Abstract
We examined DNA polymorphisms in the nonrecombining portion of the Y-chromosome to investigate the contribution of distinct patrilineages to the present-day white Brazilian population. Twelve unique-event polymorphisms were typed in 200 unrelated males from four geographical regions of Brazil and in 93 Portuguese males. In our Brazilian sample, the vast majority of Y-chromosomes proved to be of European origin. Indeed, there were no significant differences when the haplogroup frequencies in Brazil and Portugal were compared by means of an exact test of population differentiation. Y-chromosome typing was quite sensitive in the detection of regional immigration events. Distinct footprints of Italian immigration to southern Brazil, migration of Moroccan Jews to the Amazon region, and possible relics of the 17th-century Dutch invasion of northeast Brazil could be seen in the data. In sharp contrast with our mtDNA data in white Brazilians, which showed that ⩾60% of the matrilineages were Amerindian or African, only 2.5% of the Y-chromosome lineages were from sub-Saharan Africa, and none were Amerindian. Together, these results configure a picture of strong directional mating between European males and Amerindian and African females, which agrees with the known history of the peopling of Brazil since 1500.
The Amerindians originally peopled South America in the Pleistocene. Centuries later (on April 22, 1500), the Portuguese “discovered” Brazil, by then inhabited by ∼2.4 million Amerindians (IBGE 2000). Colonization of the new country initially involved men only; the immigration of European women during the first centuries was insignificant (Ribeiro 1995). Thus, the first Brazilians arose by mating between European males and Amerindian females. During the period 1500–1808, ∼500,000 Portuguese, mostly men, arrived in Brazil. With the exception of an unknown number of colonizers who arrived during the Dutch 30-year domination of the northeast of Brazil in the 17th century, Portugal was the only significant source of European immigrants to Brazil until 1808. Starting in the mid-16th century and continuing until 1855, ∼4 million African slaves were sent to Brazil (IBGE 2000). In 1808 the Portuguese court, fleeing Napoleon’s army, moved to Brazil and opened its seaports to trade with all nations. This was soon followed by the arrival of settlers from other countries. During the period 1820–1975, 5,686,133 immigrants, mostly Europeans, arrived officially in Brazil (IBGE 2000). Portuguese and Italian immigrants arrived in almost equal numbers (comprising almost 70% of the total), followed by immigrants from Spain, Germany, Syria, Lebanon, and Japan.
Brazilians, therefore, form one of the most heterogeneous populations in the world. The question that arises is how much did the three different groups—Amerindian, European, and African—actually contribute to the gene pool of present-day Brazilians? We decided to answer this question by using DNA markers to study matrilineages (mtDNA) and patrilineages (Y-chromosome polymorphisms) in the white Brazilian population. The mtDNA analysis of Brazilian mtDNAs for HVS-I and selected RFLP sites permitted the assignment of their ancestry to different continents (Alves-Silva et al. 2000). The total sample showed nearly equal amounts of Amerindian, African, and European matrilineal genetic contribution, with regional differences. In the present article, we describe our studies that use Y-chromosome DNA polymorphisms to evaluate the geographical distribution and ancestry of patrilineages in white Brazilian males.
DNA was obtained from 200 unrelated Brazilian males self-defined as “whites,” randomly drawn from paternity casework. This is essentially a subsample of the 247 Brazilians described elsewhere (Alves-Silva et al. 2000). The samples were from the following Brazilian regions: 49 from the north (states of Amazonas, Acre, Rondônia, and Pará), 49 from the northeast (state of Pernambuco), 50 from the southeast (state of Minas Gerais), and 52 from the south (states of Rio Grande do Sul, Santa Catarina, and Paraná) (fig. 1). Discrepancies in numbers were caused by geographical reclassification of two samples. We also typed another 10 white Brazilian males from a poor rural area in the northeast region of Minas Gerais (fig. 1). As a reference group, we studied 93 unrelated Portuguese individuals from north Portugal (primarily the Porto district), the place of origin of most of the Portuguese immigrants to Brazil. All studies were anonymous, and participants provided written consent.
Figure 1.
Geographic localization of Brazilian regions selected for study: Amazonas, Acre, Rondônia, Pará (northern region, N), Pernambuco (northeast region, NE), Minas Gerais (southeast region, SE), and Paraná, Santa Catarina, Rio Grande do Sul (south region, S).
Twelve Y-chromosome unique-event polymorphisms (UEPs) were studied. The Y Alu-insertion polymorphism (YAP) at DYS287 and the single-nucleotide polymorphisms (SNPs) SRY-8299, SRY-2627, sY81, and DYS199 were typed exactly as described in Santos et al. (1999). The marker 92R7 was typed according to Hurles et al. (1999), the C→T transition at SRY+465 was scored as described by Shinka et al. (1999), the G→T transversion at M34 was typed using primers described by Underhill et al. (2000), and the procedure for typing the 12f2 deletion at DYS11 has been published by Rosser et al. (2000).
We developed new typing methods for two markers. PN2 (Hammer et al. 1997) was studied by means of the ARMS (Amplification Refractory Mutation System) technique (Newton et al. 1989). Common primers (giving rise to a 536-bp amplicon) were those described by Hammer et al. (1997), and allele-specific primers were PN2.1 (gag cat taa taa aac taa tac c) for the C allele (ancestral) and PN2.2 (gag cat taa taa aac taa tac t) for the T allele (derived), both producing a 405-bp segment in the presence of the common primers. All samples were tested in separate reactions with the common primers, together with either the PN2.1 and PN2.2 allele-specific primers. The products were visualized in 6% polyacrylamide gels stained with silver salts, as described by Santos et al. (1993). The locus SRY-1532 (in this study we evaluated two SNPs—SRY-1532-I and SRY-1532-II—at the same locus) was amplified according to the method of Santos et al. (1999), but the alleles were identified by SSCP (Orita et al. 1989). After addition of 2.0 vol of formamide, PCR products were denatured at 95°C for 5 min, and the mixture was applied to a native 8% polyacrylamide gel (18 × 17 × 0.1 cm), TBE 0.5X with 10% glycerol, was subjected to electrophoresis in TBE 1.0X at 15 W for 3 h at room temperature, and then was silver stained.
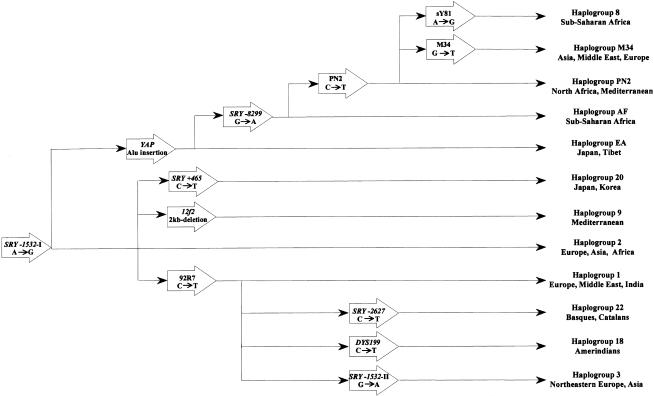
We typed the 12 Y-chromosome UEPs after a hierarchical analysis and could assign all individuals to haplogroups, the relationship and phylogeography of which are depicted in figure 2. The nomenclature of Y-chromosome haplogroups presented a problem, since it has not been standardized among the several research groups who use different sets of markers. We followed the haplogroup classification of Zerjal et al. (1999), with three exceptions, for which we had to create new nomenclature; to avoid numerical confusion, we renamed haplogroups 3A, 3G, and 4, from the study by Hammer et al. (1998), as haplogroups AF, EA, and PN2, respectively, and we classified as belonging to haplogroup M34 those individuals carrying the derived allele at the new marker M34 (Underhill et al. 2000).
Figure 2.
Phylogeography of the Y-chromosome haplogroups studied (Hammer et al. 1998; 2000; Pandya, 1998; Hurles et al. 1999; Shinka et al. 1999; Zerjal et al. 1999).
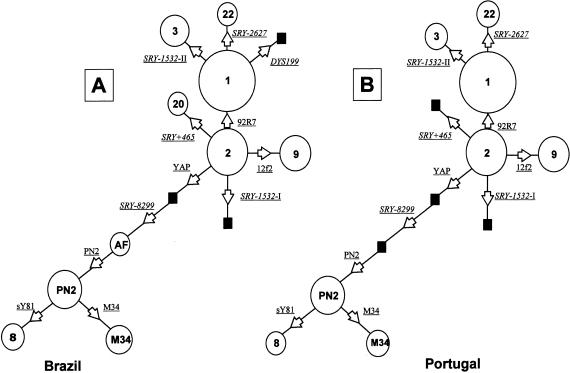
Following this methodology, we identified 10 haplogroups in Brazilians and 8 in the Portuguese (table 1). Parsimonious haplotypic networks were constructed manually for the Brazilian and Portuguese populations on the basis of ancestral/derived allelic states at all UEPs (fig. 3). Inspection of table 1 and fig. 3 reveals immediately that, in spite of centuries of interethnic admixture between Europeans, Amerindians, and Africans in Brazil, not a single instance of the Amerindian-specific haplogroup 18 and only five instances of the sub-Saharan haplogroups 8 and AF (Hammer et al. 1998) were observed among the 200 Brazilian males tested.
Table 1.
Percentage of Individuals Observed in Each Haplogroup in Brazilians and in Portuguese[Note]
|
Region of Brazil |
||||||
| Haplogroup | N(%)(n = 49) | NE(%)(n = 49) | S(%)(n = 52) | SE(%)(n = 50) | Total(%)(n = 200) | Portugal(%)(n = 93) |
| 1 | 53.1 | 65.3 | 42.3 | 56.0 | 54.0 | 63.4 |
| 2 | 12.2 | 18.4 | 28.8 | 12.0 | 18.0 | 12.9 |
| PN2 | 10.2 | 8.2 | 9.6 | 12.0 | 10.0 | 9.7 |
| 9 | 16.3 | 2.0 | 3.8 | 10.0 | 8.0 | 6.5 |
| 3 | 2.0 | 2.0 | 9.6 | .0 | 3.5 | 2.2 |
| M34 | 2.0 | .0 | 5.8 | 4.0 | 3.0 | 2.2 |
| 8 | .0 | 4.1 | .0 | 4.0 | 2.0 | 1.1 |
| 22 | .0 | .0 | .0 | 2.0 | .5 | 2.2 |
| AF | 2.0 | .0 | .0 | .0 | .5 | .0 |
| 20 | 2.0 | .0 | .0 | .0 | .5 | .0 |
| EA | .0 | .0 | .0 | .0 | .0 | .0 |
| 18 | .0 | .0 | .0 | .0 | .0 | .0 |
Note.— Haplogroups are in decreasing order of frequency in Brazilians.
Figure 3.
Y-chromosome haplotype networks in Brazilians (A) and Portuguese (B). Sampled haplogroups are indicated by circles (with area proportional to the absolute frequency), whereas haplogroups not seen in our sample but essential to drawing the network are indicated by blackened squares. Each link corresponds to one mutational event and is indicated by arrows pointing to the derived state.
The most frequent Y chromosomes in white Brazilian males belonged to haplogroup 1 (54%), which has been observed in high frequencies (40%–80%) in Europe and seems to be absent from Africa and Japan (Pandya 1998). Accordingly, this haplogroup was seen in 60% of the Portuguese tested. In Brazil, haplogroup 1 showed discrete regional variation, with the lowest frequency in the south. Second in frequency (18%) was haplogroup 2, which has a rather wide geographical range that includes Europe, Africa, and Asia. This haplogroup was also the second most common among the Portuguese (13%) and is known to be especially frequent in Italy (Previderé et al. 2000), from where 30% of the European immigrants to Brazil originated (Ribeiro 1995). Haplogroup 2 showed regional variation, having the highest frequencies in the south (29%) and in the northeast (18%). Its relatively high frequency in the south of Brazil is probably related to the large Italian immigration to this area. The increase in frequency in the northeast is very discrete and could be caused by chance, but it is interesting to observe that this region was under Dutch domination for almost three decades in the 17th century. Besides, the frequency (32%) of haplogroup 2 in the Netherlands is the highest in Western Europe (Rosser et al., 2000). Our attribution of the origin of most Brazilian haplogroup 2 chromosomes to Europe is reinforced by the low frequency (.5%) in Brazil of the Asian haplogroups EA (Hammer et al. 1998) and 20 (which has the highest frequency in Korea and Japan) (Shinka et al. 1999) and of sub-Saharan African haplogroups. The same applies to haplogroup 3, geographically distributed in northeastern Europe and Asia (Zerjal et al. 1999), which was observed in 3.5% of the Brazilian males. The fact that it was seen almost exclusively in the south of Brazil (table 1) suggests a European origin.
Haplogroup PN2 was the third in frequency, having been seen in 10% of Brazilians, with even regional distribution. This haplogroup is observed in 50% of the North Africans (Hammer et al. 2000) and in frequencies >29% in Egyptians, Greeks, Italians, and Lebanese (Hammer et al. 1998, 2000). Haplogroup PN2 is a subset of the haplogroup 21 of Zerjal et al. (1999), which has been more extensively studied (haplogroup 21=PN2+AF+M34 from our study). Haplogroup 21 has been shown to have a north-south cline in Portugal, climbing from a frequency of 10.6% in the north to 24.5% in southern Portugal (Pereira et al. 2000). This was interpreted as a footprint of North African invasions, especially of the Moorish conquest of the Iberian Peninsula in the Middle Ages, which lasted almost seven centuries. Other groups may have contributed haplogroup 21 chromosomes to Brazilians, since it can also be seen in northern (14% of Germans) and eastern Europe (15% of Hungarians and 23% of Slovakian gypsies; F. R. Santos, unpublished data). The latter may be significant in that Gitanos, one of the three main tribal groups of the gypsies, have inhabited the Iberian peninsula for centuries. At any rate, the high prevalence of haplogroup PN2 in the Portuguese suggests that these Y chromosomes did not come to Brazil directly from Africa but arrived via Portugal and perhaps also via other Mediterranean immigrants. Haplogroup M34, defined by the M34 mutation, is a subtype of haplogroup 21 and, apparently, has a low frequency but a quite broad geographical distribution in Europe, Asia, and the Middle East (Underhill et al. 2000). It was seen in 3% of the Brazilians and 2% of the Portuguese samples.
The 12f2 deletion at DYS11 defines haplogroup 9, the next most common Y-chromosome lineage observed in Brazil (8%). This haplogroup shows maximum frequency in Jews and other Middle Eastern populations (Semino et al. 1996; Hammer et al. 2000), but it is also found in North Africans (Hammer et al. 2000) and Europeans (Semino et al. 1996; Hammer et al. 2000). Portugal, where haplogroup 9 was seen in 6% of the individuals studied, seems to be the major source of these Y chromosomes in Brazil. There was a large Jewish population in Portugal until 1509, when Jews were deported during the Inquisition. To avoid expulsion, many Jews converted to Catholicism and became “New Christians,” many of whom immigrated to Brazil, carrying haplogroup 9 Y chromosomes. However, there were contributions from other populations. For example, as table 1 indicates, haplogroup 9 shows the highest frequency (16%) in the north of Brazil. Intrigued by this observation, we searched the historical records and discovered that in the early 19th century there was a significant immigration wave of Moroccan Jews to the Amazon area (Grinberg 2000), with eventual settlement in Manaus and Belem.
Thus, we have a picture of the vast majority of Brazilian Y chromosomes having a European origin. Indeed, there were no significant differences (P=.85) when the haplogroup frequencies in Brazil and Portugal were compared by means of an exact test of population differentiation (Arlequin software, version 1.1; Schneider et al. 1997). Using analysis of molecular variance, we partitioned the Brazilian haplotypic diversity into within- and among-geographical regions (Excoffier et al. 1992). Virtually all variation was found to be concentrated at the intrapopulation level, with no significant differentiation between regions (Fst=0; P=.48).
Since our sample of white Brazilian males was predominantly from the middle and upper-middle classes, it might contain a bias favoring European ancestry. To control for that, we analyzed another 10 white individuals from a poor rural region in the state of Minas Gerais (fig. 1). Again, no instances of sub-Saharan African haplogroups (8 or AF) or Amerindian Y chromosomes (haplogroup 18) were seen. Although the sample size is admittedly small, the haplogroup frequencies matched well those of the larger sample (five instances of haplogroup 1, two each of haplogroups PN2 and 9, and one of haplogroup 2). Other evidence against a significant social-class bias is the fact that >60% of the mtDNA lineages in this sample have Amerindian or African origin (Alves-Silva et al. 2000).
In 1500, Brazil was inhabited by ∼2.4 million Amerindians (IBGE 2000). Since then, ∼4 million African slaves and 6 million Europeans immigrated to the country (Ribeiro 1995). Now, after 500 years, it is worthwhile to ask about the genetic composition of the Brazilian population. We decided to try to partially answer this question by using lineage markers to ascertain the ancestry from different continents. In the first part of this project we analyzed 247 Brazilian mtDNAs and were surprised to find the high Amerindian (33%) and African (28%) contributions to the total mtDNA pool of white Brazilians (Alves-Silva et al. 2000). In the present study, we analyzed Y-chromosome UEPs in 200 Brazilian males from four different regions in Brazil (a subset of the 247 individuals typed for mtDNA). Our data demonstrate that the vast majority of Y chromosomes in white Brazilian males, regardless of their regional source, is of European origin, with a very low frequency of sub-Saharan African chromosomes and a complete absence of Amerindian contributions. Together, our results configure a picture of strong directional mating in Brazil involving European males and Amerindian and African females. This is in consonance with the known history of the peopling of Brazil since 1500.
Acknowledgments
This research was supported by grants from FAPEMIG, CNPq, and CAPES of Brazil, and FCT of Portugal. We are grateful to H.-J. Bandelt, Chris Tyler-Smith, and Eduardo Tarazona-Santos for advice. We thank Michael Hammer for providing us the PN2 sequence as well as the segment 3 sequence; P. Underhill for providing information before publication on the M34 marker; and M. Shlumukova, M. E. Hurles, and M. A. Jobling for the 12f2 PCR typing method. We are also grateful to Carlos Maurício Antunes and Roberto Campos Amado, from the Department of Parasitology of the Universidade Federal de Minas Gerais, for providing the rural Brazilian samples.
References
- Alves-Silva J, Santos MS, Guimarães PEM, Ferreira ACS, Bandelt H-J, Pena SDJ, Prado VF (2000) The Ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg K (2000) Nova língua interior: os judeus no Brasil. In: IBGE–Instituto Brasileiro de Geografia Estatística (ed) Brasil: 500 Anos de Povoamento. IBGE, Rio de Janeiro, pp. 125–139 [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Spurdle AB, Karafet T, Bonner MR, Wood ET, Novelletto A, Malaspina P, Mitchell RJ, Horai S, Jenkins T, Zegura SL (1997) The geographic distribution of human Y chromosome variation. Genetics 145:787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Pérez-Lezaun A, Bosch E, Shlumukova M, Cambon-Thomsen A, McElreavey K, López De Munain A, Röhl A, Wilson IJ, Singh L, Pandya A, Santos FR, Tyler-Smith C, Jobling MA (1999) Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 65:1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBGE–Instituto Brasileiro de Geografia Estatística (2000) Brasil: 500 Anos de Povoamento. IBGE, Rio de Janeiro [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF (1989) Analysis of any point mutation in DNA: the amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516 [DOI] [PMC free article] [PubMed]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya A (1998) Human Y-chromosomal DNA variation. DPhil thesis, Oxford University [Google Scholar]
- Pereira L, Brión MJ, Prata MJ, Jobling MA, Carracedo A, Amorim AM (2000) Gradient of Y chromosome haplogroup 21 across the western Iberia. In: Sensabaugh GF, Lincoln PJ, Olaisen B (eds) Progress in forensic genetics 8. Elsevier Science BV, Amsterdam, pp. 281–283 [Google Scholar]
- Previderé C, Tyler-Smith C, Grignani P, Peloso G (2000) Y chromosomal haplotypes and haplogroups in male identification. In: Sensabaugh GF, Lincoln PJ, Olaisen B (eds) Progress in forensic genetics 8. Elsevier Science BV, Amsterdam, pp. 257–259 [Google Scholar]
- Ribeiro D (1995) O povo brasileiro: a formação e o sentido do Brasil. Companhia das Letras, São Paulo [Google Scholar]
- Rosser Z, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FR, Carvalho-Silva DR, Pena SDJ (1999) PCR-based DNA profiling of human Y chromosomes. In: Epplen JT, Lubjuhn T (eds) DNA profiling and DNA fingerprinting. Birkhäuser Verlag AG, Basel, pp. 133–152 [Google Scholar]
- Santos FR, Pena SDJ, Epplen JT (1993) Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum Genet 90:655–656 [DOI] [PubMed] [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excoffier L (1997) Arlequin ver. 1.1: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti AS (1996) A view of the neolithic demic diffusion in Europe through two Y chromosome-specific markers. Am J Hum Genet 59:964–968 [PMC free article] [PubMed] [Google Scholar]
- Shinka T, Tomita K, Toda T, Kotliarova SE, Lee J, Kuroki Y, Jin DK, Tokunaga K, Nakamura H, Nakahori Y (1999) Genetic variations on the Y chromosome in the Japanese population and implications for modern human Y chromosome lineage. J Hum Genet 44:240–245 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner P (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Zerjal T, Pandya A, Santos FR, Adhikari R, Tarazona E, Kayser M, Evgrafov OV, Singh L, Thangaraj K, Destro-Bisol G, Thomas MG, Qamar R, Mehdi SQ, Rosser ZH, Hurles ME, Jobling MA, Tyler-Smith C (1999) The use of Y-chromosomal DNA variation to investigate population history. In: Papiha SS, Deka R, Chakraborty R (eds) Genomic diversity: applications in human population genetics. Kluwer Academic/Plenum Publishers, New York, pp. 91–101 [Google Scholar]