholar]
- [125].Jana J, Mondal S, Bhattacharjee P, Sengupta P, Roychowdhury T, Saha P, et al. Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-quadruplex structures at their promoter regions. Sci Rep 2017;7(1):40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Singh N, Sharma B. Toxicological effects of berberine and sanguinarine. Front Mol Biosci 2018;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Shi Y, Li L, Wang C, Huang J, Feng L, Chen X, et al. Developmental toxicity induced by chelerythrine in zebrafish embryos via activating oxidative stress and apoptosis pathways. Comp Biochem Physiol C Toxicol Pharmacol 2023;273:109719. [DOI] [PubMed] [Google Scholar]
- [128].Li Z, Tan JH, He JH, Long Y, Ou TM, Li D, et al. Disubstituted quinazoline derivatives as a new type of highly selective ligands for telomeric G-quadruplex DNA. Eur J Med Chem 2012;47:299–311. [DOI] [PubMed] [Google Scholar]
- [129].Lu YJ, Ou TM, Tan JH, Hou JQ, Shao WY, Peng D, et al. 5-N-methylated quindoline derivatives as telomeric G-quadruplex stabilizing ligands: effects of 5-N positive charge on quadruplex binding affinity and cell proliferation. J Med Chem 2008;51(20):6381–92. [DOI] [PubMed] [Google Scholar]
- [130].Han G, Chen L, Wang Q, Wu M, Liu Y, Wang Q. Design, synthesis, and antitobacco mosaic virus activity of water-soluble chiral quaternary ammonium salts of phenanthroindolizidines alkaloids. J Agric Food Chem 2018;66(4):780–8. [DOI] [PubMed] [Google Scholar]
- [131].Peng W, Sun ZY, Zhang Q, Cheng SQ, Wang SK, Wang XN, et al. Design, synthesis, and evaluation of novel p-(methylthio)styryl substituted quindoline derivatives as neuroblastoma RAS (NRAS) repressors via specific stabilizing the RNA G-quadruplex. J Med Chem 2018;61(15):6629–46. [DOI] [PubMed] [Google Scholar]
- [132].Shan C, Yan JW, Wang YQ, Che T, Huang ZL, Chen AC, et al. Design, synthesis, and evaluation of isaindigotone derivatives to downregulate c-myc transcription via disrupting the interaction of NM23-H2 with G-quadruplex. J Med Chem 2017;60(4):1292–308. [DOI] [PubMed] [Google Scholar]
- [133].Franceschin M, Cianni L, Pitorri M, Micheli E, Cacchione S, Frezza C, et al. Natural aromatic compounds as scaffolds to develop selective G-quadruplex ligands: from previously reported berberine derivatives to new palmatine analogues. Molecules 2018;23(6):1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Barrett MP, Gemmell CG, Suckling CJ. Minor groove binders as anti-infective agents. Pharmacol Ther 2013;139(1):12–23. [DOI] [PubMed] [Google Scholar]
- [135].Baraldi PG, Bovero A, Fruttarolo F, Preti D, Tabrizi MA, Pavani MG, et al. DNA minor groove binders as potential antitumor and antimicrobial agents. Med Res Rev 2004;24(4):475–528. [DOI] [PubMed] [Google Scholar]
- [136].Cai X, Gray PJ Jr, Von Hoff DD. DNA minor groove binders: back in the groove. Cancer Treat Rev 2009;35(5):437–50. [DOI] [PubMed] [Google Scholar]
- [137].Alniss HY. Thermodynamics of DNA minor groove binders. J Med Chem 2019;62(2):385–402. [DOI] [PubMed] [Google Scholar]
- [138].Ngoc Nguyen TQ, Lim KW, Phan AT. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res 2020;48(18):10567–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Jana J, Mohr S, Vianney YM, Weisz K. Structural motifs and intramolecular interactions in non-canonical G-quadruplexes. RSC Chem Biol 2021;2(2):338–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Vianney YM, Weisz K. Indoloquinoline ligands favor intercalation at quadruplex–duplex interfaces. Chemistry 2022;28(7):e202103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Nguyen TQN, Lim KW, Phan AT. A dual-specific targeting approach based on the simultaneous recognition of duplex and quadruplex motifs. Sci Rep 2017;7(1):11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Asamitsu S, Obata S, Phan AT, Hashiya K, Bando T, Sugiyama H. Simultaneous binding of hybrid molecules constructed with dual DNA-binding components to a G-quadruplex and its proximal duplex. Chemistry 2018;24(17):4428–35. [DOI] [PubMed] [Google Scholar]
- [143].Bazzi S, Novotny J, Yurenko YP, Marek R. Designing a new class of bases for nucleic acid quadruplexes and quadruplex-active ligands. Chemistry 2015;21(26):9414–25. [DOI] [PubMed] [Google Scholar]
- [144].Tassinari M, Zuffo M, Nadai M, Pirota V, Sevilla Montalvo AC, Doria F, et al. Selective targeting of mutually exclusive DNA G-quadruplexes: HIV-1 LTR as paradigmatic model. Nucleic Acids Res 2020;48(9):4627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Chen SB, Hu MH, Liu GC, Wang J, Ou TM, Gu LQ, et al. Visualization of RNA G-quadruplex structures in cells with an engineered fluorogenic hybridization probe. J Am Chem Soc 2016;138(33):10382–5. [DOI] [PubMed] [Google Scholar]
- [146].Liu LY, Ma TZ, Zeng YL, Liu W, Zhang H, Mao ZW. Organic-platinum hybrids for covalent binding of G-quadruplexes: structural basis and application to cancer immunotherapy. Angew Chem Int Ed Engl 2023;62(36):e202305645. [DOI] [PubMed] [Google Scholar]
- [147].Betzer JF, Nuter F, Chtchigrovsky M, Hamon F, Kellermann G, Ali S, et al. Linking of antitumor trans NHC-Pt(II) complexes to G-quadruplex DNA ligand for telomeric targeting. Bioconjug Chem 2016;27(6):1456–70. [DOI] [PubMed] [Google Scholar]
- [148].Sanchez-Martin V, David AS, Matilde OG, Ana SL, Angel LR, Virginia PC, et al. Targeting ribosomal G-quadruplexes with naphthalene-diimides as RNA polymerase I inhibitors for colorectal cancer treatment. Cell Chem Biol 2021;28(12):1807. [DOI] [PubMed] [Google Scholar]
- [149].Muller S, Sanders DA, Di Antonio M, Matsis S, Riou JF, Rodriguez R, et al. Pyridostatin analogues promote telomere dysfunction and long-term growth inhibition in human cancer cells. Org Biomol Chem 2012;10(32):6537–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Shinohara K, Sannohe Y, Kaieda S, Tanaka K, Osuga H, Tahara H, et al. A chiral wedge molecule inhibits telomerase activity. J Am Chem Soc 2010;132(11):3778–82. [DOI] [PubMed] [Google Scholar]
- [151].Zhao C, Song H, Scott P, Zhao A, Tateishi-Karimata H, Sugimoto N, et al. Mirror-image dependence: targeting enantiomeric G-quadruplex DNA using triplex metallohelices. Angew Chem Int Ed Engl 2018;57(48):15723–7. [DOI] [PubMed] [Google Scholar]
- [152].Xiong K, Ouyang C, Liu J, Karges J, Lin X, Chen X, et al. Chiral Ru(II)–Pt(II) complexes inducing telomere dysfunction against cisplatin-resistant cancer cells. Angew Chem Int Ed Engl 2022;61(33):e202204866. [DOI] [PubMed] [Google Scholar]
- [153].Yu H, Wang X, Fu M, Ren J, Qu X. Chiral metallo-supramolecular complexes selectively recognize human telomeric G-quadruplex DNA. Nucleic Acids Res 2008;36(17):5695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Zhao A, Howson SE, Zhao C, Ren J, Scott P, Wang C, et al. Chiral metallohelices enantioselectively target hybrid human telomeric G-quadruplex DNA. Nucleic Acids Res 2017;45(9):5026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wang J, Chen Y, Ren J, Zhao C, Qu X. G-quadruplex binding enantiomers show chiral selective interactions with human telomere. Nucleic Acids Res 2014;42(6):3792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bhambhani S, Kondhare KR, Giri AP. Diversity in chemical structures and biological properties of plant alkaloids. Molecules 2021;26(11):3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Daley SK, Cordell GA. Biologically significant and recently isolated alkaloids from endophytic fungi. J Nat Prod 2021;84(3):871–97. [DOI] [PubMed] [Google Scholar]
- [158].Plazas E, Avila MM, Munoz DR, Cuca SL. Natural isoquinoline alkaloids: pharmacological features and multi-target potential for complex diseases. Pharmacol Res 2022;177:106126. [DOI] [PubMed] [Google Scholar]
- [159].Zhang J, Morris-Natschke SL, Ma D, Shang XF, Yang CJ, Liu YQ, et al. Biologically active indolizidine alkaloids. Med Res Rev 2021;41(2):928–60. [DOI] [PubMed] [Google Scholar]
- [160].Chen J, Lv S, Liu J, Yu Y, Wang H, Zhang H. An overview of bioactive 1,3-oxazole-containing alkaloids from marine organisms. Pharmaceuticals 2021;14(12):1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Almeida MC, Resende D, da Costa PM, Pinto MMM, Sousa E. Tryptophan derived natural marine alkaloids and synthetic derivatives as promising antimicrobial agents. Eur J Med Chem 2021;209:112945. [DOI] [PubMed] [Google Scholar]
- [162].Thawabteh A, Juma S, Bader M, Karaman D, Scrano L, Bufo S, et al. The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins 2019;11(11):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ma YM, Liang XA, Kong Y, Jia B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J Agric Food Chem 2016;64(35):6659–71. [DOI] [PubMed] [Google Scholar]
- [164].Hu Y, Chen S, Yang F, Dong S. Marine indole alkaloids-isolation, structure and bioactivities. Mar Drugs 2021;19(12):658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Netz N, Opatz T. Marine indole alkaloids. Mar Drugs 2015;13(8):4814–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Xu K, Yuan XL, Li C, Li AX. Recent discovery of heterocyclic alkaloids from marine-derived aspergillus species. Mar Drugs 2020;18(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Tajuddeen N, Bringmann G. N,C-coupled naphthylisoquinoline alkaloids: a versatile new class of axially chiral natural products. Nat Prod Rep 2021;38(12):2154–86. [DOI] [PubMed] [Google Scholar]
- [168].Ohnmacht SA, Neidle S. Small-molecule quadruplex-targeted drug discovery. Bioorg Med Chem Lett 2014;24(12):2602–12. [DOI] [PubMed] [Google Scholar]
- [169].Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun 2017;8(1):14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Marchetti C, Zyner KG, Ohnmacht SA, Robson M, Haider SM, Morton JP, et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J Med Chem 2018;61(6):2500–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Martinez-Jimenez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20(10):555–72. [DOI] [PubMed] [Google Scholar]
- [172].Cancer WIB. Addiction to oncogenes—the Achilles heal of cancer. Science 2002;297(5578):63–4. [DOI] [PubMed] [Google Scholar]
- [173].Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 2006;3(8):448–57. [DOI] [PubMed] [Google Scholar]
- [174].Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol 2020;21(8):459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lemmens B, van Schendel R, Tijsterman M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun 2015;6(1):8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Wulfridge P, Yan Q, Rell N, Doherty J, Jacobson S, Offley S, et al. G-quadruplexes associated with R-loops promote CTCF binding. Mol Cell 2023;83(17):3064–3079.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Hou Y, Li F, Zhang R, Li S, Liu H, Qin ZS, et al. Integrative characterization of G-quadruplexes in the three-dimensional chromatin structure. Epigenetics 2019;14(9):894–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Williams JD, Houserova D, Johnson BR, Dyniewski B, Berroyer A, French H, et al. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop ‘G4 Kissing’ interaction. Nucleic Acids Res 2020;48(11):5907–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Hegyi H Enhancer–promoter interaction facilitated by transiently forming G-quadruplexes. Sci Rep 2015;5(1):9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Liano D, Chowdhury S, Di Antonio M. Cockayne syndrome B protein selectively resolves and interact with intermolecular DNA G-quadruplex structures. J Am Chem Soc 2021;143(49):20988–1002. [DOI] [PubMed] [Google Scholar]
- [181].Lee CY, McNerney C, Ma K, Zhao W, Wang A, Myong S. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat Commun 2020;11(1):3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Wu W, Rokutanda N, Takeuchi J, Lai Y, Maruyama R, Togashi Y, et al. HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-quadruplex DNA. Cancer Res 2018;78(22):6371–85. [DOI] [PubMed] [Google Scholar]
- [183].Law MJ, Lower KM, Voon HPJ, Hughes JR, Garrick D, Viprakasit V, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010;143(3):367–78. [DOI] [PubMed] [Google Scholar]
- [184].Valton AL, Prioleau MN. G-quadruplexes in DNA replication: a problem or a necessity? Trends Genet 2016;32(11):697–706. [DOI] [PubMed] [Google Scholar]


