Abstract
The molecular evolution of DAX1, SRY, and SOX9, genes involved in mammalian sex determination, was examined in six primate species. DAX1 and SRY have been added to the X and Y chromosomes, respectively, during mammalian evolution, whereas SOX9 remains autosomal. We determined the genomic sequences of DAX1, SRY, and SOX9 in all six species, and calculated Ka, the number of nonsynonymous substitutions per nonsynonymous site, and compared this with the Ks, the number of synonymous substitutions per synonymous site. Phylogenetic trees were constructed by means of the DAX1, SRY, and SOX9 coding sequences, and phylogenetic analysis was performed using maximum likelihood. Overall measures of gene and protein similarity were closer for DAX1 and SOX9, but DAX1 exhibited nonsynonymous amino acid substitutions at an accelerated frequency relative to synonymous changes, similar to SRY and significantly higher than SOX9. We conclude that, at the protein level, DAX1 and SRY are under less selective pressure to remain conserved than SOX9, and, therefore, diverge more across species than does SOX9. These results are consistent with evolutionary stratification of the mammalian sex determination pathway, analogous to that for sex chromosomes.
DAX1 is a member of the nuclear hormone receptor superfamily (Zanaria et al. 1994). Mutations in this transcription factor result in developmental and functional abnormalities of the steroidogenic axis, as well as the clinical phenotypes of adrenal hypoplasia congenita (MIM 300200) and hypogonadotropic hypogonadism (Muscatelli et al. 1994; Zanaria et al. 1994; Guo et al. 1995; Habiby et al. 1996; McCabe 2000). Since DAX1 maps to the dosage-sensitive sex reversal (DSS [MIM 300018]) critical region (Bardoni et al. 1994; Zanaria et al. 1994), and transgenic overexpression of DAX1 in mice with a Y chromosome containing a weak SRY allele can result in XY sex reversal (Swain et al. 1998), DAX1 is a candidate gene for DSS. However, experimental and clinical evidence suggests that the role of DAX1 in sex determination is more complex. Targeted deletion of DAX1 exon 2 in mice fails to give the expected sex-reversed phenotype (Yu et al. 1998), and a female human who is genotypically homozygous for a DAX1 mutation also does not exhibit the expected phenotype (Merke et al. 1999). It is possible, however, that DAX1 acts as a part of a dosage-sensitive multiprotein complex, that time of expression is important, and that DAX1 functions differently in human and mouse (Swain et al. 1998; Goodfellow and Camerino 1999). DAX1 is a relatively recent addition to the X chromosome, since it is autosomal in marsupials (Pask et al. 1997) and maps to the stratum that was acquired by the eutherian mammal X chromosome ∼80–130 million years ago (Lahn and Page 1999).
SRY encodes a protein containing a “high-mobility group” domain (HMG box), which enables it to bind DNA (Sinclair et al. 1990; Giese et al. 1994). Mutations that map to the HMG box disrupt the SRY-DNA complex and result in XY sex reversal (Pontiggia et al. 1994), and XX mice carrying the Sry transgene may develop as males (Koopman et al. 1991). Among eutherian mammals, the SRY sequence is rapidly evolving outside the HMG box (Tucker and Lundrigan 1993; Whitfield et al. 1993). SRY is a relatively recent addition to the sex-determination pathway, appearing for the first time in mammals (Graves and Foster 1994).
SOX9, located on human chromosome 17, has also been implicated in the sex-determination pathway (Foster et al. 1994; Wagner et al. 1994). SOX9, like SRY, is an HMG family member, and mutations in a single allele of SOX9 are responsible for XY sex reversal and campomelic dysplasia (MIM 114290) (Foster et al. 1994; Wagner et al. 1994; Morais da Silva et al. 1996). SOX9 duplications in humans (Huang et al. 1999) can result in XX males in the absence of SRY. SOX9 is much older evolutionarily than DAX1 or SRY (Morais da Silva et al. 1996; McBride et al. 1997), may be part of an ancient “core” sex-determination mechanism that predates the mammalian radiation (Koopman 1999) and has important roles in sex determination and bone development (Bi et al. 1999). These factors may explain the highly conserved nature of SOX9.
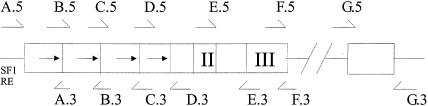
The purpose of our investigations was to examine the molecular evolution of DAX1 in relation to SRY and to SOX9. Genomic DNA was obtained from cell lines for each species ATCC numbers CRL-1857: Pan troglodytes (chimpanzee); CRL-1850: Pongo pygmaeus (orangutan); TIB-201: Hylobates species (gibbon); CRL-1773: Callithrix jacchus (marmoset); and from ClonTech Laboratories: Homo sapiens (human) and Macaca mulatta (rhesus) (fig. 1). A PCR-based approach was used to sequence the primate homologue of each gene. We used or adapted previously published primers for SRY (Whitfield et al. 1993) and SOX9 (Wagner et al. 1994). PCR primers used for DAX1 were A5 (TTGAACTACCGAGGTCATGGG) and A3 (CATGTTGTAGAGGATGCTGCC), B5 (GGGCTGCAGGAGCCGCGGGCC) and B3 (GCAGCGGTACAGGAGTGCCAC), C5 (GGGTGGGCAGAAAAGGGCTGC) and C3 (GGTGCTGCCCTGCTGCGGGTG), D5 (GGCCAGGGGGTAGAGAGGCGC) and D3 (GATCTTCTGCAGCATGCTGGG), E5 (GAGACTGTAGAAGTCTCGGAG) and E3 (GTAGGCGTACTCCTTGGTACT, F5 (AGTACCAAGGAGTACGCCTAC and F3 (CCACTGGAGTCCCTGAATGTTCTT), and G5 (ACGGGACGTGCCGGCAGTGCG) and G3 (CACTTGTGTGGCCCACATGAC). Sequences were deposited into GenBank. PCR products were sequenced using ABI 375 (PE Biosystems), and sequence data were formatted and analyzed using the GCG software package, version 10.0.
Figure 1.
Genomic sequencing strategy for DAX1. Overlapping PCR primers (sets A–G) were used to amplify primate DAX1; 5′ primers (A.5–G.5) are shown above the gene and 3′ primers (A.3–G.3) are shown below. The 5′ end of DAX1 was amplified by means of the conserved SF1-response element upstream of the start codon as a primer-anchoring site.
The ratios of nonsynonymous to synonymous (Ka/Ks) were estimated with the use of codeml, from the phylogenetic analysis using maximum likelihood (PAML) package (Goldman and Yang 1994). We imposed the commonly accepted primate topology (Pilbeam 1984) and estimated kappa (κ) and omega (ω, i.e., Ka/Ks), with alpha (α) and rho (ρ) fixed at zero. A single ω was used for the entire topology, since a model with separate ω for each branch did not give a significantly better fit. Codon frequencies were determined by means of the 12-parameter f3×4 model, but parameter estimates were largely independent of the codon-frequency model used. The tree-shape comparisons among genes were performed by likelihood-ratio tests. We compared a restricted model, having a constant ratio between respective branches of the trees for the two genes but all other evolutionary parameters unrestricted, with a more general model having separate evolutionary parameters (including the branch lengths) for the two genes. The maximum likelihood of the restricted model (18 parameters) and the maximum likelihood of the general model (26 parameters) were calculated by use of the HKY nucleotide-substitution model (Hasegawa et al. 1985), implemented in a modification of Schadt et al. (1998) (fig. 2). Our results showed that at the nucleotide level, SRY was highly divergent (92.5% average pairwise identity) and SOX9 was very conserved (98.9% average pairwise identity) (table 1). When the overall nucleotide sequence of DAX1 was examined, it was also conserved (98.1% average pairwise identity), falling closer to SOX9 than to SRY.
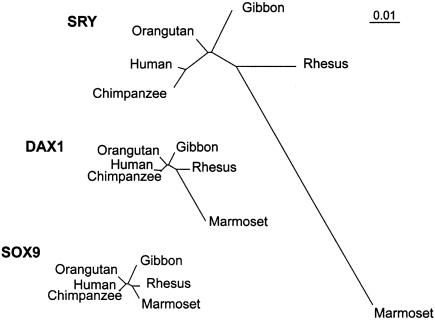
Figure 2.
Phylogenetic trees for primate SRY, DAX1, and SOX9, constructed using the HKY nucleotide-substitution model (Hasegawa et al. 1985) and the commonly accepted topology. Branch lengths correlate to relative rates of evolution.
Table 1.
Overall Measure of Sequence Divergence for Primate SRY, DAX1, and SOX9 Nucleotide Sequences
| Parameter | SRY | DAX1 | SOX9 |
| Nucleotide identitya | 92.5% | 98.1% | 98.9% |
| Amino acid identitya | 85.8% | 96.6% | 99.8% |
| Ka/Ksb | .62 | .43 | .0061 |
| Ka/Ks SEb | .11 | .08 | .0036 |
| Phylogenetic tree branch lengthc | .71 | .18 | .11 |
Average pairwise comparisons.
Ka/Ks ratios estimated by means of PAML (Goldman and Yang 1994). Imposing the topologies in figure 1 gave similar results with alternative topology for DAX1.
The sum of branch lengths in the maximum-likelihood tree, as estimated by PAML, when the topology was imposed (Goldman and Yang 1994). Similar results were obtained with the use of an alternative topology.
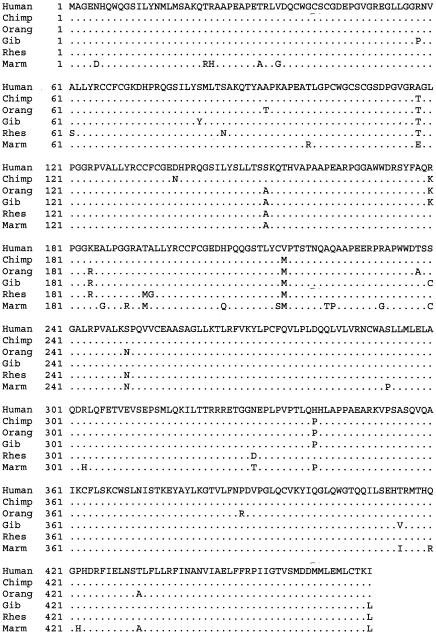
We investigated the protein products of these genes at the amino acid level. We aligned the deduced DAX1 amino acid sequences for these primate species (fig. 3). The results were consistent with the nucleotide analysis. The average pairwise identities at the amino acid level were 85.8% for SRY, 96.6% for DAX1, and 99.8% for SOX9 (table 1). Inspection of the alignment suggests a clustering of amino acid changes between residues 180 and 240 of DAX1.
Figure 3.
Amino acid alignment of primate DAX1. Amino acid identities are denoted by periods. Amino acid differences are noted.
Proteins subjected to different functional selective pressures should accumulate amino acid differences in varying proportions relative to the silent mutations. Therefore, we calculated the frequency of nonsynonymous (Ka) and synonymous (Ks) nucleotide changes for our six primate species (Li et al. 1985) (table 2). When Ka values for SRY, DAX1, and SOX9 were examined within individual species, it was clear that SRY had the highest values, with DAX1 intermediate and SOX9 lowest. Ks values were consistently lower for DAX1 than for SRY or for SOX9. Ka values, however, were consistently higher for DAX1 than for SOX9.
Table 2.
Ka and Ks Values for Nonhuman Primate to Human SRY, DAX1, and SOX9 Nucleotide Sequences[Note]
| Species and Measure | SRY | DAX1 | SOX9 |
| P. troglodytes: | |||
| Ka × 102 (95% CI) | 1.44 (.35–2.53) | .47 (.05–.88) | .00 (.00–.17)a |
| Ks × 102 (95% CI) | .82 (.00–2.46) | .30 (.00–.91) | .29 (.00–.87) |
| P. pygmaeus: | |||
| Ka × 102 (95% CI) | 2.52 (1.05–3.98) | 1.03 (.41–1.66) | .00 (.00–.17)a |
| Ks × 102 (95% CI) | 2.51 (.00–5.45) | 1.83 (.33–3.33) | 4.53 (2.15–6.91) |
| Hylobates: | |||
| Ka × 102 (95% CI) | 3.56 (1.82–5.32) | 1.60 (.82–2.37) | .08 (.00–.26) |
| Ks × 102 (95% CI) | 5.87 (1.37–10.38) | 2.46 (.71–4.21) | 5.98 (3.27–8.70) |
| M. mulatta: | |||
| Ka × 102 (95% CI) | 4.63 (2.63–6.63) | 1.12 (.47–1.77) | .00 (.00–.17)a |
| Ks × 102 (95% CI) | 13.22 (6.16–20.29) | 2.78 (.91–4.65) | 4.20 (1.91–6.49) |
| C. jacchus: | |||
| Ka × 102 (95% CI) | 12.9 (9.44–16.43) | 2.69 (1.68–3.71) | .17 (.00–.41) |
| Ks × 102 (95% CI) | 27.6 (16.52–38.59) | 6.45 (3.53–9.34) | 6.63 (3.74–9.52) |
Note.— Ka and Ks values were estimated by means of the pairwise comparison method of Li et al. (1985).
Upper bound calculated assuming a maximum of two nonsynonymous substitutions.
To examine the relative frequency of nonsynonymous nucleotide changes in comparison with the frequency of synonymous nucleotide changes, we calculated the Ka/Ks ratio for all six primate species (Goldman and Yang 1994). This ratio controls for different intrinsic rates of mutation focusing on the relative preference for amino acid changing substitutions. We found that DAX1 (Ka/Ks=.43±.08) was not significantly different from SRY (Ka/Ks=.62±.11) and was much higher than SOX9 (Ka/Ks=.0061±.0036) (table 1).
Phylogenetic trees were constructed by means of the DAX1, SRY, and SOX9 coding sequences. PAML was performed (Goldman and Yang 1994) (fig. 2). Homo sapiens (human) and P. troglodytes (chimpanzee) were always most closely related, and M. mulatta (rhesus) and C. jacchus (marmoset) were also sister taxa. The relationship of Hylobates species and P. pygmaeus could not definitely be resolved for the two genes DAX1 and SOX9.
The shapes of the phylogenetic trees for DAX1, SRY, and SOX9, however, were distinct. We compared the tree shapes under the commonly accepted primate topology (Pilbeam 1984). Tree shape under a given topology was compared by testing whether the ratio of branch length i, for gene 1 (ti), and branch length i, for gene 2 (si), is the same for all branches (ti/si=r for all branches i) (Goldman and Yang 1994). DAX1 and SRY did not differ (χ2=8.10, df 8, P=.424), whereas DAX1 differed from SOX9 (χ2=16.68, df 8, P=.0336), and SRY differed from SOX9 (χ2=30.120, df 8, P=.0002). Therefore, the SOX9 tree had a distinctive shape, differing from the tree shapes for DAX1 and SRY, which were similar to each other. These results were consistent with a discontinuity in the pressures driving the evolution of SOX9 versus SRY and DAX1.
An evaluation of rates of primate DAX1 evolution indicates that DAX1 may be under selective pressure for its amino acid sequence to evolve at a rate faster than SOX9. The Ka/Ks ratio would increase when the number of nonsynonymous changes (Ka) are well tolerated or selected, relative to the number of synonymous substitutions (Ks). Using the maximum-likelihood method for estimating Ka/Ks ratios (Goldman and Yang 1994), we found that, for the six primates that we examined, DAX1 has a Ka/Ks ratio that was statistically indistinguishable from SRY and was much higher than SOX9 (table 1). Therefore, whereas DAX1 is more highly conserved than SRY at the nucleotide level, the Ka/Ks ratio distinguished DAX1 and SOX9 as being under different evolutionary pressures. Analysis of the phylogenetic tree shapes for these three genes confirmed this distinction. Our data suggest that, although DAX1 is a relatively conserved protein overall, it is, nevertheless, experiencing amino acid substitutions at a rate higher than is SOX9. We speculate that this selective pressure may be a consequence of the relatively recent evolutionary addition of DAX1 to the sex-determination pathway (Pask et al. 1997; Lahn and Page 1999) and/or to a role in spermatogenesis (Yu et al. 1998), since male reproductive genes evolve rapidly (Wyckoff et al. 2000). The more highly conserved nature of DAX1, relative to SRY, may reflect additional constraints on DAX1 stemming from its role in adrenal development.
DAX1 and SRY may be recent additions to an evolutionary ancient “core” sex-determination program that involves SOX9. Our data on the molecular evolution of SRY, DAX1, and SOX9 support this hypothesis for a stepwise evolution of the sex-determination pathway. Just as the human sex chromosomes may be evolving by punctuated sequential events (Lahn and Page 1997, 1999), our results are consistent with evolutionary stratification of the sex-determination pathway. We speculate that SRY and DAX1 may be evolving dynamically as more recent additions to an evolutionarily stratified sex-determination pathway.
Acknowledgment
This work was supported, in part, by grants R01 HD22563, R01 HD39322, and P30 HD34610 from the National Institute of Child Health and Human Development.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for P. troglodytesSRY [accession number X86380], DAX1 [accession number AF322892], and SOX9 [accession number AF322902]; P. pygmaeusSRY [accession number X86383], DAX1 [accession number AF322893], and SOX9 [accession number AF322898]; HylobatesSRY [accession number X86384], DAX1 [accession number AF322894], and SOX9 [accession number AF322897]; M. MulattaSRY [accession number AF322901], DAX1 [accession number AF322896], and SOX9 [accession number AF322900]; and C. jacchusSRY [accession number X86386], DAX1 [accession number AF322895], and SOX9 [accession number AF322899])
- Online Mendelian Inheritance in MAN (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for adrenal hypoplasia congenita [MIM 300200], DSS [MIM 300018], SRY [MIM 480000], and campomelic dysplasia [MIM 114290])
References
- Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, Tonini G, Ferrante E, Chiumello G, McCabe ERB, Fraccaro M, Zuffardi O, Camerino G (1994) A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet 7:497–501 [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B (1999) Sox9 is required for cartilage formation. Nat Genet 22:85–89 [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525–530 [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R (1994) Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc Natl Acad Sci USA 91:3368–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736 [DOI] [PubMed] [Google Scholar]
- Goodfellow PN, Camerino G (1999) DAX-1, an `antitestis' gene. Cell Mol Life Sci 55:857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM, Foster JW (1994) Evolution of mammalian sex chromosomes and sex-determining genes. Int Rev Cytol 154:191–259 [DOI] [PubMed] [Google Scholar]
- Guo W, Mason JS, Stone CG, Jr, Morgan SA, Madu SI, Baldini A, Lindsay EA, Biesecker LG, Copeland KC, Horlick MNB, Pettigrew AL, Zanaria E, McCabe ERB (1995) Diagnosis of X-linked adrenal hypoplasia congenita by mutation analysis of the DAX1 gene. JAMA 274:324–330 [PubMed] [Google Scholar]
- Habiby RL, Boepple P, Nachtigall L, Sluss PM, Crowley WF Jr, Jameson JL (1996) Adrenal hypoplasia congenita with hypogonadotropic hypogonadism: evidence that DAX-1 mutations lead to combined hypothalamic and pituitary defects in gonadotropin production. J Clin Invest 98:1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174 [DOI] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J (1999) Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet 87:349–353 [DOI] [PubMed] [Google Scholar]
- Koopman P (1999) Sry and Sox9: mammalian testis determining genes. Cell Mol Life Sci 55:839–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351:117–121 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1997) Functional coherence of the human Y chromosome. Science 278:675–679 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Four evolutionary strata on the human X chromsome. Science 286:964–967 [DOI] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC (1985) A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol 2:150–174 [DOI] [PubMed] [Google Scholar]
- McBride D, Sang H, Clinton M (1997) Expression of Sry-related genes in the developing genital ridge/mesonephros of the chick embryo. J Reprod Fertil 109:59–63 [DOI] [PubMed] [Google Scholar]
- McCabe ERB (2000) Adrenal hypoplasias and aplasias. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York [Google Scholar]
- Merke DP, Tajima T, Baron J, Cutler GB Jr (1999) Hypogonadotropic hypogonadism in a female caused by an X-linked recessive mutation in the DAX1 gene. N Engl J Med 340:1248–1252 [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R (1996) Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet 14:62–68 [DOI] [PubMed] [Google Scholar]
- Muscatelli F, Strom TM, Walker AP, Zanaria E, Recan D, Meindl A, Bardoni B, Guiolo S, Zehetner G, Rabl W, Schwarz HP, Kaplan J-C, Camerino G, Meitinger T, Monaco AP (1994) Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372:672–676 [DOI] [PubMed] [Google Scholar]
- Pask A, Toder R, Wilcox SA, Camerino G, Graves JAM (1997) The candidate sex reversing DAX-1 gene is autosomal in marsupials: implication for the evolution of sex determination in mammals. Genomics 41:422–426 [DOI] [PubMed] [Google Scholar]
- Pilbeam D (1984) The descent of hominoids and hominids. Sci Am 250:84–96 [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME (1994) Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J 13:6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Sinsheimer JS, Lange K (1998) Computational advances in maximum likelihood methods for molecular phylogeny. Genome Res 8:222–233 [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkin JR, Griffiths BL, Smith MJ, Foster JM, Frischauf AM, Lovell-Badge R, Goodfellow PN (1990) A gene from the human sex determining region encodes a protein with homology to a conserved DNA binding motif. Nature 346:240–244 [DOI] [PubMed] [Google Scholar]
- Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R (1998) Dax1 antagonizes Sry action in mammalian sex determination. Nature 391:761–767 [DOI] [PubMed] [Google Scholar]
- Tucker PK, Lundrigan BL (1993) Rapid evolution of the sex determining locus in Old World mice and rats. Nature 364:715–717 [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79:1111–1120 [DOI] [PubMed] [Google Scholar]
- Whitfield LS, Lovell-Badge R, Goodfellow PN (1993) Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature 364:713–715 [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu C-I (2000) Rapid evolution of male reproductive genes in the descent of man. Nature 403:304–309 [DOI] [PubMed] [Google Scholar]
- Yu RN, Ito M, Saunders TL, Camper SA, Jameson JL (1998) Role of Ahch in gonadal development and gametogenesis. Nat Genet 20:353–357 [DOI] [PubMed] [Google Scholar]
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ERB, Meitinger T, Monaco AP, Sassone-Corsi P, Camerino G (1994) An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635–641 [DOI] [PubMed] [Google Scholar]