Abstract
Hereditary nonpolyposis colorectal cancer (HNPCC) describes the condition of a disparate group of families that have in common a predisposition to colorectal cancer in the absence of a premalignant phenotype. The genetic basis of this disease has been linked to mutations in genes associated with DNA mismatch repair. A large proportion of families harbor changes in one of two genes, hMSH2 and hMLH1. Approximately 35% of families in which the diagnosis is based on the Amsterdam criteria do not appear to harbor mutations in DNA-mismatch-repair genes. In this report we present data from a large series of families with HNPCC and indicate that there are subtle differences between families that harbor germline changes in hMSH2 and families that harbor hMLH1 mutations. Furthermore, there are differences between the mutation-positive group (hMSH2 and hMLH1 combined) of families and the mutation-negative group of families. The major findings identified in this study focus primarily on the extracolonic disease profile observed between the mutation-positive families and the mutation-negative families. Breast cancer was not significantly overrepresented in the hMSH2 mutation-positive group but was overrepresented in the hMLH1 mutation-positive group and in the mutation-negative group. Prostate cancer was not overrepresented in the mutation-positive groups but was overrepresented in the mutation-negative group. In age at diagnosis of colorectal cancer, there was no difference between the hMSH2 mutation–positive group and the hMLH1 mutation–positive group, but there was a significant difference between these two groups and the mutation-negative group.
Introduction
Hereditary nonpolyposis colorectal cancer (HNPCC [MIM 120435 and MIM 120436]) is an autosomal dominantly inherited disorder that is characterized primarily by the development of early-onset colorectal cancer (CRC) and that is also associated with the development of a variety of epithelial tumors that include endometrial cancer, stomach cancer, and ovarian cancer (Watson and Lynch 1993). The genetic basis of HNPCC has been linked to errors in DNA mismatch repair (Fishel et al. 1993; Leach et al. 1993; Bronner et al. 1994; Nicolaides et al. 1994), which leaves a characteristic tumor signature, of DNA microsatellite instability, that can be used as a surrogate marker for this syndrome (Ionov et al. 1993; Thibodeau et al. 1993). At least five genes have been associated with DNA mismatch repair and HNPCC; they are hMSH2, hMLH1, hMSH6, hPMS1, and hPMS2 (for review, see Papadopoulos and Lindblom 1997). Together, hMSH2 and hMLH1 account for 50%–60% of all mutations in families in which diagnosis is based on the Amsterdam criteria—three relatives with CRC, one of whom must be a first-degree relative of the other two; CRC present in at least two generations; CRC diagnosed in one relative at age <50 years; and familial adenomatous polyposis (FAP) not present. Of families with HNPCC, 2%–20% of those which are defined on the basis of the Bethesda criteria (excluding the Amsterdam-criteria families) show association with mutations in these two genes (Buerstedde et al. 1995; Wijnen et al. 1998; Heinimann et al. 1999; Syngal et al. 1999). The contribution of hPMS1 (now considered to be a pseudogene), hPMS2, and hMSH6 remains undefined; however, on the basis of current evidence, hPMS2 and hMSH6 appear to account for the condition in ∼5% of all families with HNPCC (Liu et al. 1996; Peltomaki and Vasen 1997). The remaining 30%–40% of Amsterdam-criteria families that do not evidently harbor germline mutations in the genes described either contain cryptic alterations within their coding regions, harbor changes in promoter/enhancer regions, or have mutations in other genes, which await identification (Papadopoulos and Lindblom 1997).
The identification of families with HNPCC remains problematic, despite knowledge about the genetic basis of the disease. Several criteria have been proposed, which are aimed at identification of families with a high probability of harboring germline mutations in DNA-mismatch-repair genes. To date, the accumulated evidence indicates that by far the most accurate method of family identification is use of the Amsterdam criteria. Deviation from these criteria appears to result in a poor mutation-detection rate, even when other tumors that have been clearly defined within the spectrum of HNPCC substitute for CRC (Wijnen et al. 1998; Heinimann et al. 1999).
Unlike FAP, in which genotype-phenotype correlations have been described, no such obvious relationship appears to be apparent in HNPCC. More-subtle differences have been described, indicating that there may be histological differences between hMSH2 mutation–associated tumors and hMLH1 mutation–associated tumors in HNPCC (Shashidharan et al. 1999)—and, indeed, that there may be clinical differences as well (Vasen and Wijnen 1999).
In this report we further characterize the relationship between HNPCC and the likelihood of identification of a germline mutation in either hMSH2 or hMLH1, and we compare the features displayed in families that are either mutation positive or mutation negative. Furthermore, a comparison between hMSH2 mutation–positive families and hMLH1 mutation–positive families is made that suggests that there may be some subtle differences between these two HNPCC groups. Finally, in comparing differences between the mutation-positive groups and the mutation-negative group, we were able to establish that, in disease phenotype, the mutation-negative group resembles the hMLH1 mutation–positive group more closely than it resembles the hMSH2 mutation–positive group. This evidence suggests that proteins, which interact with and affect hMLH1, could be good candidates for genetic analysis.
Patients and Methods
Index patients from 95 families enrolled in this study signed an informed-consent document (which, prior to use, was assessed by the appropriate institutional review body) authorizing genetic testing of genes associated with HNPCC. Cascade testing was performed on family members after they too had given informed consent for testing when a germline mutation had been identified in the family. Each pedigree was classified as either Amsterdam-criteria positive or Bethesda-criteria positive. Genetic analysis was performed on a fresh blood specimen from the youngest living affected proband in each family. Disease verification in affected individuals was based on either examination of pathology reports or death certificates.
hMSH2 and hMLH1 Molecular Analysis
DNA was isolated from 20 ml of peripheral blood lymphocytes, by the salting-out method first described by Miller et al. (1988). All 35 exons constituting hMSH2 and hMLH1 were analyzed by denaturing gradient gel electrophoresis, as described by Wijnen et al. (1995; 1996). In brief, each exon was amplified by PCR using specific primers, which have incorporated into either one of their 5′ ends a 40-bp GC clamp. The resulting PCR product was applied to a denaturing gradient gel and was subjected to electrophoresis for ⩾15 h. Then the gels were stained with ethidium bromide (1 mg/ml) and were photographed on a UV-Trans-illuminator. All polymorphisms were subjected to DNA sequencing by a semiautomated sequencing unit (PE Biosystems model 310), to determine the precise genetic change. Sequence alterations were analyzed for the presence of pathogenic mutations (nonsense, insertion, or deletion mutations). Sequence changes with a frequency of >5% were considered to be normal variants and were described as such. Polymorphisms that occurred at a frequency of <5% were considered to represent changes that were most likely to be associated with an increased predisposition to develop early-onset HNPCC.
Statistical Analysis
Logistic regression with a correction for clustering was used to determine whether there were differences between the groups (Royall 1986). This allows the detection of differences, with regard to the occurrence of cancer, between mutation groups. In addition to the individual records on file, the records of each family’s disease-free individuals were included in the analysis, thus reflecting the total number of individuals in each family. A similar analysis was performed for each cancer type. If there were no cases of a certain type of cancer, Fisher’s exact test was performed to obtain some measure of the degree of association. Data analyses were performed by means of the STATA statistical package (version 6). The typical STATA command for a logistic regression was xi: logit ycrc i.mutation [fweight=n], cluster (id), where ycrc is a binary variable indicating whether an individual has had a history of the cancer type (in this case, CRC). All other statistical analysis was performed by means of Student’s t-test.
Results
Mutation-Positive Families
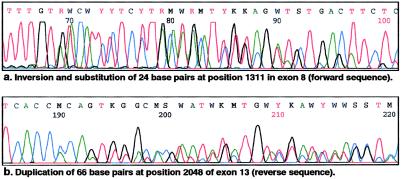
Of the 95 families, 12 harbored a genetic change in the hMSH2 gene, 22 harbored a change in the hMLH1 gene, and 61 were found not to harbor any change in either gene. The precise genetic changes identified in the hMLH1 gene and the hMSH2 gene are shown in table 1. No particular types of genetic change were overrepresented in our mutation/polymorphism data set. The ratio of observed hMSH2 genetic changes to hMLH1 genetic changes was almost 1:2. A spectrum of changes was identified in the hMLH1 gene; these included four nonsense mutations and three substitutions that resulted in stop codons, seven splice-site changes that resulted in exon skipping, six missense mutations, one loss of an ATG start site, and one 2-bp substitution that led to an Ala→Lys change. Similar changes were identified in the hMSH2 gene, which included five nonsense mutations and one substitution, all of which resulted in stop codons, one missense mutation; three 1-bp substitutions that did not result in any amino acid change; and two complex mutations. The two complex changes identified in hMSH2 were (a) an inversion of bases 1338–1361, substituting for bases 1311–1334 inclusive in exon 8, and (b) a large duplication of 66 bp in exon 13, at position 2048, both of which are predicted to result in a segment of nonfunctional peptide within the hMSH2 protein. The sequence of these two changes is shown in figure 1. In neither family were there any special features that could be related to the uniqueness of either mutation.
Table 1.
Genetic Changes Identified in the hMLH1 and hMSH2 Genes
| Gene and Family | Exon/Intron | Position(s) | Mutation Type | Nucleotide Change | Consequence of Mutation | Reference |
| hMLH1: | ||||||
| 1 | 1 | 116 | 1-bp substitution | G→A | Splice site | Farrington et al. (1998) |
| 2 | 1 | 1 | 1-bp substitution | A→G | Loss of translation start site | Wehner et al. (1997) |
| 3 | 1 | 112 | 1-bp substitution | A→G | Arg→Ser | Present study |
| 4 | 3 | +2, +1, 208, 209 | 4-bp deletion | 4-bp deletion | Loss of intron/exon junction | Present study |
| 5 | 4 | 350 | 1-bp substitution | C→T | Thr→Met | Maliaka et al. (1996) |
| 6 | 4 | 381 | 1-bp substitution | G→A | Loss of intron/exon junction | Present study |
| 7 | 7 | 554 | 1-bp substitution | T→G | Val→Gly | Kohonen-Corish et al. (1996) |
| 8 | 8 | 673 | 1-bp deletion | G deletion | Frameshift/stop | Present study |
| 9 | 8 | 655 | 1-bp substitution | A→G | Ile→Val (polymorphism) | Hutter et al. (1998) |
| 10 | 9 | 790 | 1-bp substitution | A→G | Splice site | Liu et al. (1995) |
| 11 | 12 | 1164 | 4-bp insertion | Need info | Stop codon | Present study |
| 12 | 13 | +13 | 1-bp substitution | T→A | Cryptic splice site | Present study |
| 13 | 13 | 1554 | 1-bp insertion | T insertion | Stop codon | Present study |
| 14 | 13 | 1535 | 1-bp substitution | G→T | Stop codon | Present study |
| 15 | 13 | 1460 | 4-bp deletion | TGAT deletion | Stop codon | Present study |
| 16 | 16 | 1731 | 1-bp substitution | G→A | Splice site | Present study |
| 17 | 16 | 1852 | 2-bp substitution | AA→GC | Ala→Lys | Present study |
| 18 | 17 | 1959 | 1-bp substitution | G→T | Splice site | Present study |
| 19 | 17 | 1989 | 1-bp substitution | G→A | Stop | Present study |
| 20 | 17 | 1975 | 2-bp deletion | GA deletion | Stop codon | Present study |
| 21 | 19 | 2253 | 1-bp substitution | A→G | Lys→Arg | Present study |
| 22 | 19 | 2147 | 1-bp substitution | G→A | Val→Met | Wehner et al. (1997) |
| hMSH2: | ||||||
| 23 | 6 | 1009 | 1-bp substitution | C→T | Stop codon | Present study |
| 24 | 6 | 965 | 1-bp substitution | G→A | Gly→Asp | Maliaka et al. (1996) |
| 25 | 6 | 984 | 1-bp substitution | C→T | Ala→Ala (polymorphism) | |
| 26 | 8 | 1381 | Inversion and substitution | Complexa | Nonsense segment | Present study |
| 27 | 9 | 1408 | 1-bp deletion | G deletion | Stop codon | Present study |
| 28 | 9 | 1510 | 1-bp substitution | G→C | Splice site change | Present study |
| 29 | 10 | −12 from exon 10 | 1-bp substitution | A→G | Cryptic splice site | Present study |
| 30 | 11 | 1669 | 1-bp substitution | T→C | Leu→Leu (polymorphism) | Wijnen et al. (1995) |
| 31 | 12 | 1968 | 1-bp substitution | G→C | Stop codon | Kohonen-Corish et al. (1996) |
| 32 | 13 | 2157 | 1-bp substitution | A→G | Gln→Gln (polymorphism) | |
| 33 | 13 | 2051 | 66-bp duplication | Complexb | Reiteration | Present study |
| 34 | 15 | 2502 | 7-bp deletion | Loss of TAATTTC | Stop codon | Present study |
Figure 1.
Complex DNA sequence changes identified in hMSH2
The polymorphisms identified in this study were checked against The International Collaborative Group on Hereditary NonPolyposis Colorectal Cancer mutation database, to determine whether they could be assigned causative status. Of the seven missense changes in hMLH1 (see table 1), four had been identified in previous reports, with three having been reported as being causative (Kohonen-Corish et al. 1996; Maliaka et al. 1996; Wehner et al. 1997) and one having been reported as being polymorphic (Hutter et al. 1998). The three remaining changes in hMLH1 are of unknown status. In a previous report (Maliaka et al. 1996), three of the missense mutations identified in hMSH2 (table 1) had been shown to be polymorphisms of no apparent consequence, and one had been described as pathogenic. Since tumor tissue was not available from all patients harboring missense mutations, microsatellite-instability analysis for assignment of causation could not be performed. Nevertheless, none of the new missense mutations were detected in 50 control subjects, indicating that these changes may be associated with a change of protein function.
One family was identified as having Muir-Torre syndrome, since (a) one of its members presented with a sebaceous adenoma and (b) there was a strong family history of CRC, which fulfilled the Amsterdam criteria. The proband harbored a silent polymorphism in exon 11 of hMSH2 but did not appear to harbor any other change in either hMSH2 or hMLH1.
When mutation status based on the Amsterdam criteria was compared with that based on the Bethesda criteria, differences between the two groups were observed. As expected, 60% of Amsterdam-criteria families were mutation positive, compared with 20% of Bethesda-criteria families (see table 2). There appeared to be no relationship, between the sites of mutations, that could be used to establish a genotype-phenotype correlation, for either hMLH1 or hMSH2. Within each of the two groups, a comparison between the age at onset of CRC, the spectrum of extracolonic disease, and how this related to the mutation-negative group was made.
Table 2.
Mutation Status of Bethesda- and Amsterdam-Criteria Families
|
No. of Families |
||||
| Mutation Positive |
||||
| Criterion | Mutation Negative | hMSH2 | hMLH1 | Proportion of Families That Are Positive |
| Bethesda | 50 | 6 | 6 | 19.4% |
| Amsterdam | 13 | 9 | 11 | 60.6% |
| Overall | 63 | 15 | 17 | 33.7% |
Cancer Occurrence
The overall percentage of tumors observed—in the total population, in the mutation-negative group, and in the hMSH2 mutation–positive and hMLH1 mutation–positive groups—is shown in table 3. In the total population, the most frequently observed tumor was CRC, followed by breast cancer, endometrial cancer, and stomach cancer. Other cancers common to HNPCC were also overrepresented. The relative standardized incidence rates (SIRs), including 95% confidence intervals (95% CIs), for all cancers identified in each group are shown in table 4. When the study population was subdivided into hMSH2 mutation–positive, hMLH1 mutation–positive, and mutation-negative groups, similarities and differences between the three groups could be identified.
Table 3.
Relative Percentage of Tumor Types in the Study Population
|
No. (Percentage) in Study Population |
||||
| Mutation Positive |
||||
| Cancer Type | Total | Mutation Negative | hMSH2 | hMLH1 |
| CRC | 316 (9.86) | 213 (9.80) | 73 (8.92) | 66 (12.15) |
| Endometrial/ovarian | 50 (1.37) | 36 (1.65) | 7 (.79) | 7 (1.29) |
| Stomach | 41 (1.12) | 26 (1.1) | 9 (1.02) | 6 (1.19) |
| Brain/CNS tumors | 14 (.38) | 11 (.5) | 3 (.34) | … |
| Breast | 55 (1.5) | 44 (2.02) | 2 (.22) | 9 (1.66) |
| Renal/renal-tract | 18 (.50) | 9 (.410 | 7 (.8) | 2 (.37) |
| Lymphoproliferativea | 18 (.50) | 13 (.60) | 2 (.23) | 3 (.55) |
| Prostate | 11 (.30) | 10 (.46) | 1 (.11) | … |
Includes all lymphoproliferative cancers except Hodgkin leukemia.
Table 4.
SIRs of Cancers Identified in All Families, as a Function of Mutational Status
|
Mutation-Positive Patients |
||||||||
|
All Patients |
hMSH2 |
hMLH1 |
Mutation-Negative Patients |
|||||
| Cancer Type | No. | SIR (95% CI) | No. | SIR (95% CI) | No. | SIR (95% CI) | No. | SIR (95% CI) |
| CRC | 361 | 159.54 (139.2–182.9) | 73 | 134.24 (99.1–181.8) | 66 | 196.76 (143.0–270.7) | 213 | 158.61 (132.8–189.4) |
| Endometrial/ovarian | 50 | 53.35 (37.0–77.0) | 7 | 31.08 (11.7–82.9) | 7 | 50.39 (18.9–134.3) | 36 | 64.72 (42.0–99.7) |
| Stomach | 41 | 102.2 (68.2–153.28) | 9 | 93.35 (39.3–221.6) | 6 | 100.89 (35.0–290.8) | 26 | 109.20 (65.7–181.6) |
| Brain/CNS tumors | 14 | 56.02 (28.0–112.0) | 3 | 49.95 (11.2–223.2) | … | … | 11 | 74.16 (33.9–162.0) |
| Breast | 55 | 13.38 (9.4–19.0) | 2 | 2.02 (.3–12.7)a | 9 | 14.77 (6.2–35.0) | 44 | 18.03 (12.2–26.7) |
| Renal/renal-tract | 18 | 42.17 (22.9–77.7) | 7 | 68.24 (25.7–181.8) | 2 | 31.6 (5.1–197.6) | 9 | 35.53 (15.0–84.3) |
| Lymphoproliferativeb | 18 | 13.93 (7.6–25.7) | 2 | 6.44 (1.0–40.3) | 3 | 15.66 (3.5–290.1) | 13 | 16.95 (8.3–34.8) |
| Prostate | 11 | 2.69 (1.2–5.8) | 1 | 1.02 (.1–13.6)a | … | … | 10 | 4.12 (1.8–9.4) |
Not significantly different from the expected value.
Includes all lymphoproliferative cancers except Hodgkin leukemia.
CRC
Of the 95 families, 32 fulfilled the Amsterdam criteria and 63 fulfilled the Bethesda criteria. The clinical features of each of the probands are shown in table 5. The age at diagnosis of CRC in the three groups of patients is shown in figure 2 (mutations of uncertain significance were not included). The age distributions of the hMSH2 mutation–positive group and the hMLH1 mutation–positive group were essentially identical, but there was an ∼5-year shift in the distribution seen in the mutation-negative group. The average age (±SD) at diagnosis in hMSH2 mutation-positive families was 45.77 (±24.35) years, and that in hMLH1 mutation-positive families was 47.16 (±18.65) years. There was no statistical difference between the two groups (P=.747) when we adjusted for familial clustering. The average age at diagnosis in the mutation-negative group was 52.68 (±17.65) years, which was significantly different from that in the hMSH2 mutation-positive group but was not statistically different from that in the hMLH1 mutation-positive group (P=.031 and .135, respectively). Compared with the median age of diagnosis of CRC (peak incidence being diagnosed during the 7th and 8th decades of life), all three groups had an age at onset that was significantly younger than would have been expected (Cancer in New South Wales: Incidence and Mortality 1997 database). When the hMLH1 mutation–positive group was compared with the hMSH2 mutation–positive group, there was a trend toward a lower rate of CRC in the latter group, although this trend was not statistically significant (P=.087).
Table 5.
Clinical Features of Probands Selected for Mutation Analysis
| Proband | Age atDiagnosis(years) | Tumor Type | FamilyHistory of Cancer?a | AmsterdamCriteriaPositive? |
| 1 | 27 | CRC | Yes | Yes |
| 2 | 48 | CRC | Yes | No |
| 3 | 65 | CRC | Yes | No |
| 4 | 46 | CRC | No | No |
| 5 | 45 | CRC | Yes | Yes |
| 6 | 55 | Stomach | Yes | Yes |
| 7 | 47 | CRC | Yes | No |
| 8 | 52 | CRC | Yes | Yes |
| 9 | 39 | CRC | Yes | No |
| 10 | 51 | CRC | Yes | No |
| 11 | 59 | CRC | Yes | No |
| 12 | 37 | CRC | Yes | Yes |
| 13 | 31 | CRC | No | No |
| 14 | 46 | CRC | Yes | No |
| 15 | 55 | CRC | Yes | No |
| 16 | 36 | CRC | Yes | Yes |
| 17 | 52 | CRC | Yes | No |
| 18 | 41 | CRC | Yes | Yes |
| 19 | 48 | CRC | Yes | No |
| 20 | 26 | CRC | Yes | No |
| 21 | 25 | CRC | Yes | No |
| 22 | 64 | CRC | Yes | Yes |
| 23 | 58 | Adenomas | Yes | No |
| 24 | 32 | CRC | Yes | Yes |
| 25 | 47 | CRC | Yes | Yes |
| 26 | 17 | CRC | Yes | No |
| 27 | 40 | Stomach | Yes | No |
| 28 | 44 | CRC | Yes | No |
| 29 | 41 | CRC | Yes | No |
| 30 | 46 | CRC | Yes | No |
| 31 | 19 | Adenomas | Yes | Yes |
| 32 | 52 | CRC | Yes | No |
| 33 | 43 | CRC | Yes | No |
| 34 | 37 | Endometrial cancer | Yes | Yes |
| 35 | 66 | CRC | Yes | No |
| 36 | 38 | CRC | Yes | Yes |
| 37 | 48 | CRC | Yes | Yes |
| 38 | 47 | Endometrial cancer | Yes | Yes |
| 39 | 35 | Adenomas | Yes | No |
| 40 | 66 | CRC | Yes | No |
| 41 | 74 | CRC | Yes | No |
| 42 | 49 | CRC | Yes | Yes |
| 43 | 31 | CRC | Yes | No |
| 44 | 44 | CRC | Yes | Yes |
| 45 | 27 | CRC | Yes | No |
| 46 | 51 | CRC | Yes | No |
| 47 | 58 | CRC | Yes | Yes |
| 48 | 56 | CRC | Yes | No |
| 49 | 28 | CRC | Yes | Yes |
| 50 | 45 | CRC | No | No |
| 51 | 31 | CRC | Yes | No |
| 52 | 25 | CRC | Yes | Yes |
| 53 | 53 | CRC | Yes | No |
| 54 | 69 | CRC | Yes | No |
| 55 | 62 | CRC | Yes | Yes |
| 56 | 45 | CRCb | Yes | Yes |
| 57 | 50 | CRC | Yes | Yes |
| 58 | 24 | CRC | Yes | Yes |
| 59 | 48 | CRC | Yes | Yes |
| 60 | 35 | CRC | Yes | No |
| 61 | 54 | CRC | Yes | No |
| 62 | 43 | CRC | Yes | No |
| 63 | 57 | Adenomas | Yes | No |
| 64 | 41 | CRC | Yes | Yes |
| 65 | 56 | CRC | Yes | Yes |
| 66 | 62 | CRC | Yes | No |
| 67 | 68 | Adenomas | Yes | No |
| 68 | 32 | CRC | Yes | No |
| 69 | 55 | CRC | Yes | Yes |
| 70 | 55 | Stomach | Yes | Yes |
| 71 | 29 | Stomach | Yes | No |
| 72 | 40 | Endometrial cancer | Yes | Yes |
| 73 | 45 | Adenomas | Yes | No |
| 74 | 45 | CRC | Yes | Yes |
| 75 | 57 | Adenomas | Yes | No |
| 76 | 13 | CRC | Yes | No |
| 77 | 54 | Adenomas | Yes | No |
| 78 | 40 | CRC | Yes | No |
| 79 | 33 | CRC | Yes | No |
| 80 | 36 | CRC | Yes | No |
| 81 | 61 | CRC | Yes | No |
| 82 | 65 | Adenomas | Yes | No |
| 83 | 38 | CRC | No | No |
| 84 | 32 | CRC | Yes | No |
| 85 | 30 | CRC | Yes | No |
| 86 | 52 | CRC | Yes | No |
| 87 | 63 | CRC | Yes | No |
| 88 | 44 | CRC | Yes | No |
| 89 | 54 | CRC | Yes | No |
| 90 | 78 | Ovarian cancer | Yes | No |
| 91 | 51 | CRC | Yes | No |
| 92 | 51 | CRC | Yes | Yes |
| 93 | 44 | Stomach | Yes | No |
| 94 | 50 | CRC | Yes | No |
| 95 | 30 | CRC | Yes | No |
All patients without a family history had multiple primary colon cancers; for these individuals, the age at diagnosis is that when CRC was first diagnosed.
Muir-Torre syndrome.
Figure 2.
Age at diagnosis in 292 patients with familial colorectal carcinoma, according to mutation status. One to six affected mutation-positive family members were included in this analysis, and only hMSH2 and hMLH1 causative mutations were included.
Extracolonic Cancers
The relative frequency of other malignancies associated with HNPCC is shown in figure 3. The frequencies of the following cancer types were found not to differ between the hMSH2 mutation–positive group, the hMLH1 mutation–positive group, and mutation-negative group: lymphoproliferative disease, renal/renal-tract cancers, endometrial/ovarian cancers, and stomach cancer. All these malignancies were overrepresented in all three groups, compared with the expected frequency in the general population (see table 4). Differences in SIR trends were apparent between the groups. Endometrial cancer incidence was significantly increased, compared with that in the general population, as were stomach cancer, CNS tumors (in the mutation-negative group and the hMSH2 mutation–positive group only), renal/renal-tract cancer, and lymphoproliferative disease. Breast cancer was not overrepresented in the hMSH2 mutation–positive group, but it was overrepresented in both the hMLH1 mutation–positive group and the mutation-negative group, which was a highly significant difference. The SIR of breast cancer in the mutation-negative group was similar to that observed in the hMLH1 mutation–positive group. Differences in the SIR of prostate cancer were observed between the mutation-negative group and the hMSH2 mutation–positive group (no cases were observed in the hMLH1 mutation–positive group). This difference was significant, as determined by Fisher’s exact test (P=.038), which indicated that there was a relatively high number of prostate cancer patients in the mutation-negative group, compared with both mutation-positive groups.
Figure 3.
Relative percentage of malignancies within the three groups of families with HNPCC. The tumor spectrum was determined on the basis of 559 patients with cancer, all of whom were from families with a clustering of CRC.
The age at diagnosis of breast cancer in the study population does appear to be younger than expected. The average age at diagnosis of breast cancer diagnosis was 54.27 years in the overall population in this study; in the mutation-negative group, it was 55.55 years; in the hMLH1 mutation–positive group, 51.33 years; and in the hMSH2 mutation–positive group, 54 years. This age is ∼6 years younger than the average age, 60 years, in New South Wales (Cancer in New South Wales: Incidence and Mortality 1997 database). Furthermore, we determined that this malignancy was significantly more likely to be present in the mutation-negative group and in the hMLH1 mutation–positive group than in the hMSH2 mutation–positive group (P=.006). Also, breast cancer was more likely to be identified in the hMLH1 mutation–positive group than in the hMSH2 mutation–positive group (P=.024).
When the occurrence of the cancer types was compared overall, some similarities between the groups could be identified, as shown in table 4. The spectrum and frequency of malignancies in the hMLH1 mutation–positive group more closely matched those in the mutation-negative group than did those in the hMSH2 mutation–positive group. Interestingly, in the hMSH2 mutation–positive group the incidence of kidney and renal-tract cancers was greater than that in either the hMLH1 mutation–positive group or the mutation-negative group; however, this difference did not reach significance.
Discussion
The identification of germline mutations in families with HNPCC remains difficult, despite the fact that there have been improvements in the technology of mutation detection. In this report we have analyzed a large group of patients (and their families, when available) for mutations in the DNA-mismatch-repair genes hMLH1 and hMSH2. Selection of families on the basis of the Amsterdam citeria remains the best method in which the probability of mutation detection remains high, at ∼60%. When the criteria are extended to include a number of other facets, which are included among the Bethesda criteria, the mutation-detection rate falls to ∼34%. Given that there have been several reports (Wijnen et al. 1998; Heinimann et al. 1999) indicating that relaxation of the Amsterdam criteria results in a poor rate of identification of mutations, many groups are adhering to the Amsterdam criteria when they select families for genetic-testing purposes. However, since our mutation-detection rate identified mutations in patients who fulfilled the Bethesda criteria but not the Amsterdam criteria, we would advocate that this group of patients should be screened for DNA-mismatch-repair–gene alterations. Of particular note, two separate causative mutations were identified in two Bethesda-criteria patients who presented with multiple colonic primaries over a period of many years and who did not have a family history of disease. There remain, however, in both categories, a substantial proportion of families that have an autosomal dominant disease pattern, which is best accounted for by a single, highly penetrant disease allele. Studies using denaturing gradient gel electrophoresis to detect mutations in various genes suggest detection rates of 90%–95% (Wahlberg et al. 1999). Therefore, we do not believe that, in the identification of genetic change within coding regions of the genes under investigation, methodologies used in the present study are so deficient that many changes in hMSH2 or hMLH1 have been missed. Nevertheless, some changes will not be detectable when the strategy is employed; these are likely to be changes in promoter or enhancer regions, cryptic changes within noncoding sequences, and large deletions encompassing either whole exons or the entire gene.
Two mutations in hMSH2 were of special interest: (1) the 24-bp inversion and substitution, at positions 1311–1334 inclusive in exon 8, and (2) the 66-bp duplication at positions 2048–2114 in exon 13. Neither of these mutations led to any unique phenotype—and, presumably, neither results in altered hMSH2 function. The best explanation for both of these mutations is a recombination event that resulted in the inadvertent duplication of sequence. There was no apparent disease phenotype that could be readily identified with any particular mutation in this study, confirming the notion that a breakdown in DNA mismatch repair is not associated with overt differences in disease phenotype. Disease-spectrum differences associated with hMSH2 or hMLH1 were, however, identified on examination of the types of tumors present in families that harbored either a hMLH1 mutation, a hMSH2 mutation, or no mutation.
On average, the age at diagnosis of CRC in the mutation-negative group was 5 years older than that in the mutation-positive group, but the difference was significant only in the hMSH2 mutation–positive group. In the hMLH1 mutation–positive group, there was an apparent trend toward ages at diagnosis that were younger than those in the mutation-negative group, but the difference did not reach statistical significance. Interestingly, in terms of the age at disease penetrance, there was no difference between the hMSH2 mutation–positive group and the hMLH1 mutation–positive group, which indirectly supports the concept that DNA mismatch repair is a caretaker function and not a gatekeeper function (Kinzler and Vogelstein 1997). This does not mean, however, that differences do not exist between the two groups. The protein MSH2 has been shown to be important in the recognition of DNA mispairs and is an energy-driven process, whereas MLH1 binds to MSH2:DNA complexes and helps to orchestrate endonuclease activity (Fishel 1998). In addition, hMLH1 appears to be involved in meiotic recombination, giving it a multifunctional role (Anderson et al. 1999). Given these subtle differences between the two proteins, we studied the disease profiles of the hMSH2 mutation–positive group and the hMLH1 mutation–positive group. There was a tendency for the frequency of CRC in the hMLH1 mutation–positive group to be greater than that in the hMSH2 mutation–positive group, with the relative percentages being 12.15 and 8.92, respectively. This suggests that, with regard to CRC risk, there may be some differences between the two groups. A possible explanation for this difference is the greater potential for allelic silencing in hMLH1, by hypermethylation of the wild-type allele (Cunningham et al. 1998). Nevertheless, except for breast cancer and prostate cancer, the rates of other cancers were similar in both groups, suggesting the possibility of colon-specific differences in disease expression. The frequency of breast cancer in the hMSH2 mutation–positive group was significantly less than that observed in either the hMLH1 mutation–positive group or the mutation-negative group, suggesting a real difference between the three groups. There are conflicting reports as to whether breast cancer risk is increased in HNPCC (Itoh et al. 1990; Nelson et al. 1993; Watson and Lynch 1993; Risinger et al. 1996). Breast cancer incidence overall in our study population was overrepresented within both the hMLH1 mutation–positive group and the mutation-negative group but was not overerepresented in the hMSH2 mutation–positive group. Why breast cancer should be underrepresented in the hMSH2 mutation–positive group is not known, but it may be associated with the proposed roles of hMLH1 and hMSH2 in the BRCA1-associated genome surveillance complex supercomplex involved in the recognition and repair of aberrant DNA structures (Wang et al. 2000). The finding that breast cancer does not appear to be associated with hMSH2 mutations may explain some of the discrepancies that have been reported in the literature, with respect to breast cancer risk in HNPCC.
As expected, the incidence of prostate cancer was not overrepresented in the mutation-positive groups considered together. Interestingly, a feature of the mutation-negative group did appear to be the increased incidence of prostate cancer, and this may therefore be a feature of the families with HNPCC that are mutation negative.
Finally, the overall spectrum of disease within the mutation-negative group more closely resembled that observed in the hMLH1 mutation–positive group than it resembled that in the hMSH2 mutation–positive group. This, coupled with the similar ages at diagnosis of CRC, suggests either that there are more-subtle changes occurring in the hMLH1 gene that are not being identified by mutation analysis or that other proteins may interact with hMLH1, such that their disruption results in a disease spectrum similar to that observed in hMSH2 mutation–positive families.
In conclusion, use of the Bethesda criteria for the selection of families for gene analysis results in a reduced probability of mutation detection. Nevertheless, families or patients that can be identified on the basis of these criteria should be offered genetic testing for genes associated with HNPCC, since this will result in an increased rate of identification of gene-mutation carriers. To improve the probability of mutation detection if the Bethesda criteria are adopted, we would suggest that mutation analysis be performed in conjunction with DNA microsatellite testing and, possibly, immunohistochemical staining for DNA-mismatch-repair proteins. Subdivision of the mutation-positive and mutation-negative groups makes it possible to tease out subtle differences between the various populations, such as similar ages at onset of disease in the mutation-negative and hMLH1 mutation–positive groups and disease-spectrum differences within each mutation-positive group. Better knowledge of the disease spectrum associated with mutation status will aid in the management of these families. Better classification of the mutation-negative group will aid in identification of additional genes associated with this disorder.
Acknowledgments
The authors would like to thank the Hunter Area Pathology Service and the Hunter Area Health Service for supporting this work and would like to thank all clinicians who supplied samples for this study.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- New South Wales Cancer Council, http://www.nswcc.org.au/pages/ccic/stats/index.htm
- International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer, The, http://www.nfdht.nl/database/mdbchoice.htm (for the mismatch-repair-gene–mutation database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for HNPCC [MIM 120435 and MIM 120436])
References
- Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag RJ, Godwin AR, Ward DC, Nordenskjold M, Fishel R, Kolodner R, Liskay RM (1994) Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature 368:258–261 [DOI] [PubMed] [Google Scholar]
- Buerstedde JM, Alday P, Torhorst J, Weber W, Muller H, Scott R (1995) Detection of new mutations in six out of 10 Swiss HNPCC families by genomic sequencing of the hMSH2 and hMLH1 genes. J Med Genet 32:909–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim C-Y, Roche PC, Burgart LJ, Thibodeau SN (1998) Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58:3455–3460 [PubMed] [Google Scholar]
- Farrington SM, Lin-Goerke J, Ling J, Wang Y, Burczak JD, Robbins DJ Dunlop MG (1998) Systematic analysis of hMSH2 and hMLH1 in young colon cancer patients and controls. Am J Hum Genet 63:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R (1998) Mismatch repair, molecular switches, and signal transduction. Genes Dev 12:2096–2101 [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75:1027–1038 [DOI] [PubMed] [Google Scholar]
- Heinimann K, Scott RJ, Buerstedde JM, Weber W, Siebold K, Attenhofer M, Muller Hj, Dobbie Z (1999) Influence of selection criteria on mutation detection in patients with hereditary nonpolyposis colorectal cancer. Cancer 8 [DOI] [PubMed] [Google Scholar]
- Hutter P, Couturier A, Membrez V, Joris F, Sappino AP, Chappuis PO (1998) Excess of hMLH1 germline mutations in Swiss families with hereditary non-polyposis colorectal cancer. Int J Cancer 78:680–684 [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363:558–561 [DOI] [PubMed] [Google Scholar]
- Itoh H, Houlston RS, Harocopas C, Slack J (1990) Risk of cancer deaths in first degree relatives of patients with hereditary non-polyposis cancer syndrome (Lynch type II): a study of 130 kindreds in the United Kingdom. Br J Surg 77:1367–1370 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B (1997) Cancer-susceptibility genes: gatekeepers and caretakers. Nature 386:761–763 [DOI] [PubMed] [Google Scholar]
- Kohonen-Corish M, Ross VL, Doe WF, Kool DA, Edkins E, Faragher I, Wijnen J, Meera Khan P, Macrae F, St John JB (1996) RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet 59:818–824 [PMC free article] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, et al (1993) Mutations in a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215–1225 [DOI] [PubMed] [Google Scholar]
- Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopoulos N, Peltomaki P de la Chapelle A, Hamilton SR, Kinzler KW, Vogelstein B (1995) Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet 9:48–55 [DOI] [PubMed] [Google Scholar]
- Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P, de la Chapelle A, Hamilton SR, Vogelstein B, Kinzler KW (1996) Analysis of mismatch repair genes in hereditary nonpolyposis colorectal cancer patients. Nat Med 2:169–174 [DOI] [PubMed] [Google Scholar]
- Maliaka YK, Chudina AP, Belev NF, Alday P, Bochkov NP, Buerstedde J-M (1996) CpG dinucleotides in the hMSH2 and hMLH1 genes are hotspots for HNPCC mutations. Hum Genet 97:251–255 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CL, Sellers TA, Rich SS, Potter JD, McGovern PG, Kushi LH (1993) Familial clustering of colon breast, uterine, and ovarian cancers as assessed by family history. Genet Epidemiol 10:235–244 [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei Y-F, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser GM, Adams MD, Venter JC, Dunlop MG, Hamilton SR, Petersen GM, de la Chapelle A, Vogelstein B, Kinzler K (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371:75–80 [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Lindblom A (1997) Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat 10:89–99 [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen HF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study: The International Collaborative Group on Hereditary Non Polyposis Colorectal Cancer. Gastroenterology 113:1146–1158 [DOI] [PubMed] [Google Scholar]
- Risinger JI, Barrett JC, Watson P, Lynch HT, Boyd J (1996) Molecular genetic evidence of the occurrence of breast cancer as an integral tumor in patients with the hereditary nonpolyposis colorectal carcinoma syndrome. Cancer 77:1836–1843 [DOI] [PubMed] [Google Scholar]
- Royall RM (1986) Model robust confidence intervals using maximum likelihood estimators. Int Stat Rev 54:221–226 [Google Scholar]
- Shashidharan M, Smyrk T, Lin KM, Ternent CA, Thorson AG, Blatchford GJ, Christensen MA, Lynch HT (1999) Histological comparison of hereditary nonpolyposis colorectal cancer associated with MSH2 and MLH1 and colorectal cancer from the general population. Dis Colon Rectum 42:722–726 [DOI] [PubMed] [Google Scholar]
- Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, Garber JE (1999) Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA 282:247–253 [DOI] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D (1993) Microsatellite instability in cancer of the proximal colon. Science 260:816–819 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Wijnen J (1999) Clinical implications of genetic testing of hereditary nonpolyposis colorectal cancer. Cytogenet Cell Genet 86:136–139 [DOI] [PubMed] [Google Scholar]
- Wahlberg S, Liu T, Lindblom P, Lindblom A (1999) Various mutation screening techniques in the DNA mismatch repair genes hMSH2 and hMLH1. Genet Test 3:259–264 [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14:927–939 [PMC free article] [PubMed] [Google Scholar]
- Watson P, Lynch HT (1993) Extracolonic cancer in hereditary non polyposis colorectal cancer. Cancer 71:677–685 [DOI] [PubMed] [Google Scholar]
- Wehner M, Buschhausen L, Lamberti C, Kruse R, Caspari R, Propping P Friedl W (1997) Hereditary nonpolyposis colorectal cancer (HNPCC): eight novel germline mutations in hMSH2 or hMLH1 genes. Hum Mutat 10:241–244 [DOI] [PubMed] [Google Scholar]
- Wijnen J, Meera Khan P, Vasen H, Menko FH, van der Klift H, van der Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer EJ, Lindhout D, Griffioen G, Cats A, Kleibeuker J, Varesco L, Bertario L, Bisgaard ML, Mohr J, Kolodner R, Fodde R (1996) Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer (HNPCC) cluster at the exonic region 15-16. Am J Hum Genet 58:300–307 [PMC free article] [PubMed] [Google Scholar]
- Wijnen J, Vasen H, Meera Khan P, Menko FH, van der Klift H, van Leeuwen C, van den Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer A, Lindhout D, Griffioen G, Cats A, Kleibeuker J, Varesco L, Bertario L, Bisgaard ML, Mohr J, Fodde R (1995) Seven new mutations in hMSH2, an HNPCC gene, identified by denaturing gradient gel electrophoresis. Am J Hum Genet 56:1060–1066 [PMC free article] [PubMed] [Google Scholar]
- Wijnen JT, Vasen HFA, Meera Khan P, Zwindermann AH, van der Klift H, Mulder A, Tops C, Moller P, Fodde R (1998) Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med 339:511–518 [DOI] [PubMed] [Google Scholar]