Abstract
Mutation screening of the major autosomal dominant polycystic kidney disease (ADPKD) locus, PKD1, has proved difficult because of the large transcript and complex reiterated gene region. We have developed methods, employing long polymerase chain reaction (PCR) and specific reverse transcription–PCR, to amplify all of the PKD1 coding area. The gene was screened for mutations in 131 unrelated patients with ADPKD, using the protein-truncation test and direct sequencing. Mutations were identified in 57 families, and, including 24 previously characterized changes from this cohort, a detection rate of 52.3% was achieved in 155 families. Mutations were found in all areas of the gene, from exons 1 to 46, with no clear hotspot identified. There was no significant difference in mutation frequency between the single-copy and duplicated areas, but mutations were more than twice as frequent in the 3′ half of the gene, compared with the 5′ half. The majority of changes were predicted to truncate the protein through nonsense mutations (32%), insertions or deletions (29.6%), or splicing changes (6.2%), although the figures were biased by the methods employed, and, in sequenced areas, ∼50% of all mutations were missense or in-frame. Studies elsewhere have suggested that gene conversion may be a significant cause of mutation at PKD1, but only 3 of 69 different mutations matched PKD1-like HG sequence. A relatively high rate of new PKD1 mutation was calculated, 1.8×10-5 mutations per generation, consistent with the many different mutations identified (69 in 81 pedigrees) and suggesting significant selection against mutant alleles. The mutation detection rate, in this study, of >50% is comparable to that achieved for other large multiexon genes and shows the feasibility of genetic diagnosis in this disorder.
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is the most common genetic nephropathology (population frequency of ∼0.1%), which accounts for 5%–8% of end-stage renal disease (ESRD). The disease is progressive, resulting in bilaterally enlarged polycystic organs and typically causing ESRD in late middle age. The disorder is genetically heterogeneous, with two genes, PKD1 (∼85% of ADPKD pedigrees [MIM 60131]) and PKD2 (∼15% [MIM 173910]), identified (European Polycystic Kidney Disease Consortium 1994; Mochizuki et al. 1996), plus rare unlinked families described elsewhere (Daoust et al. 1995). PKD1 encodes a large multidomain protein, polycystin-1, with a variety of characterized domains and regions of homology with other proteins (Hughes et al. 1995; International Polycystic Kidney Disease Consortium 1995; Moy et al. 1996; Sandford et al. 1997). Polycystin-1 may play a role in cell:cell/matrix interactions (Huan and van Adelsberg 1999; Wilson et al. 1999), whereas polycystin-2 (the PKD2 protein) has homology to an ion-channel subunit (Mochizuki et al. 1996; Chen et al. 1999).
PKD1 accounts for most cases of ADPKD associated with ESRD, because it is more common and results in significantly more-severe disease (average age at onset of ESRD is 53 years, compared with 69.1 years for PKD2; Hateboer et al. 1999). However, genetic diagnosis at this locus has proceeded slowly, because (1) PKD1 contains a 12,906-bp coding sequence (Hughes et al. 1995) divided into 46 exons, and (2) the 5′ region of the gene, from upstream of exon 1 to exon 33, is embedded in a complex genomic area reiterated >4 times further, proximally, on the same chromosome (European Polycystic Kidney Disease Consortium 1994). Analysis of two copies of these PKD1-like homologous genes (HG) has shown a number of specific deletions and a low level of substitutions (∼2%), compared with PKD1 (Loftus et al. 1999). The presence of the HG loci has significantly complicated analysis of PKD1, with routine PCR amplification and hybridization detecting those loci as well as PKD1. Consequently, the number of identified PKD1 mutations is still limited, with 82 changes described in the Online Human Gene Mutation Database (HGMD) (Krawczak and Cooper 1997). Although a number of methods have been employed to screen the reiterated region (Peral et al. 1997; Roelfsema et al. 1997; Watnick et al. 1997, 1998a, 1999; Thomas et al. 1999), the 3′ area has received disproportionate attention, with 57.3% of all mutations found in the single-copy area covering ∼20% of the coding region. It has not been possible to assess whether some areas are more prone to mutation, since no systematic study of the entire gene has been described. Analysis of the less-complex PKD2 has yielded 41 mutations (see HGMD), which are found throughout the gene and are mainly predicted to truncate and probably inactivate the protein (Veldhuisen et al. 1997). The majority of PKD1 mutations are also predicted to truncate the protein, although a significant number of missense changes have been described elsewhere (Peral et al. 1997; Daniells et al. 1998; Perrichot et al. 1999; Thomas et al. 1999).
Inactivating germline mutations are consistent with the hypothesis that cystogenesis occurs by a two-hit mechanism in which a cyst only forms after the normal ADPKD allele has been inactivated by a somatic mutation. Evidence for this mechanism comes from the finding of somatic mutations in the PKD1 gene in cystic epithelia isolated from single cysts in PKD1 renal and liver tissue (Qian et al. 1996; Watnick et al. 1998b) and corresponding results in material from patients with PKD2 (Koptides et al. 1999; Pei et al. 1999; Torra et al. 1999). Recently, it has been suggested that cysts may evolve by a transheterozygous method, with a germline mutation to PKD2 and a somatic mutation to PKD1 or vice versa (Koptides et al. 2000; Watnick et al. 2000). Further evidence that germline mutations may be inactivating comes from knockout models of Pkd1 and Pkd2; homozygotes generally die in the mid- to late-fetal stages, with renal and pancreatic cysts developing from ∼E14.5d, whereas heterozygotes develop occasional cysts in later life (Lu et al. 1997, 1999; Kim et al. 2000). Nevertheless, there are questions of whether mutant polycystin-1 molecules, at least in some cases, may have residual function or even an altered role. Targeted disruption of Pkd1 in the 3′ region has a more severe phenotype than disruption of the gene in exon 34, suggesting a dominant negative role for the larger mutant protein (Lu et al. 1997; Kim et al. 2000). Studies of human cystic epithelia have also found strong polycystin-1 staining in many cyst linings, suggesting the presence of normal or mutant polycystin-1 protein and not entirely consistent with the predicted two-hit model (Geng et al. 1996; Ward et al. 1996; Ong et al. 1999).
The necessity for numerous somatic mutations to explain the formation of multiple cysts and evidence of a significant rate of new germline mutations (Peral et al. 1997) have led to the proposal that unusual mechanisms promote a high rate of PKD1 mutation. First, a long polypyrimidine tract in IVS21, which could potentially form triplex DNA structures (Van Raay et al. 1996; Blaszak et al. 1999), has been implicated in inducing mutations in the downstream exons (Watnick et al. 1997). Subsequently, these multiple substitutions, and other changes, were found to match HG sequence, suggesting gene conversion with the distantly located HG loci (Watnick et al. 1998a; Phakdeekitcharoen et al. 2000). At this stage, the importance of these unusual mechanisms for causing mutation at PKD1 are unclear, as is the question of whether mutation is occurring at a higher than average level.
In this study, we describe techniques for mutation analysis of the entire PKD1 gene. We have employed these methods to screen a cohort of 155 apparently unrelated ADPKD pedigrees, to generate a clearer view of the pattern of mutation at PKD1 and the prospects for molecular diagnostics.
Subjects and Methods
Patients with ADPKD
Patients were recruited through the Oxford Renal Unit (135 pedigrees), other adult nephrology centers (7 pedigrees), or pediatric nephrology units (13 pedigrees). In each case, the proband had ADPKD, as defined by the Ravine criteria for ultrasound examination (Ravine et al. 1994). Most cases had typical ADPKD, but the group also included 10 pedigrees that comprised a case of early-onset ADPKD. Informed consent was obtained and blood samples collected for DNA isolation from the proband and from all available family members. At-risk undiagnosed individuals wishing to take part in the study were examined by abdominal ultrasound. The study had approval from the University of Oxford Ethics Committee.
Genetic Linkage Analysis
Pedigrees with more than six informative meioses were analyzed for linkage to PKD1 or PKD2 with microsatellite markers. The markers KG8, SM6, 16AC2.5, and CW2 were routinely employed for PKD1 linkage (Peral et al. 1994) and haplotype analysis of recurrent mutations. The markers D4S1534, D4S1563, D4S423, and JSTG3 were employed for analysis of PKD2 (Gyapay et al. 1994; Mochizuki et al. 1996). The markers were amplified and analyzed on native polyacrylamide gels, as described elsewhere (Harris et al. 1991; Peral et al. 1994).
Amplification of the PKD1 transcript
RNA isolation from lymphoblast cell lines and leukocytes and reverse transcription (RT)–PCR for cDNA synthesis were performed as described elsewhere (European Polycystic Kidney Disease Consortium 1994; Peral et al. 1997). Primer pairs for specific amplification of PKD1 cDNA (exons 7–23; see fig. 1) are shown in table 1 (top). These were designed to match regions where the PKD1 and HG sequence differ, with the mismatches positioned at the 3′ ends of the primers to maximize specificity. Patient cDNA was amplified at high annealing temperatures (see table 1, top) in a DMSO-containing buffer (Dodé et al. 1990) with a 90-s extension time and the addition of Taq Extender (1 U/kb amplified; Stratagene). The specificity of fragments was tested with HG-only somatic cell hybrids, P-MWH2A and 77-2/1 (containing the der16 chromosome of patient 77-2; European Polycystic Kidney Disease Consortium 1994), and the PKD1-only radiation hybrid, Hy145.19 (European Polycystic Kidney Disease Consortium 1994). Details of the primer pairs to amplify exons 22–46 (Spec 5–7) have been described elsewhere (Peral et al. 1997).
Table 1.
Primers for Specific Amplification of PKD1
| Fragment | Primer Sequencea | AnnealingTemperature(°C) | Position | Size(bp) | Exon(s) |
| cDNA: | |||||
| Spec 1 | (F) CTACGTCTGCGAGCTGCAGCCC | 66 | 1792–4000 | 2,209 | 7–15 |
| (R) CACATGCTCCACTGTTGCCTCC | |||||
| Spec 2 | (F) CCGTCACCTTCTACCCGCACCC | 68 | 3639–5519 | 1,881 | 15 |
| (R) TATGGGTGGTAAATGGCTCGGA | |||||
| Spec 3 | (F) TGGTTAGGGATGGCACCAACG | 65 | 5175–7121 | 1,947 | 15 |
| (R) TCGAAGCCACACAGGCCCAG | |||||
| Spec 4 | (F) GGCGATCACAGCGCAACTACT | 66 | 6696–8901 | 2,206 | 15–23 |
| (R) ACGGAGTTGGCGGAGTTGGC | |||||
| Genomic: | |||||
| Gen 1b | (F) CGCAGCCTTACCATCCACCT | 64 | 2033–4310 | 2,278 | 1 |
| (R) TCATCGCCCCTTCCTAAGCA | |||||
| Gen 2–10b | (F) CCAGCTCTCTGTCTACTCACCTCCGCATCGCCTGCA | 68 | 17647–24008 | 6,362 | 2–10 |
| (R) GGCTGGGTGTGTCTGGTGCA | |||||
| Gen 22–33b | (F) GAGCCAGGTGAGGACCCGTGTA | 68 | 36868–44310 | 7,442 | 22–33 |
| (R) TGAGCTTCAGAGCCCCCTCCTC |
Underlined nucleotides differ between PKD1 and all characterized HG sequence.
One primer absent in all known HG sequence.
Amplification of the PKD1 gene
Genomic DNA was isolated from peripheral blood by standard phenol/chloroform extraction methods. The primer pairs used to amplify PKD1 genomic fragments are shown in table 1 (bottom), as is their PKD1 specificity, tested employing the somatic cell hybrid panel. Long PCR to amplify Gen 2–10 and Gen 22–33 was performed using the Gene Amp XL Kit (PE Biosytems) and a hot-start protocol. In brief, each reaction containing 60 ng genomic DNA, 5 pmol each primer, 200 μM each dNTP, 1 mM Mg(OAc), and the supplied buffer was heated to 93°C for 3 min. Subsequently, 1 U rTth enzyme was added, and the reaction was incubated for 35 cycles of 93°C, 60 s; 68°C, 60 s; and 70°C, 6 min.
Because of the extreme GC richness of exon 1, a different protocol was employed: DNA (120 ng), primers (8 pmol each), dNTPs (200 mM each), and MgCl2 (2.5 mM) was heated to 100°C for 5 min. On cooling to 95°C, the DMSO buffer, Taq Extender (4 U), and Amplitaq (2 U) were added before 15 cycles of 95°C for 60 s, 64°C for 60 s, and 72°C for 3 min. Subsequently, 20 further cycles were completed using the conditions as described above, with the addition of 10 s to each annealing step and of 20 s per extension step and, finally, 72°C for 10 min.
Protein Truncation Test (PTT)
A ∼1:1000 dilution of the appropriate Spec PCR product was amplified using the PTT primer sets (see table 2, top, and Peral et al. [1997] for details of fragments PTT 6–9). Each upstream PTT primer had additional 5′ sequence added containing the T7 promoter and translation-initiation codon (Roest et al. 1993). Details of the transcription, translation, and analysis of the resulting polypeptide products have been described elsewhere (Peral et al. 1997). Smaller constant bands were sometimes seen (see fig. 2) because of translational initiation from internal start codons (Rowan and Bodmer 1997).
Table 2.
PTT and Sequencing Primers
| Fragment | Sequence | AnnealingTemperature(°C) | Position(cDNA) | Size(bp) | Exon(s) |
| cDNA: | |||||
| PTT 1 | (F) GATGCCGAGAACCTCCTCG | 63 | 1829–3869 | 2,041 | 8–15 |
| (R) TCATGTCCACGCTGAGTCCG | |||||
| PTT 2 | (F) GCCAACCACACCTATGCCTCG | 62 | 3740–5431 | 1,692 | 15 |
| (R) GGCACTGAGGGTGACGCTTGT | |||||
| PTT 3 | (F) AGCGGCAAAGGCTTCTCGCTC | 66 | 5246–7052 | 1,807 | 15 |
| (R) ACTCGCTCCCATCCAGCACC | |||||
| PTT 4 | (F) ACTGAGTACCGCTGGGAGGTGTATC | 66 | 6758–7942 | 1,185 | 15–20 |
| (R) GGGCTCTGGGAGGGTGATG | |||||
| PTT 5 | (F) AAGACGCTGGTGCTGGATGAG | 62 | 7448–8644 | 1,197 | 18–23 |
| (R) AGCCTCGGGGATGGAGAAG | |||||
| cSeq 1 | (F) TGAGTACCGCTGGGAGGTGTATC | 62 | 6760–7336 | 577 | 15–17 |
| (R) TGCCTTGCAGGACACACACTC | |||||
| cSeq 2 | (F) TGTCCCTGAGGGTCCACACTG | 62 | 8163–8681 | 519 | 22–23 |
| (R) GCACCACGTCACTGAGGTTGG | |||||
| cSeq 3 | (F) CTCAACGAGGAGCCCCTGAC | 62 | 8516–9126 | 611 | 23–24 |
| (R) TCAGCACCCTGGAGTGACTCTG | |||||
| cSeq 4 | (F) CAGTCTACCTACACTCGGAGCCC | 62 | 9039–9648 | 610 | 24–27 |
| (R) TCCACCCCATACAGCATGATG | |||||
| Genomic: | |||||
| GenSeq 1 | (F) CCTGAGCTGCGGCCTCCG | 64 | 3580–3799 | 220 | 1 |
| (R) CAGTTGACGCGGCAGGCG | |||||
| GenSeq 2 | (F) TGCGAGCCCCCCTGCCTC | 66 | 3744–3923 | 180 | 1 |
| (R) AACCCGCCCACGCCCGCCCGTCC | |||||
| GenSeq 3 | (F) CTTGGGGATGCTGGCAATG | 59 | 19810–20239 | 430 | 2–3 |
| (R) AACTGGGAGGGCAGAAGGG | |||||
| GenSeq 4 | (F) AGTGGGGGGCTGGCATAGAC | 64 | 20371–21017 | 647 | 4–5 |
| (R) GCAAAGGAGGCACTGGAGGG | |||||
| GenSeq 5 | (F) CTTCTAGGTGAGGAGTATGTCGCC | 64 | 20812–21593 | 782 | 5 |
| (R) AACGAGGGTGTCAACGGTCAG | |||||
| GenSeq 6 | (F) ACCGTTGACACCCTCGTTCC | 60 | 21576–21855 | 280 | 6 |
| (R) TCTCTGCCCCAGTGCTTCAG | |||||
| GenSeq 7 | (F) CTGTGAGGGTGGGAGGATGG | 64 | 22166–22787 | 622 | 7–8 |
| (R) GGAGGGCAGGTTGTAGAACGTG |
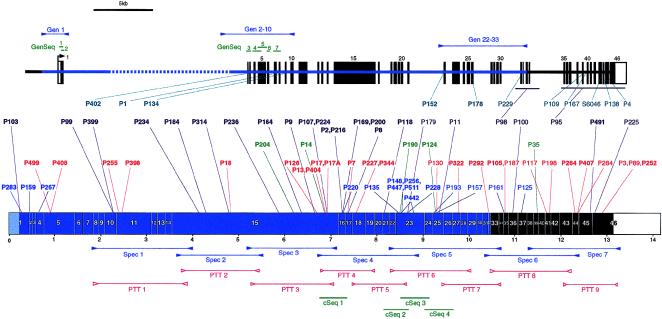
Figure 1 .
Diagram showing the fragments employed for mutation screening of the PKD1 gene and the mutations detected. The PKD1 gene (top) and transcript (bottom), showing the intron/exon structure, duplicated region (blue), single-copy coding exons (solid boxes), and 5′ and 3′ UTRs (open boxes). The positions of fragments to amplify the transcript specifically (Spec; blue), plus the PTT fragments (PTT; red) and cDNA sequencing products (cSeq; green), are shown at the bottom. The locations of the PKD1-specific and anchored genomic fragments (Gen; blue), plus the genomic sequencing products (GenSeq; green), are illustrated at the top. The sites of the mutations detected in the 81 different PKD1 pedigrees (P) are shown in the center. Different mutation types are color coded and grouped: splicing mutations, turquoise; frameshifting deletions or insertions, purple; in-frame deletions or insertions, green; nonsense mutations, red; and missense mutations, dark blue. Newly described mutations are shown in boldface type.
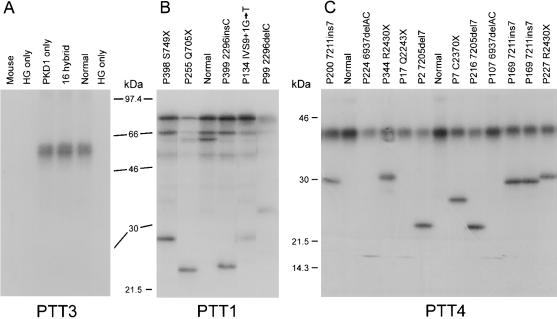
Figure 2 .
Examples of PTT gels. A, Fragment PTT3, transcripted and translated from cDNA of mouse; human (Normal); somatic-cell hybrids, 77-2/1 and P-MWH2A (HG only; lane 2 and 6, respectively); P-SHH1H; whole chromosome 16 (16 hybrid); and the radiation hybrid, Hy145.19 (PKD1 only). The product is only generated in PKD1-containing samples showing the specificity of this PTT assay. Analysis of PKD1 patient and normal cDNA by PTT assays, PTT1 (B) and PTT4 (C). The pedigree number and detected mutation are shown for each sample. Mutant polypeptides are seen as smaller products on the gel.
DNA Sequencing
Sequencing of cDNA was used to analyze four fragments (cSeq 1–4) (fig. 1) and to characterize mutations detected by PTT and other methods. Genomic sequencing was used to screen the 5′ region of the gene (exons 1–8) and to confirm other mutations. The method of dye primer sequencing was employed with primers (see table 2) modified by addition to the 5′ end with the −21 M13 universal primer sequence: 5′-TGTAAAACGACGGCCAGT-3′ (upstream) and the −28 M13 reverse primer sequence: 5′-AGGAAACAGCTATGACCAA-3′ (downstream). Appropriate PKD1-specific DNA was diluted ∼1:1000 and was amplified with the modified sequencing primer using the protocol 94°C for 1 min; 30 cycles of 94°C for 30 s, 59°C–66°C (see table 2) for 30 s, and 72°C for 30 s; and, finally, 72°C for 10 min. Amplification of exon 1 was as described above, except that a denaturation step of 100°C for 5 min was added before addition of the enzyme, and annealing and elongation times were extended as for the primary amplification of exon 1. Products from these PCR reactions (200 ng) were used for sequencing employing the BigDye Primer Cycle Sequencing Reaction Kit with the appropriate M13 primer (PE Biosystems). The cycle sequencing consisted of 15 cycles of 95°C for 30 s, 55°C for 30 s, and 70°C for 60 s and 15 cycles of 95°C for 30 s and 70°C for 60 s. The four different primer reactions were mixed and precipitated with ethanol, and the pellet was resuspended in loading buffer (95% formamide, 0.01% bromophenol blue). The reactions were run on an ABI PRISM 377 sequencer, and the resulting sequence was analyzed with SEQUENCER 3.0 software.
Heteroduplex Analysis
The PCR product to be assayed was denatured at 95°C for 5 min and then was incubated at 37°C for 2 h. These products were separated on a 10% PAGE at 200 V for 20 h and were visualized after staining with ethidium bromide.
Allele-Specific Oligonucleotides (ASOs)
To confirm the mutations, 7205del7 and 6937delAC, oligonucleotides were designed matching either the normal or mutant sequence, with the mismatch in the middle of the primer. The oligonucleotides were end labeled with 32P; hybridized to a dot blot containing the amplified target sequence from the patient, family members, and normal controls; washed; and exposed to x-ray film (Wood et al. 1985). Differential hybridization of the oligonucleotides was used to confirm the mutation and trace the change within families.
Signal Peptide Prediction
The SPSCAN program in the SeqWeb package (GCG) was used to predict the probability of signal peptides, using the eukaryotic scoring matrix (Nielsen et al. 1997). Hydrophobicity measurements were made with MacVector, using the Kyle-Doolittle plot with a window size of 20 residues.
Sequence Designation
The positions of primers and mutations are shown relative to the PKD1 genomic (GenBank accession number L39891), cDNA (GenBank accession number L33243), and protein sequence (GenBank accession number AAC37576).
Results
Strategy for Mutation Screening
To overcome the problem of duplication of the 5′ region of PKD1, we employed different strategies to specifically amplify the gene. For the region from exons 9 to 46, PKD1-specific fragments were amplified from cDNA generated from lymphoblast or leukocyte RNA (fig. 1). The duplicated area from exons 23–32 and the single-copy 3′ region was amplified as described elsewhere (Peral et al. 1997), with one or both primers in the single-copy area to provide PKD1 specificity. For exons 9–23, a method of PKD1-specific amplification was employed, using primers that match the rare differences between PKD1 and HG sequence. HG sequence was obtained from HG cDNAs (European Polycystic Kidney Disease Consortium 1994) and from genomic sequence of HG loci (Loftus et al. 1999; authors' unpublished data). The rare sequence differences were positioned at the 3′ ends of primers, to maximize their specifying effect (see table 1). The PKD1 specificity of each fragment was tested using somatic cell hybrids containing just the PKD1 or HG loci (see, e.g., fig. 2A). For the region from exons 1–8, PKD1-specific amplification of cDNA proved impossible, because of the lack of differences between the PKD1 and HG transcripts. Consequently, this region was amplified by long PCR employing a genomic DNA template. Specificity for the exon 2–8 region was obtained by positioning the 5′ primer in a region of IVS 1 deleted in the HG1 and HG2 loci (Loftus et al. 1999) and with a specific 3′ primer (Gen 2–10; table 1, bottom, and fig. 1). A novel HG locus has recently been sequenced (HG3) that contains the IVS 1 primer but is deleted for the 3′ primer and so is also not amplified with this primer pair (data not shown). The GC-rich exon 1 was amplified from genomic DNA using one primer 5′ to the duplicated region, to provide PKD1 specificity, and a second primer in IVS 1 (Gen 1 fig. 1 and table 1, bottom). A long PCR linking the single copy area to IVS 21 (Gen 22–33; table 1, bottom, and fig. 1) was also developed to test the validity of mutations found in exons 22–32.
Exons 9–46 were screened for mutations using the PTT, with nine overlapping fragments (for details, see table 2 and fig. 1). Exons 1–9 were screened by direct sequencing of PKD1 exons using seven sets of modified primers (GenSeq 1–7; table 2, top, and fig. 1). The areas from exons 21–26 and the 3′ ends of exons 15 and 16 also were analyzed by direct sequencing of cDNA.
Each identified mutation was confirmed using an independent method (for details, see table 3). Where PKD1 genomic DNA was readily specifically amplified, the confirmation was by genome sequencing, restriction digest, or other method. In other cases, PTT, sequencing, or restriction digest employed cDNA from a relative or leukocyte cDNA from the proband. When samples were available, segregation within the family was tested (see table 3). This was especially important for potential missense mutations and subtle in-frame changes that were also screened in ⩾98 normal chromosomes (see table 3). Only if the change segregated with the disease (except P285; see below) and was not seen in the normal population was it considered a possible missense mutation. All probands with putative missense changes were included in the screen of subsequent fragments, but no further changes were found in these samples in the rest of the gene.
Table 3.
Details of Newly Identified PKD1 Mutations
| Pedigree | MutationDesignation | Method ofDetectiona | Confirmationb | SegregationDemonstrated | PedigreeSizec | PKD1Linkedd | FamilyHistorye | NormalChromosomes |
| P1 | IVS7+1G→A | Gen seq | Gen digest: MnlI | Yes | 54 | Yes | Yes | |
| P2 | 7205del7 | PTT | Gen ASO | Yes | 3 | Yes | De novo | |
| P7 | C2370X | PTT | Leuk PTT | NP | 4 | NP | Yes | |
| P8 | 7324delGT | PTT | Leuk PTT | Yes | 5 | Yes | De novo | |
| P9 | 6645del28 | PTT | Gen hetero | Yes | 6 | NP | Yes | |
| P13 | C2229X | PTT/cDNA seq | Gen Seq | Yes | 23 | Yes | Yes | |
| P14 | 6868del15 | cDNA seq | Gen Gel | Yes | 10 | Yes | Yes | |
| P17 | Q2243X | PTT | Rel-cDNA seq | Yes | 29 | Yes | Yes | |
| P17A | Q2243X | PTT | Rel-cDNA seq | Yes | 15 | Yes | Yes | |
| P18 | E1537X | PTT | Rel-PTT | Yes | 12 | Yes | Yes | |
| P99 | 2296delC | PTT | Gen seq | NP | 2 | NP | ? | |
| P103 | 224delC | Gen seq | Gen hetero | Yes | 8 | Yes | Yes | |
| P105 | Q3513X | Gen digest, AlwNI | Gen seq | Yes | 4 | NP | Yes | |
| P107 | 6937 delAC | PTT | Gen hetero/ASO | NP | 1 | NP | No | |
| P118 | 8126dup20 | PTT | Rel-PTT/hetero | Yes | 2 | NP | De novo/EO | |
| P124 | 9245del18 | Gen gel | Gen seq | Yes | 5 | NP | Yes | |
| P126 | R2163X | PTT | Gen digest, BclI | Yes | 2 | NP | ? | |
| P134 | IVS9+1G→T | PTT | Rel-PTT | Yes | 4 | NP | Yes | |
| P135 | A2752D | cDNA seq | Gen digest, StuI | Yes | 4 | NP | Yes | 0/250 |
| P148 | E2771K | cDNA seq | Gen digest, BseRI | Yes | 6 | Yes | Yes/EO | 0/230 |
| P152 | IVS21−2delAG | cDNA seq | Gen seq | Yes | 2 | NP | De novo | |
| P159 | S75F | Gen seq | Rel-Gen seq | Yes | 2 | NP | Yes/EO | 0/98 |
| P164 | 6356insG | PTT | Leuk PTT | NP | 3 | NP | Yes | |
| P169 | 7211dup7 | PTT | Rel-PTT | Yes | 6 | NP | Yes/EO | |
| P178 | IVS25−16G→A | PTT | Leuk PTT/gen digest, PvuII | NP | 1 | NP | ? | |
| P184 | 4291delG | PTT | Gen seq | NP | 2 | NP | ? | |
| P190 | 8507ins12 | cDNA seq | Gen gel | Yes | 2 | NP | No/EO | |
| P200 | 7211dup 7 | PTT | Leuk PTT | NP | 6 | NP | Yes | |
| P204 | F1992L + 1993delT | cDNA seq | Gen digest, DdeI | NP | 1 | NP | No | 0/106 |
| P216 | 7205del7 | PTT | Gen ASO | Yes | 3 | NP | No | |
| P220 | Y2336D | cDNA seq | Gen digest, RsaI | Yes | 3 | NP | No | 0/180 |
| P224 | 6937delAC | PTT | Gen hetero | Yes | 7 | Yes | Yes | |
| P227 | R2430X | PTT | Leuk cDNA digest, SphI | NP | 2 | NP | ? | |
| P228 | L2816P | cDNA seq | Gen digest, MscI | Yes | 5 | Yes | Yes | 0/350 |
| P234 | 4137delCT | PTT | Gen hetero | Yes | 3 | NP | ? | |
| P236 | 5870del14 | PTT | Gen hetero | Yes | 3 | NP | Yes | |
| P252 | R4227X | ARMS | Gen seq | Yes | 7 | Yes | Yes | |
| P255 | Q705X | PTT | Gen seq | Yes | 4 | Yes | Yes | |
| P256 | E2771K | cDNA seq | Gen digest, BseRI | Yes | 7 | NP | Yes | 0/230 |
| P264 | R4020X | Gen digest, Ddel | Gen seq | Yes | 5 | NP | Yes | |
| P267 | W139C | Gen seq | Rel-gen seq | Yes | 7 | NP | Yes | |
| P285 | L13Q | Gen seq | Gen seq | NP | 2 | NP | Yes | 0/98 |
| P292 | Q3394X | PTT | Gen seq | NP | 3 | NP | Yes | |
| P314 | 4784delG | PTT | Gen seq | NP | 1 | NP | ? | |
| P322 | W3180X | PTT | Leuk PTT | NP | 4 | NP | ? | |
| P344 | R2430X | PTT | Leuk cDNA digest, SphI | NP | 1 | NP | ? | |
| P398 | S749X | PTT | Gen seq | NP | 2 | NP | ? | |
| P399 | 2296insC | PTT | Gen seq | NP | 5 | Yes | Yes | |
| P402 | IVS2−2AG | Gen seq | Gen digest, MspI | NP | 4 | NP | Yes | |
| P404 | C2229X | PTT/cDNA seq | Gen seq | NP | 2 | NP | Yes | |
| P407 | Y4039X | PTT | Gen digest, DdeI | NP | 1 | NP | No | |
| P408 | Q227X | Gen seq | Gen seq | NP | 4 | NP | Yes | |
| P442 | V2768M + G2858S | cDNA seq | Gen digest, SphI and BsaHI | Yes | 2 | NP | No | 0/250 |
| P447 | E2771K | cDNA seq | Gen seq, BseRI | NP | 3 | Yes | Yes | 0/230 |
| P491 | 12617delC | PTT | Gen digest, BspMI | NP | 2 | NP | ? | |
| P499 | S225X | Gen seq | Rel-gen seq | Yes | 2 | NP | No | |
| P511 | E2771K | cDNA seq | Gen seq, BseRI | NP | 1 | NP | ? | 0/230 |
Gen seq = genomic sequencing; PTT = protein truncation test; cDNA seq = cDNA sequencing; Gen digest = genomic digestion; and ARMS = amplification refractory mutation system.
Gen ASO = genomic-allele specific oligonucleotide hybridization; Leuk PTT = PTT using leukocyte-isolated RNA; Gen gel = genomic gel electrophoresis; Rel = change confirmed in affected relative; Hetero = heteroduplex analysis.
No. of traced affected cases.
Prior to mutation detection; NP = not possible to test because of insufficient family members/samples.
EO = early onset.
Population Screened for PKD1 Mutations
A total of 131 patients from apparently unrelated families were screened for PKD1 mutations in this study. These included 73 patients that were negative in previous rounds of mutation detection from exons 23–46 (European Polycystic Kidney Disease Consortium 1994; Peral et al. 1995, 1996a, 1996b, 1997; Torra et al. 1998), in which a total of 24 mutations were characterized. Of the ADPKD pedigrees in the present study, 34 were linked to PKD1, and the remainder were from families that were too small (or too few samples were available) for linkage analysis. Six families found to be linked to PKD2, or to have PKD2 mutations (authors' unpublished data), were eliminated from the study population. Analysis for larger rearrangements by hybridization of field inversion gels of EcoRI-digested DNA identified no further rearrangements than the two described elsewhere (European Polycystic Kidney Disease Consortium 1994).
Mutations Detected by PTT
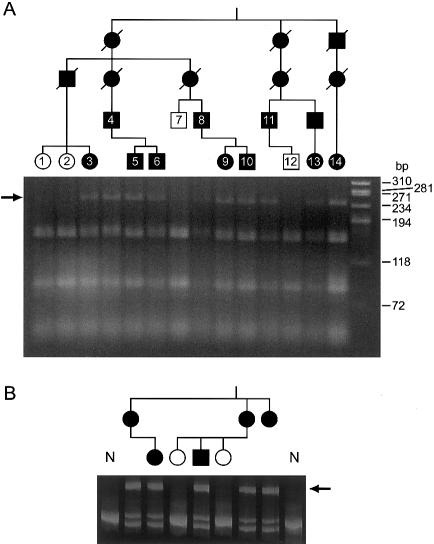
PTT was employed as a screening method for most of the gene, because relatively large fragments could be assayed and practically all detected changes were pathogenic mutations, overcoming the problem of significant polymorphism in PKD1. In the nine PTT fragments (covering 87.5% of the coding area), 27 different mutations were found in 33 families (for details, see table 4 and fig. 1). PTT gels showing PKD1 specificity of amplification and examples of mutations are shown in figure 2. The mutation in each case was identified by direct sequencing of cDNA, with splicing changes characterized in genomic DNA. An example of segregation of the splicing change IVS7+1G→A, which was detected in a large family (P1) that was originally used to map PKD1 to 16p13.3 (Reeders et al. 1985; NDM-A), is illustrated in figure 4A. The PTT changes consisted of 10 frame-shifting deletions, 4 insertions, 11 nonsense mutations, and 2 frame-shifting splicing changes. In P178, exon 26 was skipped, but the only detected genomic change was IVS25−16G→A. It is not clear how this causes exon skipping, but the introduction of an A nucleotide may disrupt branch-site formation. The mutation associated with one aberrant PTT fragment detected in two pedigrees was not identified initially by sequencing but was rediscovered as the nonsense mutation C2229X during direct sequencing of the area.
Table 4.
Summary of PKD1 Mutation Changes Identified in the Study Population
| Designation | Location | cDNAChange(s) | AminoAcidChangea | Comments | Pedigree(s) | Reference(s) |
| Nonsense: | ||||||
| S225X | EX5 | 885C→A | 225↓ | At CpG | P499 | Present study |
| Q227X | EX5 | 890C→T | 227↓ | P408 | Present study | |
| Q705X | EX11 | 2324C→T | 705↓ | P255 | Present study | |
| S749X | EX11 | 2457C→G | 749↓ | P398 | Present study | |
| E1537X | EX15 | 4820G→T | 1537↓ | At CpG | P18 | Present study |
| R2163X | EX15 | 6698C→T | 2163↓ | C→T at CpG | P126 | Present study |
| C2229X | EX15 | 6898C→A | 2229↓ | P13, P404 | Present study | |
| Q2243X | EX15 | 6938C→T | 2243↓ | P17, P17A | Present study | |
| C2370X | EX17 | 7321T→A | 2370↓ | P7 | Present study | |
| R2430X | EX18 | 7499C→T | 2430↓ | C→T at CpG, matches HG 1, 2, and 3 | P227, P344 | Present study |
| E3020X | EX25 | 9269G→T | 3020↓ | At CpG | P130 | Peral et al. (1997) |
| W3180X | EX27 | 9751G→A | 3180↓ | P322 | Present study | |
| Q3394X | EX32 | 10391C→T | 3394↓ | P292 | Present study | |
| Q3513X | EX35 | 10748C→T | 3513↓ | P105, P187 | Peral et al. (1997); present study | |
| Y3818X | EX41 | 11665C→A | 3818↓ | At CpG | P117 | Peral et al. (1996a) |
| Q3837X | EX41 | 11720C→T | 3837↓ | P198 | Peral et al. (1996b) | |
| R4020X | EX44 | 12269C→T | 4020↓ | C→T at CpG | P264 | Present study |
| Y4039X | EX44 | 12328C→A | 4039↓ | At CpG | P407 | Present study |
| Q4041X | EX44 | 12332C→T | 4041↓ | P284 | Torra et al. (1998) | |
| R4227X | EX46 | 12890C→T | 4227↓ | C→T at CpG | P3, P89, P252 | Peral et al. 1996b, 1997); present study |
| Deletion or insertion: | ||||||
| Frameshift: | ||||||
| 224del13 | EX1 | 224del13 | 4↓ | 8-bp repeat 13 bp apart | P103 | Present study |
| 2296delC | EX10 | 2296delC | 695↓ | 6 × C nt | P99 | Present study |
| 2296insC | EX10 | 2296insC | 695↓ | 6 × C nt | P399 | Present study |
| 4137delCT | EX15 | 4137delCT | 1308↓ | P234 | Present study | |
| 4291delG | EX15 | 4291delG | 1360↓ | P184 | Present study | |
| 4784delG | EX15 | 4784delG | 1524↓ | P314 | Present study | |
| 5870del14 | EX15 | 5870del14 | 1886↓ | 7-bp repetition, 14 bp apart | P236 | Present study |
| 6937delAC | EX15 | 6937delAC | 2242↓ | 2 × AC repeats | P107, P224 | Present study |
| 7205del7 | EX15 | 7205del7 | 2331↓ | 7-bp direct repeat | P2, P216 | Present study |
| 7211ins7 | EX16 | 7211ins7 | 2333↓ | 7-bp direct repeat | P169, P200 | Present study |
| 7324delGT | EX17 | 7324delGT | 2371↓ | 4 × GT repeats | P8 | Present study |
| 8126ins20 | EX21 | 8126ins20 | 2638↓ | 9×bp repeat, 20 bp apart | P118 | Present study |
| 8657delC | EX23 | 8657delC | 2815↓ | 3 × C nt | P179 | Peral et al. (1997) |
| 9299delC | EX25 | 9299delC | 3029↓ | P11 | Peral et al. (1997) | |
| IVS30del2 kb | IVS30−IVS34 | 10262del446 | 3350↓ | P98 | European Polycystic Kidney Disease Consortium (1994) | |
| IVS34del5.5 kb | IVS34−3′UTR | 10708del to 3′UTR | 3499↓ | P95 | European Polycystic Kidney Disease Consortium (1994) | |
| 10947insT | EX36 | 10947insT | 3578↓ | P100 | Peral et al. (1996a) | |
| 12617delC | EX45 | 12617delC | 4135↓ | P491 | Present study | |
| 12739delA | EX46 | 12739delA | 4176↓ | Flanked by 5 × C/3 × C | P225 | Peral et al. (1997) |
| In frame: | ||||||
| F1992L, 1993delT | EX15 | 6187delC | 1992*Δ1993 | P204 | Present study | |
| 6868del15 | EX15 | 6868del15 | Δ2220–2224 | 13-bp repeat, 15 bp apart | P14 | Present study |
| 8507ins12 | EX23 | 8507dup12 | +2762–2765 | 10-bp repeat, 12 bp apart | P190 | Present study |
| 9245del18 | EX25 | 9245del18 | Δ3012–3017 | Incomplete repeat (13/18), 18 bp apart | P124 | Present study |
| 11457del15 | EX39 | 11457del15 | Δ3749–3753 | 7-bp repeat, 15 bp apart | P35 | Peral et al. (1996b) |
| Splicing: | ||||||
| Frameshift: | ||||||
| IVS7+1G→A | IVS7 | 1597del221 | 536↓ | Skip exon 7 | P1 | Present study |
| IVS9+1G→T | IVS9 | 1934del127 | 574↓ | Skip exon 9 | P134 | Present study |
| IVS25−16G→A | IVS25 | 9413del196 | 3067↓ | Skip exon 26, C→T at CpG | P178 | Present study |
| IVS31+25del9 | IVS31 | 10378ins71 | 3389↓ | Inclusion IVS 31, matches HG1, 12-bp repeat, 19 bp apart | P229 | Peral et al. (1997) |
| IVS39+1G→C | IVS39 | 11365del113 | 3717↓ | Skip exon 39 | P109 | Peral et al. (1997) |
| In frame: | ||||||
| IVS2−2A→G | IVS2 | 499del72 | Δ97–120 | Skip exon 3 | P402 | Present study |
| IVS21−2delAG | IVS21 | 8228del42 | Δ2673–2686 | Cryptic splice exon 22 | P152 | Present study |
| IVS39−25del72 | IVS39 | 11478del51 | Δ3756–3772 | 5-bp repeat 67 bp apart; cryptic splice exon 40 | P167 | Peral et al. (1996b) |
| IVS43+17del18 | IVS43 | 11921del291 + others | Δ3904–4000 + others | 9-bp repeat 20 bp apart | S6046 | Peral et al. (1995) |
| IVS43+14del20 | IVS43 | 11921del291 + others | Δ3904–4000 + others | 9-bp repeat 20 bp apart | P138 | Peral et al. (1995) |
| IVS44+1G→C | IVS44 | 12212del135 | Δ4001–4045 | Skip exon 44 | P4 | European Polycystic Kidney Disease Consortium (1994) |
| Missense: | ||||||
| Nonconservative: | ||||||
| L13Q | EX1 | 249T→A | L13Q | P285 | Present study | |
| S75F | EX2 | 435C→T | S75F | P159 | Present study | |
| W139C | EX4 | 628G→T | W139C | P267 | Present study | |
| Y2336D | EX16 | 7217T→G | Y2336D | P220 | Present study | |
| A2752D | EX23 | 8466C→A | A2752D | P135 | Present study | |
| V2768M | EX23 | 8513G→A | V2768M | C→T at CpG | P442 | Present study |
| G2858S | EX23 | 8783G→A | G2858S | C→T at CpG | P442 | Present study |
| E2771K | EX23 | 8522G→A | E2771K | C→T at CpG | P148, P256, P447, P511 | Present study |
| L2816P | EX23 | 8658T→C | L2816P | Matches HG1, 2, and 3 | P228 | Present study |
| L2993P | EX25 | 9189T→C | L2993P | P193 | Peral et al. (1997) | |
| Q3016R | EX25 | 9258A→G | Q3016R | P157 | Peral et al. (1997) | |
| Conservative: | ||||||
| L3510V | EX35 | 10739C→G | L3510V | P161 | Peral et al. (1997) | |
| E3631D | EX37 | 11104G→C | E3631D | P125 | Peral et al. (1996b) |
↓ = Frameshift after indicated residue; Δ = deletion of indicated residues (inclusive); + = duplication of indicated residues (inclusive).
Figure 4 .
Segregation of PKD1 mutations in pedigrees. A, IVS7+1G→A segregates in pedigree 1 (P1). Restriction digest of PKD1 specifically amplified DNA (GenSeq 7) digested with MnI1. A 234-bp fragment (arrow) is found in the affected individuals because of loss of a restriction site. Samples numbered in the pedigree are in the corresponding gel lane. B, Genomic heteroduplex analysis of the mutation 224del13 in pedigree 103 (P103). Heteroduplexes (arrow) and two homoduplex fragments, normal and deleted, can be seen in the affected individuals.
Mutations Detected by Direct Sequencing
The region from exons 1–8 was analyzed by direct sequencing in genomic DNA, and eight different mutations were detected: two stop codons, two splicing changes, one deletion, and three missense changes (for examples of sequencing, see table 4 and fig. 3). The most 5′ mutation detected was a 13-bp deletion located just 13 bp after the translational start, and segregation of this change with the disease is illustrated in figure 4B. A second putative mutation was also identified in the first exon—a missense change, L13Q, located within the region encoding the signal peptide. Although it has not been possible to show whether this change segregates with the disease, because of lack of family members, this nonconservative change significantly affects the hydrophobicity of the signal peptide. The hydrophobicity score falls from a normal maximum of 1.54–1.15, and the signal peptide probability score drops from a normal of 9.2–6.7 (below the normal threshold of 7.0), suggesting that this mutation would significantly impair the function of this region. Two putative missense mutations disrupt the structure of the leucine-rich repeat (LRR) or flanking region. In pedigree 159, serine 75, in the first LRR, is replaced by phenylalanine. This residue is conserved from humans to Fugu in polycystin-1 and in many other LRRs (Kobe and Deisenhofer 1994). The bulky hydrophobic phenylalanine may disrupt the β-sheet structure of this part of the motif. The second mutation (in P267) replaces tryptophan 139 with a cysteine residue in the LRR C-flanking region (Hughes et al. 1995). The introduction of an additional cysteine may be disruptive by formation of inappropriate disulfide bonds in an area that contains four highly conserved cysteines.
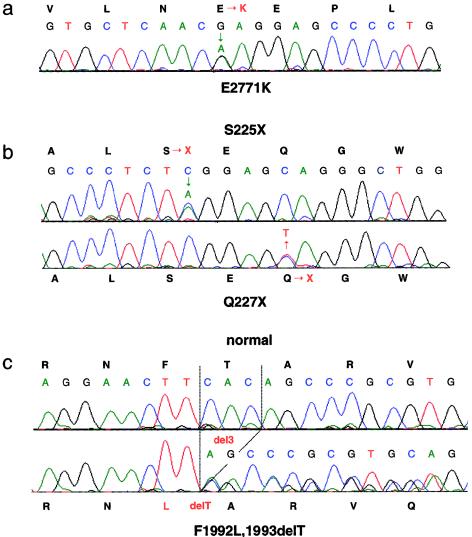
Figure 3 .
Examples of direct sequence analysis of patient DNA. A, The missense substitution E2771K, caused by G→A transition at position 8522 nt. B, Two nonsense mutations S225X and Q227X, caused by C→A and C→T substitutions at positions 885 and 890 nt, respectively. C, Normal and patient DNA with the mutation F1992L, 1993delT, caused by a single codon deletion, 6187del3. Note the double peak at the site of a substitution (A and B) and continued double peaks after the deletion (C).
Direct sequencing of 2,062 bp of cDNA from exons 22–26 and 572 bp from exons 15–17 revealed an in-frame deletion (6868del15), an insertion (8507ins12), and a splicing event (IVS21−2delAG). In addition, six nonconservative missense changes were detected in eight different pedigrees (e.g., see fig. 3). Interestingly, in one case, two segregating changes were detected within 90 amino acids (see table 4, pedigree P442), and one putative missense mutation, E2771K, was found in four unrelated pedigrees. These missense changes were detected in the region homologous to the suREJ protein and a related human protein, PKDREJ (Moy et al. 1996; Hughes et al. 1999). The function and structure of this area are unknown, so the consequences of the substitutions are difficult to predict. Each of the substitutions is at a position that is identical or highly conserved in human, murine, and Fugu polycystin-1; four of the six are also conserved in suREJ or PKDREJ. All of the changes are nonconservative (see table 4), except V2768M, where an additional change, G2858S, was detected in the patient. Despite the segregation data and the fact that no other changes were detected in these patients, it will require further population analysis, a better understanding of REJ structure, and, ultimately, functional studies to determine whether these are pathogenic mutations.
One additional 3-bp deletion, resulting in a substitution and single amino acid loss (F1992L and 1993delT), was found fortuitously during sequencing to confirm another mutation in exon 15. In addition to the missense mutations, 20 polymorphisms were detected by direct sequencing of cDNA, genomic DNA, and when confirming other mutations (table 5). Ten of these changes resulted in an amino acid substitution, and, although several of these were nonconservative, we were able to demonstrate by segregation analysis or from the screen of normal individuals that they were not pathogenic changes.
Table 5.
Polymorphisms Identified in the Study Population
| Designation | Location | cDNA Change or Amino AcidPosition | Comments | Enzyme | Frequency (%) | Reference |
| Amino acid change: | ||||||
| P/S2674 | EX22 | 8231C/T | AvaII | 7/348 (2.0) | Present study | |
| T/M2708 | EX22 | 8334C/T | BsaHI | 5/318 (1.8) | Present study | |
| P/T2734 | EX23 | 8411C/A | BanI | 1/340 (.3) | Present study | |
| Q/L 2735 | EX23 | 8415A/T | 1/190 (.5) | Present study | ||
| R/C2765 | EX23 | 8504C/T | C→T at CpG | BsiHKAI | 5/410 (1.2) | Present study |
| V/M2782 | EX23 | 8556G/A | C→T at CpG | NlaIII | 1/210 (.5) | Present study |
| G/R2814 | EX23 | 8651G/A | C→T at CpG, matches HG1 and 2 | MspAlI | 4/422 (.9) | Present study |
| R/G2888 | EX23 | 8873C/G | At CpG/de novo change | DsaI | 1/190 (.5) | Present study |
| V/I2905 | EX23 | 8924G/A | C→T at CpG | XmnI | 2/296 (.7) | Present study |
| E/D2966 | EX24 | 9109G/C | 1/190 (.5) | Present study | ||
| F/L3066 | EX25 | 9406GT/CC | 20/190 (10.5) | Peral et al. 1997 | ||
| T/M3509 | EX35 | 10737C/T | C→T at CpG | SfaNI | 2/384 (.5) | Peral et al. 1997 |
| A/V3511 | EX35 | 10743C/T | C→T at CpG | HhaI | 14/300 (4.7) | Peral et al. 1997 |
| I/V4044 | EX44 | 12341A/G | MscI | 18/88 (20.5) | Rossetti et al. 1996 | |
| A/V4058 | EX45 | 12384C/T | AvaII | 15/188 (8.0) | Rossetti et al. 1996 | |
| S/F4189 | EX46 | 12777C/T | Rare | Peral et al. 1997 | ||
| No amino acid change: | ||||||
| 487G/A | EX2 | A92 | C→T at CpG | 1/146 (.7) | Present study | |
| 1234C/T | EX5 | A341 | C→T at CpG | 3/146 (2.1) | Present study | |
| 1330T/C | EX5 | L373 | 17/146 (11.6) | Present study | ||
| 1420C/T | EX6 | H403 | 1/146 (.7) | Present study | ||
| 1921C/T | EX7 | H570 | C→T at CpG/matches HG2 | 1/146 (.7) | Present study | |
| 7138C/T | EX16 | A2309 | C→T at CpG | 4/190 (2.1) | Present study | |
| 7147G/A | EX16 | A2312 | C→T at CpG | 1/190 (.5) | Present study | |
| 8650C/T | EX23 | S2813 | C→T at CpG | MspAII | 3/190 (1.6) | Present study |
| 8890C/G | EX23 | S2893 | At CpG | 1/190 (.5) | Present study | |
| 9541T/C | EX26 | P3310 | Matches HG1, 2, and 3 | 20/190 (10.5) | Peral et al. 1997 | |
| 9880G/A | EX28 | T3223 | C→T at CpG | 1/128 (.8) | Peral et al. 1997 | |
| 11521G/A | EX40 | A3371 | C→T at CpG | 1/90 (1.1) | Present study | |
| 11584G/C | EX40 | S3791 | At CpG | 2/90 (2.2) | Peral et al. 1996b | |
| 12484A/G | EX45 | A4091 | HhaI | 27/100 (2.7) | Peral et al. 1996b | |
| 12838C/T | EX46 | P4209 | Rare | Peral et al. 1997 | ||
| 12973C/T | EX46 | P4254 | C→T at CpG | AciI | 1/90 (1.1) | Peral et al. 1996b |
Screening by Restriction Enzyme or Allele-Specific PCR
Prior to screening patients with PTT, samples were analyzed with restriction enzymes and allele-specific PCR (Peral et al. 1997) for known changes within the single-copy region. In three pedigrees, mutations were detected in this way: in P105, Q3513X; in P264, R4020X; and in P252, R4227X (for details, see table 3). One further in-frame deletion (9245del18) was detected by agarose gel electrophoresis during a screen to determine the frequency of another polymorphism.
Discussion
We have described the first mutation screen of the entire PKD1 gene, overcoming the problem of duplication at this locus by anchored and PKD1-specific PCR and screening for mutations by PTT and direct sequencing. These data allow an objective analysis of the type and position of pathogenic mutations, and the implications for genetic diagnosis and understanding of the mutational mechanism can be considered.
The first conclusion is that mutations are found throughout the gene, from 13 bp 3′ to the initiating codon to 228 bp 5′ to the stop codon (fig. 1). No clear hotspot for mutation was found, with 69 different changes characterized in 81 pedigrees. One missense mutation, E2771K, was found in four pedigrees and the stop mutation R4227X in three, whereas seven other changes were identified in two families (table 4). Haplotype analysis with PKD1 markers (see Subjects and Methods section) showed in three cases (C2229X, Q2243X, and Q3513X) that the mutation was on the same or a nearly identical haplotype, indicating a probable common origin, whereas the other six appeared to be recurrent changes. Analysis of independent truncating events showed no significant frequency difference between the single-copy region and the duplicated segment (14 in 20.2%, compared with 38 in 79.8% of the translated region, respectively). However, the frequency of changes in the 5′ half of the gene (212–6665 nt) was significantly lower than that of the 3′ half (16–36; χ2=7.69, P=.006). The area of the gene with the highest density of changes was 1 kb (7.7% of coding DNA) 3′ to the halfway point (exons 15–18), accounting for 26.9% of independent truncating events. It does not appear that the uneven spread of mutations was due to the methods used for screening, since lower levels were detected in the 5′ region by PTT and sequencing. These data suggest regions that may be screened initially for mutations, but the functional significance, if any, will require detailed analysis of the phenotypes in the different mutation groups.
The full range of disease associated mutations can be evaluated in the 28.6% of the coding region that was sequenced from genomic DNA (exons 1–8) or cDNA (exons 15–17 and 22–26). In these areas, 17 missense and in-frame mutations were identified in 19 pedigrees, compared with 13 frame-shifting changes in 16 independent pedigrees. Despite the evidence from segregation analysis, the screen of normal individuals, the lack of other changes in the rest of the gene, and the nonconservative nature of the substitutions (see the Results section), it is likely that some of these missense changes will ultimately be revealed as nonpathogenic polymorphisms. Nevertheless, it appears that as many as half of all mutations will be missense or in-frame; therefore, it seems reasonable to suspect that many of the undiscovered mutations in this ADPKD population are missense changes to PKD1.
Studies of other genes have indicated that mRNAs with nonsense or frameshifting mutations are often rapidly degraded by the RNA surveillance mechanism, nonsense-mediated mRNA decay (Culbertson 1999; Hentze and Kulozik 1999). However, in PKD1 we have detected most of this type of mutation by analysis of lymphoblast or leukocyte RNA, with equal peak intensity seen by PTT (fig. 2) and sequencing, indicating that such PKD1 mutant RNAs are not rapidly degraded. Furthermore, the DNA screened region (exons 1–8) did not reveal a higher level of truncating mutations. It is not clear whether these mutated mRNA are stable because functional alternatively spliced or cleaved products are generated from these transcripts or whether they may generate stable mutant proteins (Hughes et al. 1995; International Polycystic Kidney Disease Consortium 1995; Ponting et al. 1999).
The results of the mutation screen, showing changes throughout the gene, including a frameshifting mutation within the signal-peptide-encoded region, are consistent with the theory that PKD1 mutations are inactivating and that disease occurs through a loss, or dosage reduction, of polycystin-1 protein. Nevertheless, the uneven distribution of mutations along the gene, the significant level of in-frame and missense events, and the stability of mutant mRNAs indicates that this will be an interesting population to analyze phenotype/genotype correlations.
By analyzing the pedigrees with PKD1 mutations, we can estimate the frequency of new mutations at this locus. Of a total of 411 affected individuals in 81 pedigrees, de novo mutation has been demonstrated in 7 cases by molecular analysis and has been suspected in a further 8 because of documented negative imaging studies or clinical indications in the parents. Overall, this indicates a new mutation in 1 of 27.4 cases, and if we assume a PKD1 population frequency of .001, the new mutation rate is 1.8×10-5 per gamete, per generation. This estimated figure is higher than the average for X-chromosomal disease genes (Stevenson and Kerr 1967) and the rate recently calculated for hemophilia B (Green et al. 1999). It is, however, lower than previous estimates for ADPKD, 6.9×10-5 (Dobin et al. 1993) and 6.5×10-5 (Dalgaard 1957), and is in line with other dominant disorders, especially those associated with large genes (Vogel and Motulsky 1997).
This significant level of new mutation suggests a steady increase in the disease frequency unless it is balanced by a reduction in fitness. The many different PKD1 mutations we have detected, mainly in a single geographically defined PKD1 population (from a total Oxford Renal Unit catchment of ∼0.6 M), and the lack of common ancestral changes indicates that most mutations have arisen in the last few generations, with significant selection applied to eliminate these changes. Early death (or renal death) before or during reproductive age can occur, due to subarachnoid hemorrhage or severe renal cystic disease, for instance. A second means by which PKD1 mutations could be selected against is if individuals (especially women) have a lower reproductive fitness. Interestingly, data from Dalgaard (1957) indicated a significantly lower level of reproduction for women with ADPKD, compared with the age-matched Danish population (60%–80% expected). It seems possible that a combination of reduced survival and reduced reproductive fitness explains the observed selection against PKD1 mutant alleles.
There have been suggestions that special factors promote mutation at PKD1 that may explain the relatively high mutation rate (see Introduction section). The frequency of mutations in the sequenced exons flanking the polypyrimidine tract in IVS21 was analyzed. Exons 22–26 revealed putative mutations in 14 pedigrees, a change every 80 bp, compared with an independent change every 58 bp in exons 15–17 and one every 215 bp in the exon 1–8 region. In addition to disease-related mutations, a number of polymorphisms were detected in these fragments (table 5). If these substitutions are also included, the frequency is 1/49 bp, exons 22–26; 1/48 bp, exons 15–17, and 1/132 bp, exons 1–8. Hence, the frequency is higher both 3′ to and further 5′ of IVS21 than at the 5′ end of the gene. It is possible that the polypyrimidine tract is having a long-range effect, reflecting the higher rate of mutation overall in the 3′ part of the gene. However, a high level of multiple changes, as found in experimental systems analyzing triplex structures (Wang et al. 1996) was not found, and most mutations were at sequences known to promote mutation in other genes (see below).
The second suggested unusual mutational mechanism at PKD1, conversion events with the HG loci, was tested by seeing how many mutations match HG sequence. Analysis shows that 3/69 disease associated mutations (4/81 pedigrees) and 3/22 polymorphisms match the sequence of at least one HG sequence (for details, see tables 4 and 5). So, although the HG-related changes do not represent a large proportion of the total, this level seems slightly higher than expected by chance, with substitution levels of only ∼2%–3% found between the HG and PKD1 gene (Loftus et al. 1999). However, four of the changes are at CpG dinucleotides and one flanked by tandem repeats, known sites of enhanced mutation (see below), reflecting that they are sites of divergence between PKD1 and the HG. Moreover, in five cases we have analyzed the sequence flanking the PKD1 change for other substitutions found in HG sequence, and, out of 45 readable sites, no further HG-matching changes were detected. These results indicate that these DNA changes are not due to conversion with a significant stretch of HG sequence, but the most likely explanation is that they are recurrent events at sites with higher-than-average rates of mutation. Because all HG loci have not been sequenced, it is possible that other mutations will match uncharacterized HG sequence. However, our mapping data indicate that the structure of the uncharacterized loci are similar to HG1 or HG2 and so are unlikely to have many novel substitutions (authors' unpublished data). It is possible that the methods we have employed, especially PKD1-specific PCR, may not amplify PKD1 loci with mutations caused by larger conversion events. However, higher rates of HG-matching changes were not found in areas examined by anchored PCR, and the level of detected changes, considering that many areas have not been screened for missense changes, does not leave a large reservoir of undetected mutations that might be due to conversion events.
If the positions and precise sites of PKD1 mutations are examined, the mechanisms underlying the changes mainly appear to be ones known to promote mutations in other genomic regions. Of the 39 different substitutions associated with mutation, 13 are at CpG dinucleotides, including 8 C→T transversions, that account for all the recurrent substitutions, making a total of 20 of 46 independent substitutions at CpGs. These sites are hotspots for mutation, because the cytosine is often methylated and susceptible to spontaneous deamination to give a thymidine, with a mutation rate ∼8.5 times greater than average (Cooper et al. 1995). Consequently, CpGs are significantly underrepresented in vertebrate DNA (20% expected)—although CpG dinucleotides constitute 7.2% of the PKD1 coding region (higher than the genomic average of 0.8% because of the GC richness of this area), they account for 43.5% of the substitutions, an enrichment of sixfold. In an analysis of the 31 deletions and insertions, 20 (including the three recurrent events) are flanked by short stretches of tandem-repeated sequence (for details, see table 4), suggesting that slippage events during DNA replication are the major cause of this type of change. Slippage between tandem repeats is a well-characterized cause of DNA deletions and duplications (Cooper et al. 1995).
We have shown that PKD1 has a relatively high mutation rate but also that there is no clear evidence for an unusual mechanism that can account for this. However, if we consider that a large number of different mutations cause PKD1, possibly any inactivating mutation, and that the PKD1 transcript is a large mutational target (12,906-bp coding region), these factors may in themselves explain the observed rate. If we conservatively estimate that 1,000 different mutations will cause the disease, we can calculate a mutation rate, per nucleotide, per generation, of 1.8×10-8, similar to that recently calculated from the factor IX gene (Giannelli et al. 1999). In addition, PKD1 is unusually CpG-rich (see above) and thus contains more highly mutable sequences. Hence, overall, it is not clear that germline mutation at the nucleotide level in PKD1 is occurring at a higher level than expected, and, probably, no special mechanisms are required to explain the relatively high gene-mutation rate. It remains to be seen whether unusual mechanisms are required to explain somatic mutation at this locus.
In this study we have primarily used two methods for mutation detection: PTT and direct sequencing. For PTT, cDNA from cell lines or patient leukocytes is required to maintain an open reading frame, relatively large fragments can be screened (∼2 kb of cDNA), and almost all detected changes are pathogenic mutations. This method could be improved if larger cDNA fragments were amplified, as has been described for PKD1 (Thongnoppakhun et al. 1999). The major disadvantage is that missense or small in-frame changes are not detected, which are a significant source of mutation at PKD1. Direct sequencing should detect every base-pair mutation and any polymorphism in the sequence. Recently, direct sequencing has been used to analyze the COL4A5 gene in Alport syndrome (Martin et al. 1998) and recommended as a primary mutation-screening tool. To be effective however, the sequence quality has to be very good, with heterozygotes detected as doublet peaks and a low level of background, which is aided by the Big Dye primer-labeling method. Also, it is relatively expensive, since many small fragments have to be sequenced for a large gene such as PKD1. For cost-effective large-scale screening of PKD1 in the future, it may be beneficial to introduce a rapid, inexpensive, and semiautomated screening step, such as denaturing high-performance liquid chromatography (Liu et al. 1998), to flag fragments with base-pair changes for sequence analysis.
The level of detection of PKD1 mutations in the ADPKD pedigrees was 52.3%, and it was 61.7% in known PKD1 pedigrees, indicating that some families with PKD2 or unlinked ADPKD were represented in the population. These figures compare well with primary screens of other large disease genes, such as COL4A5 in Alport syndrome (Knebelmann et al. 1996) and the fibrillin-1 gene in Marfan syndrome (Hayward et al. 1997). Furthermore, nearly 75% of the gene was not screened for missense and in-frame changes. These results show the feasibility of molecular diagnosis of PKD1. Although the demand for presymptomatic screening is, at present, low, if and when therapies are developed to treat this disorder, it will be essential that diagnosis is possible in younger individuals, for whom imaging methods are less reliable, so that treatment can start at an early age, before significant renal damage has occurred.
Acknowledgments
We wish to thank S. Butler, J. Sloane-Stanley, and K. Clark for technical assistance; K. Zerres and S. Ozen for supplying samples; the patients and their families for taking part in the study; and V. E. Torres and D. J. Weatherall for support and encouragement. The work was supported by the Medical Research Council (United Kingdom), the Mayo Foundation, Telethon (Italy), NATO/Royal Society (United Kingdom), EMBO, Oxford Kidney Unit Trust Fund, and the Wellcome Trust.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Database/index.html (for PKD1 genomic sequence [L39891], PKD1 cDNA sequence [L33243], and polycystin-1 [AAC377576])
- Online Human Gene Mutation Database (HGMD), http://www.uwcm.ac.uk/uwcm/mg/hgmd0.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim
References
- Blaszak RT, Potaman V, Sinden RR, Bissler JJ (1999) DNA structural transitions within the PKD1 gene. Nucleic Acids Res 27:2610–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-Z, Vassilev PM, Basora N, Peng J-B, Nomura H, Segal Y, Brown EM, Reeders ST, Hediger MA, Zhou J (1999) Polycystin-L is a calcium-regulated cation channel permeable to calcium ions. Nature 401:383–386 [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Antonorakis SE (1995) The nature and mechanisms of human gene mutation. In: Scriver C, Beaudet AL, Sly WS, Valle D (eds) Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, pp 259–291 [Google Scholar]
- Culbertson MR (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet 15:74–80 [DOI] [PubMed] [Google Scholar]
- Dalgaard OZ (1957) Bilateral polycystic disease of the kidneys: a follow-up of two hundred and eighty-four patients and their families. Acta Med Scand Suppl 328:1–255 [PubMed] [Google Scholar]
- Daniells C, Maheshwar M, Lazarou L, Davies F, Coles G, Ravine D (1998) Novel and recurrent mutations in the PKD1 (polycystic kidney disease) gene. Hum Genet 102:216–220 [DOI] [PubMed] [Google Scholar]
- Daoust MC, Reynolds DM, Bichet DG, Somlo S (1995) Evidence for a third genetic locus for autosomal dominant polycystic kidney disease. Genomics 25:733–736 [DOI] [PubMed] [Google Scholar]
- Dobin A, Kimberling WJ, Pettinger W, Bailey-Wilson JE, Shugart YY, Gabow P (1993) Segregation analysis of autosomal dominant polycystic kidney disease. Genet Epidemiol 10:189–200 [DOI] [PubMed] [Google Scholar]
- Dodé C, Rochette J, Krishnamoorthy R (1990) Locus assignment of human a globin mutations by selective amplification and direct sequencing. Brit J Haemat 76:275–281 [DOI] [PubMed] [Google Scholar]
- European Polycystic Kidney Disease Consortium (1994) The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77:881–894 [DOI] [PubMed] [Google Scholar]
- Geng L, Segal Y, Peissel B, Deng N, Pei Y, Carone F, Rennke HG, Glücksmann-Kuis AM, Schneider MC, Ericsson M, Reeders ST, Zhou J (1996) Identification and localization of polycystin, the PKD1 gene product. J Clin Invest 98:2674–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli F, Anagnostopoulos T, Green PM (1999) Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental human mutations inferred from hemophilia B. Am J Hum Genet 65:1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PM, Saad S, Lewis CM, Giannelli F (1999) Mutation rates in humans. I. Overall and sex-specific rates obtained from a population study of hemophilia B. Am J Hum Genet 65:1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J (1994) The 1993–94 Généthon human genetic linkage map. Nat Genet 7:246–339 [DOI] [PubMed] [Google Scholar]
- Harris PC, Thomas S, Ratcliffe PJ, Breuning MH, Coto E, Lopez-Larrea C (1991) Rapid genetic analysis of families with polycystic kidney disease by means of a microsatellite marker. Lancet 338:1484–1487 [DOI] [PubMed] [Google Scholar]
- Hateboer N, van Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D (1999) Comparison of phenotypes of polycystic kidney disease types 1 and 2. Lancet 353:103–107 [DOI] [PubMed] [Google Scholar]
- Hayward C, Porteous ME, Brock DJ (1997) Mutation screening of all 65 exons of the fibrillin-1 gene in 60 patients with Marfan syndrome: report of 12 novel mutations. Hum Mutat 10:280–289 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310 [DOI] [PubMed] [Google Scholar]
- Huan Y, van Adelsberg J (1999) Polycystin-1, the PKD1 gene product, is in a complex containing E-cadherin and the catenins. J Clin Invest 104:1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Ward CJ, Aspinwall R, Butler R, Harris PC (1999) Identification of a human homologue of the sea urchin receptor for egg jelly: a polycystic kidney disease–like protein. Hum Mol Genet 8:543–549 [DOI] [PubMed] [Google Scholar]
- Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millán JL, Gamble V, Harris PC (1995) The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nature Genet 10:151–160 [DOI] [PubMed] [Google Scholar]
- International Polycystic Kidney Disease Consortium (1995) Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81:289–298 [DOI] [PubMed] [Google Scholar]
- Kim K, Drummond I, Ibraghimov-Beskrovnaya O, Klinger K, Arnaout MA (2000) Polycystin 1 is required for the structural integrity of blood vessels. Proc Natl Acad Sci USA 97:1731–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebelmann B, Breillat C, Forestier L, Arrondel C, Jacassier D, Giatras I, Drouot L, Deschenes G, Grunfeld JP, Broyer M, Gubler MC, Antignac C (1996) Spectrum of mutations in the COL4A5 collagen gene in X-linked Alport syndrome. Am J Hum Genet 59:1221–1232 [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19:415–421 [DOI] [PubMed] [Google Scholar]
- Koptides M, Hadjimichael C, Koupepidou P, Pierides A, Constantinou Deltas C (1999) Germinal and somatic mutations in the PKD2 gene of renal cysts in autosomal dominant polycystic kidney disease. Hum Mol Genet 8:509–513 [DOI] [PubMed] [Google Scholar]
- Koptides M, Mean R, Demetriou K, Pierides A, Deltas CC (2000) Genetic evidence for a trans-heterozygous model for cystogenesis in autosomal dominant polycystic kidney disease. Hum Mol Genet 9:447–452 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Cooper DN (1997) The Human Gene Mutation Database. Trends Genet 13:121–122 [DOI] [PubMed] [Google Scholar]
- Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD (1998) Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acid Res 26:1396–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Kim U-J, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, Cao Y, Xu RX, Kang H-L, Eichler EE, Harris PC, Venter JC, Adams MD (1999) Genome duplications and other features in 12 Mbp of DNA sequence from human chromosome 16p and 16q. Genomics 60:295–308 [DOI] [PubMed] [Google Scholar]
- Lu W, Fan X, Basora N, Babakhanlou H, Law T, Rifai N, Harris PC, Perez-Atayde AR, Rennke HG, Zhou J (1999) Late onset of renal and hepatic cysts in Pkd1-targeted heterozygotes. Nat Genet 21:160–161 [DOI] [PubMed] [Google Scholar]
- Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J (1997) Perinatal lethality with kidney and pancreas defects in mice with a targeted Pkd1 mutation. Nat Genet 17:179–181 [DOI] [PubMed] [Google Scholar]
- Martin P, Heiskari N, Zhou J, Leinonen A, Tumelius T, Hertz JM, Barker D, Gregory M, Atkin C, Styrkarsdottir U, Neumann H, Springate J, Shows T, Pettersson E, Tryggvason K (1998) High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol 9:2291–2301 [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhusien B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, Kimberling WJ, Breuning MH, Deltas CC, Peters DJM, Somlo S (1996) PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272:1339–1342 [DOI] [PubMed] [Google Scholar]
- Moy GW, Mendoza LM, Schulz JR, Swanson WJ, Glabe CG, Vacquier VD (1996) The sea urchin sperm receptor for egg jelly is a modular protein with extensive homology to the human polycystic kidney disease protein, PKD1. J Cell Biol 133:809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6 [DOI] [PubMed] [Google Scholar]
- Ong ACM, Harris PC, Davies DR, Pritchard L, Rossetti S, Biddolph S, Vaux DJT, Migone N, Ward CJ (1999) Polycystin-1 expression in PKD1, early onset PKD1 and TSC2/PKD1 cystic tissue: implications for understanding cystogenesis. Kidney Int 56:1324–1333 [DOI] [PubMed] [Google Scholar]
- Pei Y, Watnick T, He N, Wang K, Liang Y, Parfrey P, Germino G, George-Hyslop PS (1999) Somatic PKD2 mutations in individual kidney and liver cysts support a “two-hit” model of cystogenesis in type 2 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 10:1524–1529 [DOI] [PubMed] [Google Scholar]
- Peral B, Gamble V, San Millán JL, Strong C, Sloane-Stanley J, Moreno F, Harris PC (1995) Splicing mutations of the polycystic kidney disease 1 (PKD1) gene induced by intronic deletion. Hum Mol Genet 4:569–574 [DOI] [PubMed] [Google Scholar]
- Peral B, Gamble V, Strong C, Ong ACM, Sloane-Stanley J, Zerres K, Winearls CG, Harris PC (1997) Identification of mutations in the duplicated region of the polycystic kidney disease 1 (PKD1) gene by a novel approach. Am J Hum Genet 60:1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peral B, Ong ACM, San Millán JL, Gamble V, Rees L, Harris PC (1996a) A stable, nonsense mutation associated with a case of infantile onset polycystic kidney disease 1 (PKD1). Hum Mol Genet 5:539–542 [DOI] [PubMed] [Google Scholar]
- Peral B, San Millán JL, Ong ACM, Gamble V, Ward CJ, Strong C, Harris PC (1996b) Screening the 3′ region of the polycystic kidney disease 1 (PKD1) gene reveals six novel mutations. Am J Hum Genet 58:86–96 [PMC free article] [PubMed] [Google Scholar]
- Peral B, Ward CJ, San Millán JL, Thomas S, Stallings RL, Moreno F, Harris PC (1994) Evidence of linkage disequilibrium in the Spanish polycystic kidney disease 1 (PKD1) population. Am J Hum Genet 54:899–908 [PMC free article] [PubMed] [Google Scholar]
- Perrichot RA, Mercier B, Simon PM, Whebe B, Cledes J, Ferec C (1999) DGGE screening of PKD1 gene reveals novel mutations in a large cohort of 146 unrelated patients. Hum Genet 105:231–239 [DOI] [PubMed] [Google Scholar]
- Phakdeekitcharoen B, Watnick TJ, Ahn C, Whang D, Burkhart B, Germino GG (2000) Thirteen novel mutations of the replicated region of PKD1 in an Asian population. Kidney Int 58:1400–1412 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Hofmann K, Bork P (1999) A latrophilin/CL-1-like GPS domain in polycystin-1. Curr Biol 9:R585–R588 [DOI] [PubMed] [Google Scholar]
- Qian F, Watnick TJ, Onuchic LF, Germino GG (1996) The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type 1. Cell 87:979–987 [DOI] [PubMed] [Google Scholar]
- Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM (1994) Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 343:824–827 [DOI] [PubMed] [Google Scholar]
- Reeders ST, Breuning MH, Davies KE, Nicholls RD, Jarman AP, Higgs DR, Pearson PL, Weatherall DJ (1985) A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature 317:542–544 [DOI] [PubMed] [Google Scholar]
- Roelfsema JH, Spruit L, Saris JJ, Chang P, Pirson Y, van Ommen G-JB, Peters DJM, Breuning MH (1997) Mutation detection in the repeated part of the PKD1 gene. Am J Hum Genet 61:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest PAM, Roberts RG, Sugino S, van Ommen G-JB, den Dunnen JT (1993) Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum Mol Genet 2:1719–1721 [DOI] [PubMed] [Google Scholar]
- Rossetti S, Bresin E, Restagno G, Carbonara A, Corrá S, De Prisco O, Pignatti PF, Turco AE (1996) Autosomal dominant polycystic kidney disease (ADPKD) in an Italian family carrying a novel nonsense mutation and two missense changes in exon 44 and 45 of the PKD1 gene. Am J Med Genet 65:155–159 [DOI] [PubMed] [Google Scholar]
- Rowan AJ, Bodmer WF (1997) Introduction of a myc reporter tag to improve the quality of mutation detection using the protein truncation test. Hum Mut 9:172–176 [DOI] [PubMed] [Google Scholar]
- Sandford R, Sgotto B, Aparacio S, Brenner S, Vaudin M, Wilson R, Chissoie S, Pepin K, Bateman A, Chothia C, Hughes J, Harris P (1997) Comparative analysis of the polycystic kidney disease 1 (PKD1) gene reveals an integral membrane glycoprotein with multiple evolutionary conserved domains. Hum Mol Genet 6:1483–1489 [DOI] [PubMed] [Google Scholar]
- Stevenson AC, Kerr CB (1967) On the distribution of frequencies of mutation to genes determining harmful traits in man. Mutat Res 4:339–352 [DOI] [PubMed] [Google Scholar]
- Thomas R, McConnell R, Whittacker J, Kirkpatrick P, Bradley J, Sandford R (1999) Identification of mutations in the repeated part of the autosomal dominant polycystic kidney disease type 1 gene, PKD1, by long-range PCR. Am J Hum Genet 65:39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongnoppakhun W, Wilairat P, Vareesangthip K, Yenchitsomanus PT (1999) Long RT-PCR amplification of the entire coding sequence of the polycystic kidney disease 1 (PKD1) gene. Biotechniques 26:126–132 [DOI] [PubMed] [Google Scholar]
- Torra R, Badenas C, Peral B, Darnell A, Gamble V, Turco A, Harris PC, Estivill X (1998) Recurrence of the PKD1 nonsense mutation Q4041X in Spanish, Italian and British families. Hum Mutat Suppl 1:S117–S120 [DOI] [PubMed]
- Torra R, Badenas C, San Millan JL, Perwz-Oller L, Estivill X, Darnell A (1999) A loss-of-function model for cystogenesis in human autosomal dominant polycystic kidney disease type 2. Am J Hum Genet 65:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Burn TC, Connors TD, Petry LR, Germino GG, Klinger KW, Landes GM (1996) A 2.5kb polypyrimidine tract in PKD1 gene contains at least 23 H-DNA-forming sequences. Microb Comp Genomics 1:317–327 [DOI] [PubMed] [Google Scholar]
- Veldhuisen B, Saris JJ, de Haij S, Hayashi T, Reynolds DM, Mochizuki T, Elles R, Fossdal R, Bogdanova N, van Dijk MA, Coto E, Ravine D, Nørby S, Verellen-Dumoulin C, Breuning MH, Somlo S, Peters DJM (1997) A spectrum of mutations in the second gene for autosomal dominant polycystic kidney disease (PKD2). Am J Hum Genet 61:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Motulsky AG (1997) Human genetics: problems and approaches. Springer-Verlag Berlin Heidelberg, New York, pp 1–851 [Google Scholar]
- Wang G, Seidman MM, Glazer PM (1996) Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science 271:802–805 [DOI] [PubMed] [Google Scholar]
- Ward CJ, Turley H, Ong ACM, Comley M, Biddolph S, Chetty R, Ratcliffe PJ, Gatter K, Harris PC (1996) Polycystin, the polycystic kidney disease 1 protein, is expressed by epithelial cells in fetal, adult and polycystic kidney. Proc Natl Acad Sci USA 93:1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick TJ, Gandolph MA, Weber H, Neumann HPH, Germino GG (1998a) Gene conversion is a likely cause of mutation in PKD1. Hum Mol Genet 7:1239–1243 [DOI] [PubMed] [Google Scholar]
- Watnick T, He N, Wang K, Liang Y, Parfrey P, Hefferton D, St George-Hyslop P, Germino G, Pei Y (2000) Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 25:143–144 [DOI] [PubMed] [Google Scholar]
- Watnick T, Phakdeekitcharoen B, Johnson A, Gandolph M, Wang M, Briefel G, Klinger KW, Kimberling W, Gabow P, Germino GG (1999) Mutation detection of PKD1 identifies a novel mutation common to three families with aneurysms and/or very-early-onset disease. Am J Hum Genet 65:1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick TJ, Piontek KB, Cordal TM, Weber H, Gandolph MA, Qian F, Lens XM, Heumann HPH, Germino GG (1997) An unusual pattern of mutation in the duplicated portion of PKD1 is revealed by use of a novel strategy for mutation detection. Hum Mol Genet 6:1473–1481 [DOI] [PubMed] [Google Scholar]
- Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, Landes G, Germino GG (1998b) Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2:247–251 [DOI] [PubMed] [Google Scholar]
- Wilson PD, Geng L, Li X, Burrow CR (1999) The PKD1 gene product, “polycystin-1” is a tyrosine-phosphorylated protein that colocalizes with a2b1-integrin in focal clusters in adherent renal epithelia. Lab Invest 79:1311–1323 [PubMed] [Google Scholar]
- Wood WI, Gitschier J, Lasky LA, Lawn RM (1985) Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci USA 82:1585–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]